Abstract
The extreme 5′-terminal sequences of the GB virus C/hepatitis G virus (GBV-C/HGV), containing elements essential for regulation of viral gene expression and replication, have not been determined. By using a RNA-ligase-mediated RACE (rapid amplification of the cDNA ends) procedure, we have cloned the extreme 5′-terminal sequences of the viral genome from the serum of three Taiwanese patients. Sequence analysis of the 5′ noncoding region in alignment with one West African and two American isolates showed that (i) a consensus 5′-end sequence was cloned; (ii) about 97% of sequences were homologous among the three Taiwan isolates and also between the two American isolates, whereas about 90% of sequences were homologous among the isolates from the three different geographic areas; (iii) the sequence heterogeneity related to geographic separation is confined mainly to three domains; and (iv) a potential hairpin structure, resembling the hairpin structure found in the 5′ end of hepatitis C virus genome, was detected in the 5′ end of the noncoding region. Our data support the hypotheses that (i) the extreme 5′ end of the hepatitis GBV-C/HGV viral genome has been cloned, (ii) there are different genotypes correlated with geographic separation, and (iii) the viral translation and replication mechanisms may be similar to that of hepatitis C virus and pestiviruses. Our data have not only shed light on the viral replication mechanism but also offer information for selection of optimal primer sequences for the detection and genotyping of the hepatitis GBV-C/HGV virus by PCR assays.
Recently, two flavivirus-like genomes, namely GB virus C (GBV-C) and hepatitis G virus (HGV), were isolated from the serum of several patients with cryptogenic hepatitis by Simons et al. (1) and by Linnen et al. (2), respectively. Both are positive-stranded RNA viruses of about 9.5 kb (2, 3). Their genetic organization resembles that of the flaviviruses, with structural proteins located in the N-terminal region and a variety of nonstructural proteins in the C-terminal region of the viral polyprotein (2, 3). The amino acid identity of the complete viral polyprotein is about 90% between GBV-C and HGV (2), whereas it is only about 30% between GBV-C and hepatitis C virus (HCV) (3). Phylogenetic analysis further showed that GBV-C and HGV did not belong to the HCV group (1–3). GBV-C and HGV should be regarded as members of a separate hepatitis virus group (provisionally, hepatitis GBV-C/G virus or GBV-C/HGV) in view of their 80% nucleotide and about 90% amino acid similarity (2, 3). GBV-C/HGV infection has been found to be worldwide and to contribute to some posttransfusion hepatitis, community acquired non-A–E acute hepatitis, cryptogenic chronic hepatitis (2), and fulminant hepatitis (4).
Theoretically, the 5′ noncoding region, as well as the 3′ noncoding region, of a viral genome contains elements crucial for regulation of viral gene expression, genome encapsidation, and replication. However, previous studies showed discrepant results in the 5′-terminal sequences and the initiation codon for the viral polyprotein (2, 3). To determine the extreme 5′-terminal sequences, we applied a PCR procedure modified from the 5′ rapid amplification of the cDNA ends (RACE) (5, 6) to cloning from the serum of three Taiwanese patients with chronic infection of GBV-C/HGV, coinfection of GBV-C/HGV and hepatitis B virus, and coinfection of GBV-C/HGV and HCV, respectively. We analyzed the sequence homology with one West African (3) and two American isolates (2) and further characterized the potential secondary structure in the 5′ noncoding region. Our data provide insights into the geographic heterogeneity and the structural conservation in the 5′ noncoding region of the GBV-C/HGV genome, which may have important ramifications regarding the replication strategy and evolution of the virus.
MATERIALS AND METHODS
Isolation of the Hepatitis GBV-C/HGV Viral RNA.
The hepatitis GBV-C/HGV viral RNA was isolated from the serum of three Taiwanese patients with chronic infection of GBV-C/HGV alone (T-138), of hepatitis B virus and GBV-C/HGV (T-338), and of HCV and GBV-C/HGV (T-094), respectively (S.-Y.H., P.-Y.Y. and Y.-F.L., unpublished results). The RNA was extracted with the single-step acid/phenol method (7).
Cloning of the 5′-Terminal Sequences of the Hepatitis GBV-C/HGV Viral Genome.
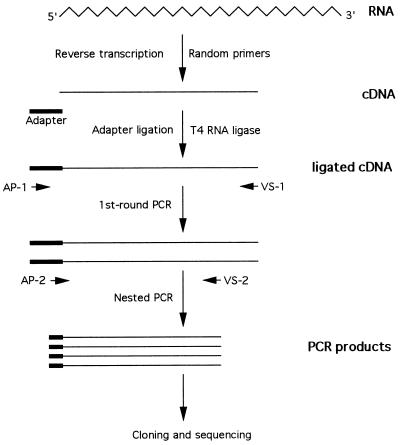
To avoid cross-contamination during manipulation, molecular cloning of the extreme 5′ noncoding region from the serum samples of three patients was done separately. We utilized a modified 5′-RACE procedure (5, 6). In brief, the RNA was converted to single-stranded cDNA with random primers by reverse transcription. A 5′-phosphorylated synthetic oligonucleotide, which was used as an adapter, was ligated to the 3′ end of the single-stranded cDNA with T4 RNA ligase by incubating overnight at room temperature. The ligation solution contained 5 pg of the synthetic oligonucleotide adapter, 50 mM Tris·HCl (pH 7.8), 10 mM MgCl2, 1 mM 2-mercaptoethanol, 1 mM ATP, 20 units of human placenta ribonuclease inhibitor (RNasin, Promega), and 20 units of T4 RNA ligase (New England BioLabs). After phenol/chloroform extraction, the adapter-ligated cDNA was extracted twice by using a Centricon 50 (Amicon). One microliter of the cDNA was then subjected to a first-round of PCR with an adapter primer (AP-1) complementary to the 3′ region of the adapter and an outer viral-specific primer (VS-1). A second-round nested PCR was done with another adapter primer (AP-2) complementary to the 5′ region of the adapter and the inner viral specific primer (VS-2). The final PCR product was examined by gel electrophoresis, cloned into pGEM-T vector (Promega), and finally subjected to sequencing analysis (Sequenase Version 2.0 sequencing kit, United States Biochemical). To further confirm the sequencing results, at least three clones were sequenced bidirectionally for each isolate. The nucleotide sequences of the adapter and the primers were as follows: adapter, 5′-GTAGGAATTCGGGTTGTAGGGAGAGGTCGACATTGCC-3′; AP-1, 5′-GGGAATGTCGACCTCCCTACA-3′; AP-2, 5′-ACCTCCCTACAACCCGAATTCCT-3′; VS-1, 5′-ATATTGCCCATTCGCTCGACA-3′; VS-2, 5′-CGCGGAGCTGGGTGGCCCGAT-3′. This RNA-ligase-mediated 5′-RACE protocol is schematically summarized in Fig. 1.
Figure 1.
The RNA-ligase-mediated 5′-RACE procedure to clone the 5′ terminus of a viral RNA genome. Viral RNA is converted to cDNA with random primers by reverse transcription. A phosphorylated synthetic oligodeoxynucleotide adapter is ligated to the 3′ end of cDNA by using T4 RNA ligase, and then two rounds of PCR amplification are performed. The first-round PCR is done with the adapter primer (AP-1), complementary to the 3′ end of the adapter, and the viral-specific primer 1 (VS-1). The second-round nested PCR is done with the other adapter primer (AP-2), complementary to the 5′ portion of the adapter, and the viral-specific primer 2 (VS-2). The PCR products are subjected to cloning and then sequencing. The RNA molecule is represented as a wavy line, cDNA molecules are straight lines, adapters are thick lines, and primers are short arrows.
Computer Analysis.
Sequence alignment was undertaken by using the Jotun Hein method, megalign, dnastar (Macintosh). The potential RNA secondary structure of 5′ noncoding region was analyzed based upon the sequence conservation by using the macdnasis program (Hitachi Software Engineering America).
RESULTS
Cloning of the 5′-Terminal Sequences of the GBV-C/HGV Genome.
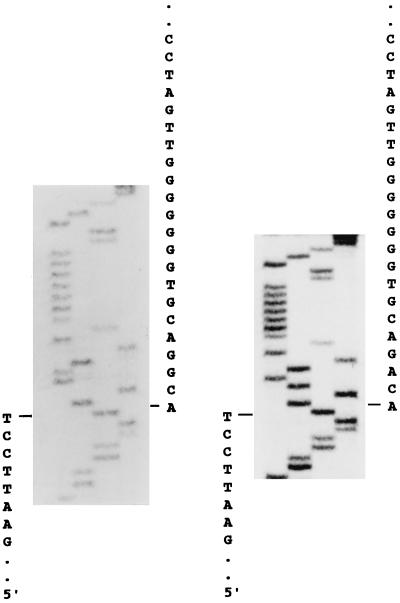
We employed the method combining the modified 5′-RACE procedure and the nested PCR to increase the sensitivity and specificity for the cloning of the 5′-terminal sequences of the GBV-C/HGV genome, as described above (Fig. 1). The nested PCR products were cloned and sequenced. Fig. 2 demonstrates the extreme 5′ sequences adjacent to the adapter sequences of two of the three isolates. As shown in Fig. 3, in alignment with the three isolates reported by Linnen et al. (ref. 2; American isolates PNF2161 and R10291, designated HGV-1 and HGV-2, respectively) and by Leary et al. (ref. 3; West African isolate designated GBV-C), all three of our isolates, T-138, T-338, and T-094, have compatible extreme 5′ sequences that have four more bases in the extreme 5′ ends than the sequence of HGV-1. Paradoxically, HGV-2 and GBV-C have much shorter and discrepant extreme 5′ ends (Fig. 3).
Figure 2.
Sequencing analysis of the extreme 5′ terminus of hepatitis GBV-C/HGV virus. RNA was extracted from 100 μl of serum from patients with chronic infection of hepatitis GBV-C/HGV virus (T-138) (Left) or chronic coinfection of hepatitis GBV-C/HGV virus and HCV (T-094) (Right). Cloning of the 5′ terminus of the viral RNA genome was done as described in Fig. 1. The adapter sequences are indicated on the left and the 5′ terminus sequences of the viral RNA are labeled on the right.
Figure 3.
Alignment of nucleotide sequences of the 5′ noncoding region of six hepatitis GBV-C/HGV virus isolates. The sequences are compared with the American isolate PNF2161 (HGV-1, ref. 2) shown in the top line. Nucleotide substitutions are in uppercase type, identical nucleotides are shown as dots, and deleted or missed nucleotides are shown as a blank. The potential initiation codon for the viral polyprotein is indicated by an arrowhead. The three variable regions are indicated as V-1 (nucleotides −399 to −369), V-2 (nucleotides −274 to −238), and V-3 (nucleotides −53 to −27).
Comparison of Sequence Homology and Divergence of the 5′ Noncoding Region.
A comparison of the sequence homology of the 5′ noncoding region among the three Taiwan isolates, the two American isolates (2), and the West African isolate (3) is shown in Fig. 3 and summarized in Table 1. Sequence homology among the three Taiwan isolates and between the two American isolates is 97% and 96%, respectively. However, the sequence homology among the isolates from different areas is relatively low. It is only about 90% between the Taiwan isolates and either the American isolates or the West African isolate. Similarly, it is only about 88% between the American isolates and the West African isolate. On the other hand, there is no significant sequence homology between any of the GBV-C/HGV isolates and the HCV.
Table 1.
Primary sequence identity (%) of the 5′ noncoding region
| T-138 | T-338 | T-094 | HGV-1 | HGV-2 | GBV-C | |
|---|---|---|---|---|---|---|
| T-338 | 97 | |||||
| T-094 | 97 | 97 | ||||
| HGV-1 | 90 | 90 | 90 | |||
| HGV-2 | 90 | 90 | 90 | 96 | ||
| GBV-C | 90 | 90 | 90 | 88 | 88 | |
| HCV | <30 | <30 | <30 | <30 | <30 | <30 |
In addition, 70–75% of the nucleotide heterogeneity among the GBV-C/HGV isolates from the different geographic areas is constrained to three regions: variable region 1 (V-1) from nucleotides −399 to −369, variable region 2 (V-2) from nucleotides −274 to −238, and variable region 3 (V-3) from nucleotides −53 to −27. However, isolates from the same geographic area have high sequence homology even in the variable region (Fig. 3). Otherwise, most of the 5′ noncoding region shows high sequence consensus among all the isolates.
Conservation of the 5′ Hairpin Structure.
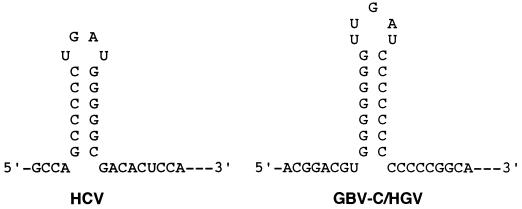
The most striking sequence consensus is the polypurine and polypyrimidine tracts in the 5′-terminal region (Fig. 3). A hairpin structure can be formed by base pairing of these two tracts (Fig. 4). Such a hairpin structure near the 5′ end of viral genome is reminiscent of that of the HCV and pestiviruses (8). It is worthy to note that both the 5′ hairpin structures of GBV-C/HGV and of HCV have very similar loop sequences (UUGAU vs. UGAU) and stem structures (seven G·C pairs vs. six G·C pairs).
Figure 4.
Proposed 5′ hairpin structures of the HCV and the hepatitis GBV-C/HGV virus. The 5′ hairpin structure of the HCV is in accordance with Brown et al. (8) and Honda et al. (11). Modulation of the potential 5′ hairpin structure of the GBV-C/HGV is based on computer analysis (macdnasis) and sequence conservation.
DISCUSSION
We have cloned the extreme 5′ noncoding region of the hepatitis GBV-C/HGV viral RNA genome by using an RNA-ligase-mediated reverse transcription-coupled PCR procedure, which was modified from the 5′ RACE (6, 7). Two lines of evidence support the hypothesis that the sequences reported herein were the authentic or near authentic 5′ end of the viral genome. (i) Consistent 5′-end sequences were cloned from all the three Taiwanese patients, and these sequences were similar to that of the American isolate PNF2161 (HGV-1). (ii) A hairpin structure was predicted near the extreme 5′ end that resembles the structure found in the 5′ end of HCV genome.
Paradoxically, the American isolate R10291 (HGV-2) and the West African isolate of GBV-C have much shorter 5′ noncoding region sequences (Fig. 3; refs. 2 and 3). We favor the interpretation that it is due to incompleteness in their 5′ ends rather than due to any major heterogeneity of the 5′-terminal sequences of the viral genome. All three Taiwan isolates have the same 5′ end, which have four more bases in the 5′ end than one of the American isolate (HGV-1). It is probable that four bases have been missed during the cloning procedure. Alternatively, there is a real four-base heterogeneity in the 5′ end of GBV-C/HGV.
Our data also reveal that the 5′ noncoding region of the hepatitis GBV-C/HGV virus is highly conserved among the isolates from the same geographic area, while it is relatively less well conserved among the isolates from different areas of the world (Table 1). Obviously, there are different genotypes that are related to geographic separation. Based upon the 5′ noncoding region sequences, there are at least three genotypes among these six hepatitis GBV-C/HGV virus isolates–i.e., the Taiwan, the American, and the West African genotypes. Geographic genome variation is also observed in the HCV groups (9, 10). Interestingly, sequence heterogeneity by geographic separation are confined mainly to the three variable domains (Fig. 3). They should be the ideal sequences for genotyping of GBV-C/HGV. In contrast, the sequence homology between the hepatitis GBV-C/HGV and the HCV is less than 30%, indicating that the hepatitis GBV-C/HGV virus does not belong to the HCV group, as suggested by others (1–3).
A potential specific hairpin structure is detected near the extreme 5′ end. The 32 residues of the 5′ termini, containing a polypurine tract and a polypyrimidine tract, can form a hairpin structure (Fig. 4). Such a characteristic hairpin structure is also found at the 5′ termini of HCV (6, 11), alphaviruses (12), enteroviruses, rhinoviruses, and picornaviruses. This structure has been postulated to be a recognition signal for replicase or capsid protein in alphaviruses (12). It has also been demonstrated that the 5′-proximal RNA hairpin may play a major role in the life cycle of poliovirus, since infectious cDNA clones mutated in that region were replication-deficient (13). It has also been shown that the 5′ hairpin structure is a regulator of viral gene expression in both HCV (14, 15) and pestiviruses (16). Most strikingly, as shown in Fig. 4, the 5′ hairpin structures of GBV-C/HGV and HCV have very similar stem structures (seven G·C pairs vs. six G·C pairs) and loop sequences (UUGAU vs. UGAU). It is probable that HCV and GBV-C/HGV have very similar protein–RNA interaction in the regulation of viral gene expression, genome encapsidation, or even genome replication. In natural human infection, about one-forth of cases of chronic hepatitis C was found to be concurrently infected with GBV-C/HGV, and vice versa (S.-Y.H., P.-Y.Y. and Y.-F.L., unpublished results). It is intriguing to speculate that viral interaction between HCV and GBV-C/HGV in the same hepatocytes may occur. On the other hand, conservation of the 5′-terminal hairpin structure further substantiates our hypothesis that the 5′-terminal sequences reported herein represent the authentic or near authentic 5′ end of the hepatitis GBV-C/HGV viral genome.
The unusual long segment with structure conservation and multiple small open reading frames of the 5′ noncoding regions of HCV, pestiviruses, and polioviruses have been proven to be related to a special translation mechanism in these viruses (10, 17). An internal ribosomal binding site directing the initiation of viral translation has been found in the 5′ noncoding region of these viruses (15, 18–21). Similar characteristics in the 5′ noncoding region of GBV-C/HGV suggest that GBV-C/HGV also utilizes a compatible cap-independent internal initiation mechanism for viral gene expression. Further studies to examine this hypothesis are currently in progress in our laboratory.
During preparation of this report, Simons et al. (22) demonstrated circumstantial evidence for internal initiation of translation of GBV-C/HGV. However, the translation efficiency of the 5′ noncoding region of GBV-C/HGV within a bicistronic context was only 2–5% of that of the HCV 5′ noncoding region in rabbit reticulocyte lysates in vitro. Though several possibilities have been proposed, two questions remained unsolved in their report. (i) The initiation codon of the viral polyprotein they mapped was 96 nucleotides or 32 amino acid residues downstream of the initiation codon reported herein and would thus eliminate the basic core protein, required for the assembly of the viral core particle, in the N terminus of the viral polyprotein. As mentioned above, in the cases of concurrent infection with HCV, HCV might provide the core protein for GBV-C/HGV packaging, probably, via interaction with the above mentioned consensus 5′ hairpin structure. In spite of that, the low translation efficiency and lack of core protein were inconsistent with the observation that serum concentration of GBV-C/HGV is usually 100-fold higher than that of HCV in natural infection (108 vs. 106 copies per ml of serum; S.-Y.H., P.-Y.Y. and Y.-F.L., unpublished data). (ii) Sixty-three nucleotides in the extreme 5′ end were missing in their clones. This missing region should contain cis-acting elements, including the potential 5′ hairpin structure reported herein, required for regulation of translation efficiency and even the translation initiation site.
In conclusion, we have cloned the extreme 5′-terminal sequences of the hepatitis GBV-C/HGV viral genome. Sequence analysis of the 5′ noncoding region revealed that the three Taiwan isolates belonged to a separate genotype, different from the West African or the American type. The sequence heterogeneity by geographic separation, confined mainly to certain variable regions, is valuable for selection of optimal primer sequences for the genotyping of the GBV-C/HGV by reverse transcription-coupled PCR assays. The conserved 5′ hairpin structure that resembles that of HCV implies a translation mechanism analogous to that of HCV and, possibly, a viral interaction with HCV in natural infection.
Acknowledgments
We thank Dr. Ken-Tek Chow and Professor Kenneth S.-S. Chang for the valuable discussion and critical reading of the manuscript. This work was supported in part by a fund from the Chang Gung Memorial Hospital and a grant from the National Health Research Institute, Taiwan (DOH-85-HR-522).
ABBREVIATIONS
- HCV
hepatitis C virus
- GBV-C
GB virus C
- HGV
hepatitis G virus
- RACE
rapid amplification of the cDNA ends
Footnotes
References
- 1.Simons J N, Leary T P, Dawson G J, Pilot-Matias T J, Muerhoss A S, Schlauder G G, Desai S M, Mushahwar I K. Nat Med. 1995;6:564–569. doi: 10.1038/nm0695-564. [DOI] [PubMed] [Google Scholar]
- 2.Linnen J, Wages J, Jr, Zhang-Keck Z Y, Fry K E, Krzysztof Z, et al. Science. 1996;271:505–508. doi: 10.1126/science.271.5248.505. [DOI] [PubMed] [Google Scholar]
- 3.Leary T P, Muerhoff S A, Simons J N, Pilot-Matias T J, Erker J C, Chalmers M L, Schlauder G G, Dawson G J, Desai S M, Muchshwar I K. J Med Virol. 1996;48:60–67. doi: 10.1002/(SICI)1096-9071(199601)48:1<60::AID-JMV10>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 4.Yoshiba M, Okamoto H, Mishiro S. Lancet. 1995;346:1131–113. doi: 10.1016/s0140-6736(95)91802-7. [DOI] [PubMed] [Google Scholar]
- 5.Frohman M A, Dush M K, Martin G R. Proc Natl Acad Sci USA. 1988;85:8898–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen Z. Trends Genet. 1996;12:87–88. doi: 10.1016/0168-9525(96)81412-0. [DOI] [PubMed] [Google Scholar]
- 7.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 8.Brown E A, Zhang H, Ping L H, Lemon S M. Nucleic Acids Res. 1992;19:5041–5045. doi: 10.1093/nar/20.19.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen P J, Lin M H, Tu S J, Chen D S. Hepatology. 1991;14:73–78. doi: 10.1002/hep.1840140113. [DOI] [PubMed] [Google Scholar]
- 10.Bukh J, Purcell R H, Miller R H. Proc Natl Acad Sci USA. 1992;89:4942–4946. doi: 10.1073/pnas.89.11.4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Honda M, Ping L H, Runbrand A, Amphlett E, Clarke B, Rowlands D, Lemon S M. Virology. 1996;222:31–42. doi: 10.1006/viro.1996.0395. [DOI] [PubMed] [Google Scholar]
- 12.Strauss E G, Strauss J H. In: Togaviridae and Flaviviridae. Schlesinger S, Schlesinger M J, editors. New York: Plenum; 1986. pp. 35–115. [Google Scholar]
- 13.Simoes E A, Sarnow P. J Virol. 1991;65:913–921. doi: 10.1128/jvi.65.2.913-921.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoo B J, Spaete R R, Geballe A P, Selby M, Houghton M, Han J H. Virology. 1992;191:889–899. doi: 10.1016/0042-6822(92)90264-p. [DOI] [PubMed] [Google Scholar]
- 15.Rijnbrand R, Bredenbeek P, van der Straaten T, Whetter L, Inchauspe G, Lemon S, Spaan W. FEBS Lett. 1995;265:115–119. doi: 10.1016/0014-5793(95)00458-l. [DOI] [PubMed] [Google Scholar]
- 16.Poole T L, Wang C, Popp R A, Potgieter L N D, Siddiqui A, Collett M S. Virology. 1996;206:750–754. doi: 10.1016/s0042-6822(95)80003-4. [DOI] [PubMed] [Google Scholar]
- 17.Han J H, Shyamala V, Richman K H, Brauer M J, Irvine B, Urdea M S, Tekamp-Olson P, Kuo G, Choo Q L, Houghton M. Proc Natl Acad Sci USA. 1991;88:1711–1715. doi: 10.1073/pnas.88.5.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsuklyyama-Kohea K, Lizuka N, Kohara M, Nomoto A. J Virol. 1992;66:1476–1478. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang C, Sarnow P, Siddiqui A. J Virol. 1993;67:3338–3344. doi: 10.1128/jvi.67.6.3338-3344.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fukushi S, Katayama K, Kurihara C, Ishiyama N, Hoshino F B, Ando T, Oya A. Biochem Biophys Res Commun. 1994;199:425–432. doi: 10.1006/bbrc.1994.1246. [DOI] [PubMed] [Google Scholar]
- 21.Reynold J E, Kaminiski A, Kettinen H J, Carroll A R, Rowlands D J, Jackson R J. EMBO J. 1995;14:6010–6020. doi: 10.1002/j.1460-2075.1995.tb00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Simons J N, Desai S M, Schultz D E, Lemon S M, Mushahwar I K. J Virol. 1996;70:6126–6135. doi: 10.1128/jvi.70.9.6126-6135.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]