Abstract
Transcriptional activation in yeast INO1 chromatin was studied using the indirect end-labeling technique. INO1 chromatin is organized into an ordered, overlapping nucleosomal array under repressing conditions. Nucleosome positions were only disrupted at the promoter region under inducing conditions in the presence of SWI/SNF and INO80. Mutants lacking either remodeler demonstrated identical positioning patterns as the wild type under repressing conditions. This indicates that these two remodelers are responsible and essential for local nucleosomal mobilization at the INO1 promoter. The area of local nucleosome movement is consistent with the previously identified region of histone deacetylation activity. In light of these findings, we suggest that nucleosomes subject to local mobilization are also targets for local histone modifications.
Keywords: INO1, SWI/SNF, INO80, indirect end-labeling, chromatin structure, local nucleosome movement
Introduction
Phospholipid structural genes play an important role in regulating yeast cellular processes. These genes encode enzymes responsible for phospholipid synthesis, which define the structural integrity of cells during mitosis. One of the major membrane phospholipids found in mitotically growing cells is phosphatidylinositol (PI). PI is a ubiquitous membrane lipid, which represents a significant portion of the total phospholipid composition in yeast. In addition to its role in membrane structural maintenance, PI also plays a part in many cellular activities such as mRNA export, signal transduction pathways and vesicular trafficking (1, 2)
The de novo synthesis of PI from glucose-6-phosphate requires the structural gene INO1. INO1 encodes inositol-1-phosphate synthase, which catalyzes the rate-limiting step in the synthesis of inositol. Its expression is transcriptionally regulated by inositol and choline, two precursor molecules in phospholipid biosynthesis. In their presence, transcription is repressed and in their absence, it is activated (3). This regulation is dictated by a cis-acting promoter element termed UASINO, known as the inositol sensitive upstream activation sequence; consensus: 5′ CATGTGAAAT 3′ (4, 5). UASINO elements are binding sites for the heterodimeric transcriptional activator, Ino2p/Ino4p, encoded by INO2 and INO4, respectively. They possess basic helix-loop-helix DNA binding motifs, which activate transcription through interactions with UASINO (6–9). Sequence analysis reveals that there may be 6 potential UASINO elements within the INO1 promoter. However, it has been shown that only two are bound by Ino2p/Ino4p, and that both are required to support transcriptional activation (4, 6, 9). Nonetheless, despite the continual binding of Ino2p/Ino4p to UASINO, inositol suppresses transcriptional activation via the Opi1p repressor. Opi1p mediates repression via its activator interaction domain, which binds to the repressor interaction domain of Ino2p (10–12).
INO1 expression is also regulated by chromatin remodeling activities. It has been shown that the histone acetylases Gcn5p and Esa1p acetylate histones H3 and H4, respectively, at the INO1 promoter (13). Furthermore, the recruitment of the Gcn5p-SAGA complex is dependent on histone H3 Ser10 phosphorylation. This histone modification is catalyzed by Snf1 kinase, which is recruited by the Ino2p activation domain (14–16). On the other hand, as with other genes, histone deacetylation negatively regulates INO1. Much evidence suggests that Ume6p binds to a URS1 (upstream repressor sequence) element within the INO1 promoter, and this interaction recruits the Sin3p-Rpd3p HDAC (histone deacetylase complex) complex to repress INO1 expression (17–21).
Chromatin remodeling complexes provide a different means for gene activation. Several lines of evidence have shown that both SWI/SNF and INO80 are required for INO1 activation (22–25), although ISWI has been shown to negatively regulate INO1 expression (26, 27). However, current studies have only demonstrated the influence of individual remodelers in INO1 activation. It is still unclear how these complexes work cooperatively at the INO1 promoter.
In the present study, we aimed to understand the structural consequences of the interplay between the SWI/SNF and INO80 remodeling complexes at the INO1 promoter. To address this interest, we studied the effect of both remodelers on INO1 chromatin structure. We compared nucleosome positions in INO1 in repressing wild-type, inducing wild-type, inducing snf2Δ, and inducing ino80Δ chromatin. Our results suggest that nucleosome movement occurs at the promoter during gene activation and that both SWI/SNF and INO80 are equally important for this activity.
Materials and methods
Yeast strains and media
WT (wild-type) yeast strain BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0), snf2Δ strain (snf2Δ, MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 snf2Δ), and ino80Δ strain (ino80Δ, MATa ino80Δ::TRP1 his3Δ200 leu2Δ0 met15Δ0 trp1Δ63 ura3Δ0) (26) were used in this study. All yeast culture was grown at 30°C in SC media (synthetic complete media) containing 2% glucose (wt/vol) with100 µM inositol (myo-inositol; Sigma I-7508)(for repressing conditions; SC) or without (for inducing conditions; SC-ino) except for ino80Δ, which was grown in SC-trp (SC medium lacking tryptophan) or SC-trp-ino (SC medium lacking both tryptophan and inositol) (3).
RNA preparation and Northern blot analysis
For WT and snf2Δ strains, cells from overnight cultures were harvested and washed three times with SC-ino. Subsequently, cells were inoculated into SC or SC-ino and 40 ml of culture were removed immediately and 1 h (hour) after inoculation for RNA preparation. For the ino80Δ strain, cells were washed in SC-trp-ino, and inoculated in either SC-trp or SC-trp-ino. Total RNA was prepared from collected cells as described previously (28). Equal amounts (3 µg) of RNA were used in the Northern blots as described previously (28). The INO1 probe was a 459-bp PCR fragment (forward primer: TAATATTGCTCCAATCACCTCC; reverse primer: GCTTCGTATAGATCTGCGTTAT) and the ACT1 probe was a 269-bp PCR fragment (forward primer: ATGGATTCTGGTATGTTCTAGC; reverse primer: CATGAGACTTAGTAACAGTAGC).
Indirect end-labeling analysis
Nuclei were prepared from yeast cells as described previously (28, 29). Briefly, WT cells were grown to mid-log phase in SC-ino or SC, harvested, and washed twice with SC-ino or SC, and spheroplasts were prepared. Spheroplasts were incubated in SM (spheroplast medium, SC with 1M D-sorbitol) or SM-ino (spheroplast medium without inositol, SC-ino with 1M D-sorbitol) for 1 h under repressing or inducing conditions. Nuclei were released from spheroplasts and digested with MNase (Micrococcal nuclease). The DNA was purified, digested with BlpI and electrophoresed in long 1.2% (w/v) agarose gels. The gels were blotted and probed with an INO1 BlpI/SnaBI fragment labeled by random priming. All data were quantified using a phosphorimager. For inducing snf2Δ and ino80Δ cells, cells were grown to mid-log phase in SC or SC-trp, harvested, washed twice with SC-ino or SC-trp-ino, and spheroplasts were prepared as described above using SM-ino or SM-trp-ino. Spheroplasts were incubated in SM-ino or SM-trp-ino for 1 h under inducing conditions followed by nuclei preparation.
Results
Both INO80 and SNF2 are required for INO1 transcriptional activation
It has been shown that INO1 is expressed only during the logarithmic phase of growth in the absence of inositol (30, 31). To study how INO80 regulates INO1 expression, both WT and ino80Δ cells were grown in SC or SC-trp. When the O.D. reached 0.1~0.3, cells were harvested by centrifugation and washed twice with SC-ino or SC-trp-ino to remove inositol completely. For inducing conditions, cells were resuspended in SC-ino or SC-trp-ino and incubated at 30°C. Inositol was added to a final concentration of 100 µM as required for repressing conditions. Aliquots of culture were removed for O.D. measurement following inositol addition. The results showed that ino80Δ cells grew poorly in the absence of inositol (Figure 1A). Addition of inositol, as a result, rescues the inositol auxotrophy of the ino80Δ cells. Therefore, INO80 is required for INO1 expression.
Figure 1.
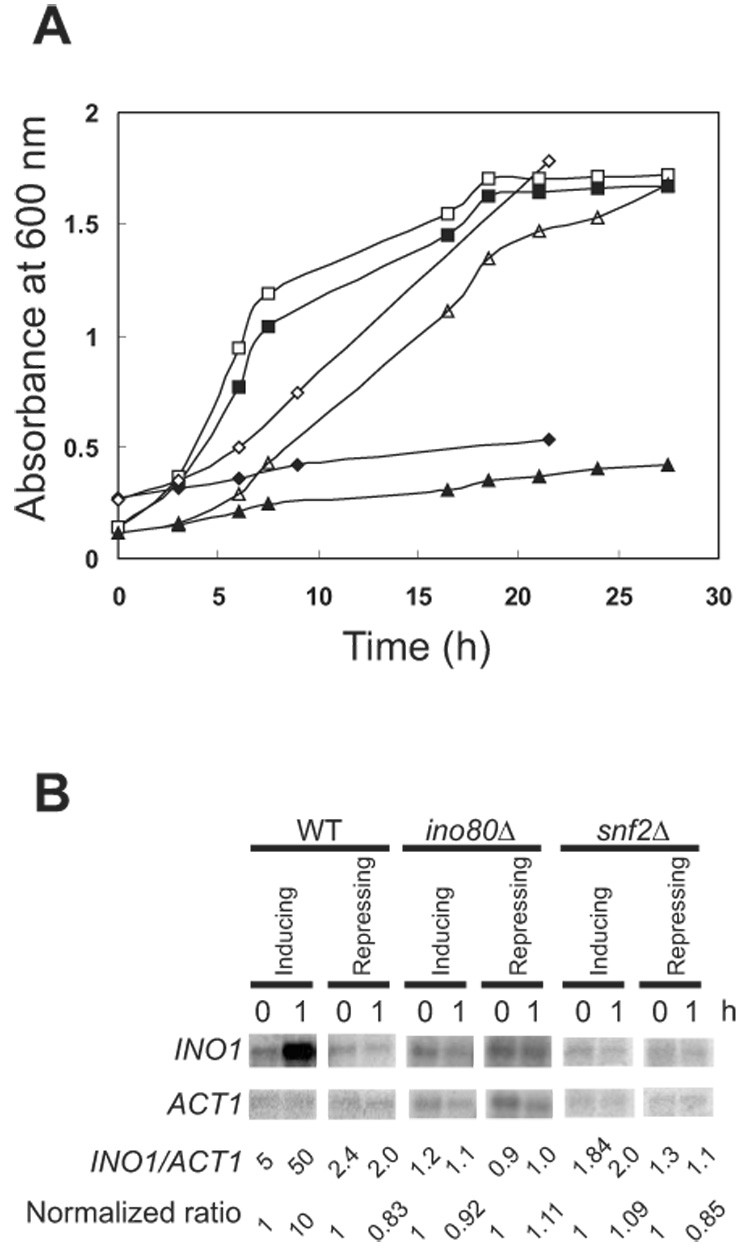
Both INO80 and SWI/SNF remodeling complexes are required for INO1 expression. (A) INO80 and SWI/SNF rescue cells from inositol auxotrophy. Yeast cell growth was monitored by A600. Cells from overnight cultures were grown in SC/SC-trp and washed three times in media without inositol. Subsequently, cells were inoculated into media with or without 100 µM inositol. WT: (□) SC + inositol, (■) SC −inositol. Mutant ino80Δ: (△) SC – trp + inositol, (▲)SC – trp − inositol. Mutant snf2Δ: (◇) SC + inositol, (◆) SC − inositol. (B) Induction of INO1 in WT, ino80Δ and snf2Δ cells. INO1 and ACT1 mRNA were detected by Northern blot hybridization with INO1 and ACT1 probes, respectively. Cells were collected immediately and 1 h after inoculation in repressing or inducing media.
To investigate whether SWI/SNF is also required for INO1 gene expression, we performed growth experiments with snf2Δ cells. We found that the growth of snf2Δ cells, as with ino80Δ cells, is severely inhibited in the absence of inositol (Figure 1A). The inositol auxotrophy is circumvented upon addition of inositol to the medium, indicating that SWI/SNF also plays a crucial role in INO1 induction. Thus, the deletion of both INO80 and SNF2 led to inositol auxotrophy due to an inability to express INO1.
To further examine the role of these remodelers on INO1 expression, INO1 mRNA levels were measured by Northern blot analysis. The normalized expression ratio of INO1/ACT1 increased 10 fold for WT cells after a 1 h induction period; the ratio was approximately 0.83 for WT cells under repressing conditions (Figure 1B). Thus, INO1 is expressed within 1 h of induction under the given conditions. For both ino80Δ and snf2Δ cells, INO1 mRNA did not significantly increase in either inducing or repressing conditions. The ratio of INO1/ACT1 was approximately 1 for both strains under both conditions (Figure 1B). This suggests that both SWI/SNF and INO80 are required for INO1 expression and that they regulate INO1 induction at the transcriptional level.
Analysis of the INO1 Chromatin structure
Indirect end-labeling analysis coupled with MNase digestion of nuclei has been widely applied for studying the in vivo translational map of nucleosome positions (32–34). We employed this technique to investigate the INO1 chromatin structure under repressing and inducing conditions. The WT cells from repressing and inducing conditions were compared and a complex but highly reproducible pattern was observed (Figure 2A, lanes 3 to 8 and lanes 11 to 14). Many bands of various intensities were observed; each band represents the upstream border of a nucleosome with respect to the BlpI restriction enzyme site downstream of the INO1 transcription start site. The intensity indicated the distribution of nucleosomes in that particular position (Figure 2B). Nucleosome positions corresponding to the bands observed in Figure 2A are shown in detail in Figure 4. Theoretically, the area of interests including the INO1 promoter and part of the ORF (−540 to +678 relative to the transcription start site; 1217 bp)(4) could accommodate a maximum of seven uniquely positioned nucleosomes if closely packed (Figure 4). However, there are many more than seven bands in repressed chromatin and the neighboring bands were significantly less than 147 bp apart. Based on our previous work, these bands probably indicate alternative overlapping nucleosome positions (28, 29, 35). Thus, our map revealed the presence of overlapping nucleosome positions on INO1. Furthermore, these positions are mutually exclusive as only one nucleosome can be present at any given moment.
Figure 2.
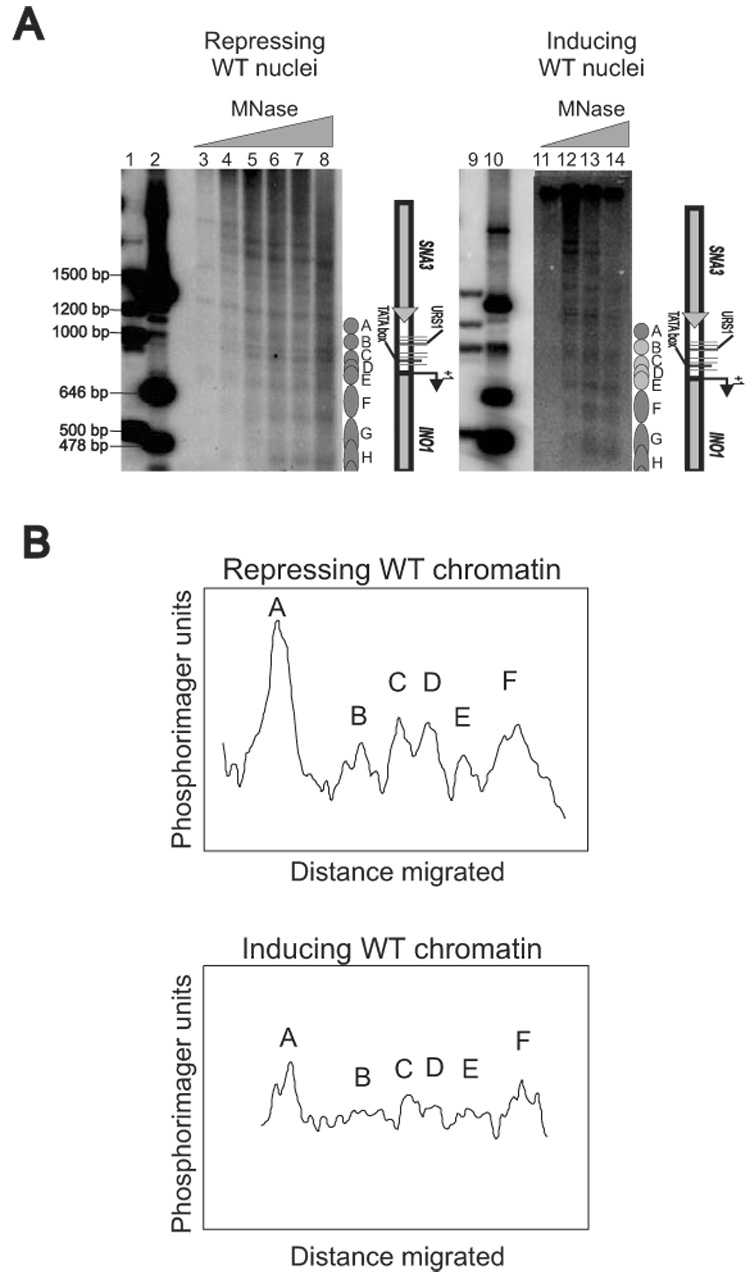
INO1 chromatin is organized into an overlapping nucleosomal array. (A) Nucleosomal organization of the INO1 chromatin in repressing WT and inducing WT cells. Nuclei were prepared from different strains and digested with increasing amounts of MNase; purified DNA was analyzed in a 1.2% agarose gel. The Southern blot was probed with a radiolabeled BlpI/SnaBI fragment. Lanes 1, 2, 9, and 10 are size markers. (B) Scan of the indirect end-labeling maps shown in panel A. The lanes corresponding to repressing WT chromatin (lane 6) and inducing WT chromatin (lane 13), were scanned using a phosphorimager.
Figure 4.
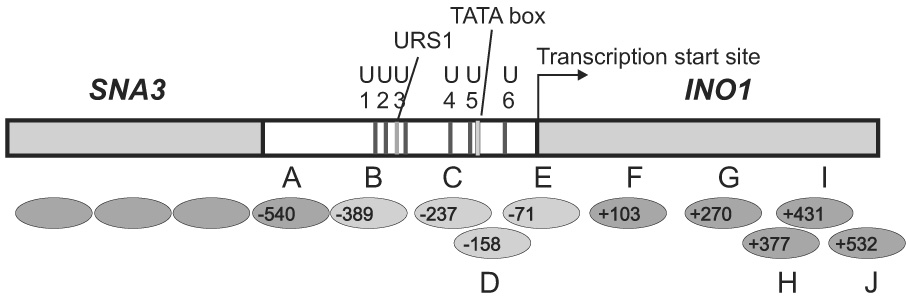
Arrays of positioned nucleosomes on INO1. The nucleosome positions mapped by indirect end-labeling in Figure 2 and Figure 3 are given with upstream borders mapped with respect to the BlpI site. INO1 coordinates are given with respect to the transcription start site at nucleotide +1 (6)(indicated by the arrow). The most important sequence elements in the INO1 promoter are indicated by the labeled lines: the TATA box, the URS1 element, and the potential UASINO1 to UASINO6 (U1 to U6). Positioned nucleosomes are shown as ovals labeled A to J. Nucleosomes shown as light gray ovals are nucleosomes subject to remodeling in induction.
Induced INO1 chromatin displayed different patterns from repressed INO1 chromatin, since some positions were disrupted under inducing conditions (Figure 2A, compare lane 7with lane 13; Figure 2B). For example, the upstream borders of Nucleosomes B, D, and E disappeared under inducing conditions, indicating that these three were remodeled or removed in the process of induction. The upstream borders of Nucleosome C became weaker during induction, suggesting that it has also been redistributed to other alternative positions or that it has been removed from its original position.
The disruption of both Nucleosomes B and C is very important. Nucleosome B covers UASINO 1, UASINO 2, UASINO 3, and URS1 sequences, while Nucleosome C covers UASINO 4, UASINO 5, and the TATA box (Figure 4). Both UASINO 3 and UASINO 4 have been confirmed as the target sites for the Ino2p/Ino4p heterodimer and appear to be functional in response to inositol deprivation (9). Thus, the removal of both nucleosomes provides the opportunity for the transcription machinery to access its binding sites. Interestingly, the TATA box element is occluded by both Nucleosomes C and D. However, Nucleosome C is not completely removed in induction. It is possible that the redistribution of nucleosomes within the alternative positions during induction allows RNA polymerase II to bind. This possibility is substantiated since Nucleosomes C, D, and E are within the same set of alternative overlapping positions (Figure 4). Nucleosomes C and D, and Nucleosomes D and E are mutually exclusive, but Nucleosomes C and E can be present simultaneously. We can only observe the average chromatin structure in a cell population under the current study. It is possible that nucleosomes are moving within these positions to provide accessibility for trans-acting factors.
Both INO80 and SWI/SNF direct the localized disruption of nucleosomal arrays at the INO1 promoter
Results obtained from growth experiments and Northern blot analysis suggested that both the Snf2p and Ino80p subunits of SWI/SNF and INO80, respectively, are required for INO1 expression. Chromatin structural analysis also demonstrated the mobilization of nucleosomes upon induction. It is possible that these two chromatin remodelers are directly responsible for the nucleosome movement. The remodeler involved would presumably be capable of mobilizing nucleosomes on a DNA template in vitro. To date, only Ino80p has been tested on reconstituted chromatin containing an INO1 promoter fragment (24). SWI/SNF has not yet demonstrated similar in vitro remodeling activity for INO1 chromatin.
To confirm that both remodelers can directly mobilize nucleosomes at the INO1 promoter in vivo, the INO1 chromatin structure was analyzed using nuclei derived from ino80Δ and snf2Δ cells grown under inducing conditions. Chromatin structures were compared with that of WT INO1 chromatin. The ino80Δ and snf2Δ chromatin were very similar to repressing WT chromatin but not to inducing WT chromatin (Figure 3A, compare lane 6 and 10 with Figure 2A lanes 7 and 13, Figure 3B). This suggests that the presence of either subunit of the chromatin remodeling complexes is insufficient for disruption of INO1 chromatin. Consequently, both SWI/SNF and INO80 are required for nucleosome mobilization under inducing conditions.
Figure 3.
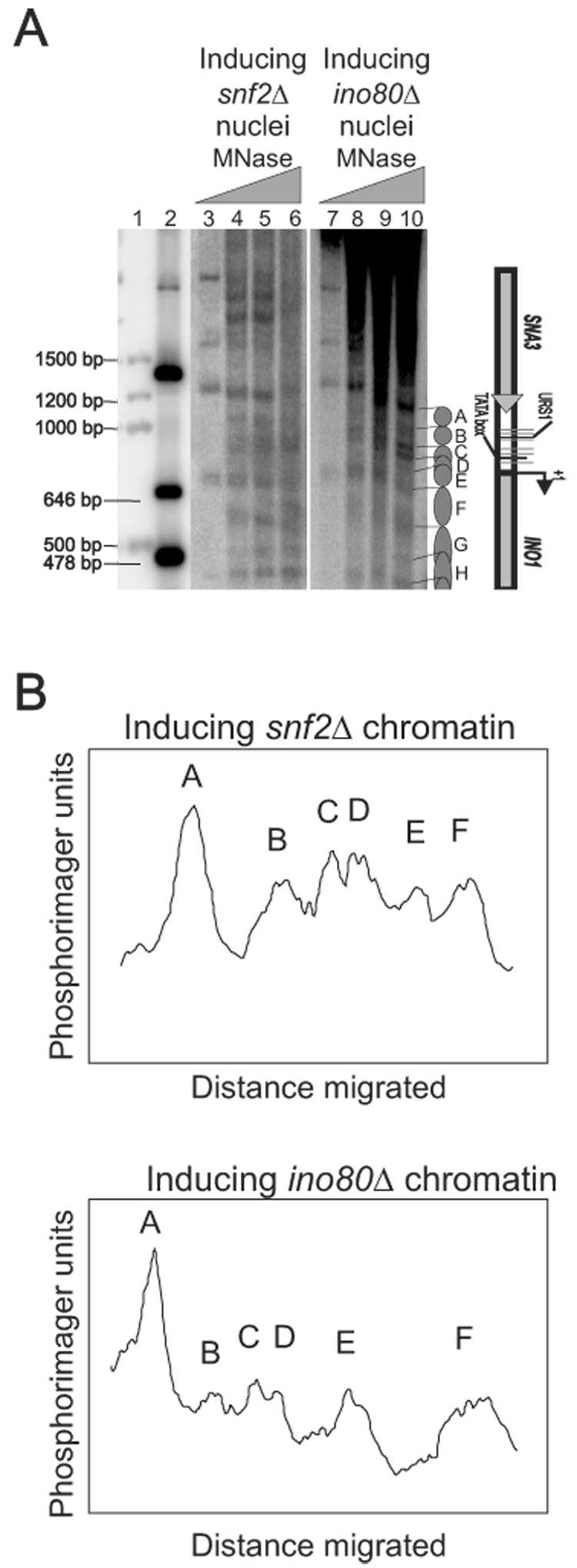
Both SWI/SNF and INO80 are required for nucleosome movement. (A) Nucleosomal organization of the INO1 chromatin in inducing snf2Δ and inducing ino80Δ cells. Samples were prepared as described in Figure 2. Lanes 1 and 2 are size markers. (B) Scan of the indirect end-labeling maps shown in panel A. The lanes corresponding to inducing snf2Δ chromatin (lane 4) and inducing ino80Δ chromatin (lane 10) were scanned using a phosphorimager.
Discussion
We have determined the INO1 chromatin structure under repressing conditions. The INO1 ORF and its promoter are organized into a set of well-positioned overlapping nucleosomes during repressing conditions. Such overlapping chromatin structures were also observed in other genes such as CUP1 (28, 35) and HIS3 (36). It is interesting to observe overlapping positions under repressing conditions. We speculate that such arrangements provide alternative choices for nucleosomes in response to various cellular activities, such as replication, transcription and repair, each may require different positions.
During induction, the orderly positioned nucleosomes were disrupted in INO1 chromatin. The disruption of nucleosomal organization strongly suggests the presence of remodeling activity at the promoter under inducing conditions. However, nucleosome movement is limited to the promoter region. These nucleosomes cover all regulatory sequences including the UAS, the URS1, and the TATA box (Figure 4). Such local remodeling is necessary for trans-acting factors to access cis-acting sequences for transcriptional activation. For instance, the TATA box is occluded by Nucleosome C and D, and the movement of both nucleosomes makes it accessible to TBP, which facilitates transcriptional activation.
In identifying the remodelers that are involved in INO1 activation, both ino80Δ and snf2Δ strains were used for indirect end-labeling analysis. Unexpectedly, we did not observe any nucleosome movement in either mutant under inducing conditions (Figure 3). This strongly suggests that both INO80 and SWI/SNF are required for remodeling during induction. Furthermore, they did not demonstrate additive effects and must be present simultaneously for nucleosome movement. This differs from previous observations in other genes. For example, in HIS3, both SWI/SNF and ISW1 act synergistically in induction (36). The ISW1 complex plays a more subtle role in nucleosome mobilization over HIS3, favoring positions different from those preferred by SWI/SNF. As a result, INO1 chromatin represents a unique transcriptional activation model.
We observed nucleosome movement only at the INO1 regulatory region ranging from −389 to +76 relative to the transcription start site, which includes Nucleosomes B, C, D, and E (Figure 4). A previous study has shown that nucleosomes in this area are subject to histone deacetylation by Rpd3p (37). In yeast, Rpd3p is recruited by the transcriptional repressor Ume6p, through an interaction with the Rpd3p-associated corepressor Sin3p. Under repressing conditions, recruitment of Rpd3p leads to histone deacetylation surrounding the URS1 motif (21). This region matches the positions of Nucleosomes B through E. Under inducing conditions, the Ume6p-Sin3p-Rpd3p complex is no longer present at the promoter, and these nucleosomes are subject to localized remodeling activities, such as histone acetylation and nucleosome movement. Therefore, Nucleosomes B, C, D and E are not only targets for nucleosome movement but also for histone modifications.
Acknowledgements
We thank Dr. Xuetong Shen for ino80 null strain. We also thank Dr. David Clark and Dr. Geoffrey Woodard for helpful comments on the manuscript. This work was supported by an NIH grant (GM67730) and PSC-CUNY awards (60084-33-34, 66243-0035 and 69182-0038).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Strahl T, Thorner J. Synthesis and function of membrane phosphoinositides in budding yeast, Saccharomyces cerevisiae. Biochim. Biophys. Acta. 2007;1771:353–404. doi: 10.1016/j.bbalip.2007.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ile KE, Schaaf G, Bankaitis VA. Phosphatidylinositol transfer proteins and cellular nanoreactors for lipid signaling. Nat. Chem. Biol. 2006;2:576–583. doi: 10.1038/nchembio835. [DOI] [PubMed] [Google Scholar]
- 3.Hirsch JP, Henry SA. Expression of the Saccharomyces cerevisiae inositol-1-phosphate synthase (INO1) gene is regulated by factors that affect phospholipid synthesis. Mol. Cell. Biol. 1986;6:3320–3328. doi: 10.1128/mcb.6.10.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lopes JM, Hirsch JP, Chorgo PA, Schulze KL, Henry SA. Analysis of sequences in the INO1 promoter that are involved in its regulation by phospholipid precursors. Nucleic Acids Res. 1991;19:1687–1693. doi: 10.1093/nar/19.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koipally J, Ashburner BP, Bachhawat N, Gill T, Hung G, Henry SA, Lopes JM. Functional characterization of the repeated UASINO element in the promoters of the INO1 and CHO2 genes of yeast. Yeast. 1996;12:653–665. doi: 10.1002/(SICI)1097-0061(19960615)12:7%3C653::AID-YEA953%3E3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 6.Lopes JM, Henry SA. Interaction of trans and cis regulatory elements in the INO1 promoter of Saccharomyces cerevisiae. Nucleic Acids Res. 1991;19:3987–3994. doi: 10.1093/nar/19.14.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwank S, Ebbert R, Rautenstrauu K, Schweizer E, Schuller HJ. Yeast transcriptional activator INO2 interacts as an Ino2p/Ino4p basic helix-loop-helix heteromeric complex with the inositol/choline-responsive element necessary for expression of phospholipid biosynthetic genes in Sacharromyces cerevisiae. Nucleic Acids Res. 1995;23:230–237. doi: 10.1093/nar/23.2.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schuller HJ, Richter K, Hoffmann B, Ebbert R, Schweizer E. DNA binding site of the yeast heteromeric Ino2p/Ino4p basic helix-loop-helix loop transcription factor: structural requirements as defined by saturation mutagenesis. FEBS Lett. 1995;370:149–152. doi: 10.1016/0014-5793(95)00818-t. [DOI] [PubMed] [Google Scholar]
- 9.Ambroziak J, Henry SA. INO2 and INO4 gene products, positive regulators of phospholipid biosynthesis in Saccharomyces cerevisiae, form a complex that binds to the INO1 promoter. J. Biol. Chem. 1994;269:15344–15349. [PubMed] [Google Scholar]
- 10.Wagner C, Dietz M, Wittmann J, Albrecht A, Schuller HJ. The negative regulatory Opi1 of phospholipid biosynthesis in yeast contacts the pleiotropic repressor Sin3 and the transcriptional activator Ino2. Mol. Microbiol. 2001;44:155–166. doi: 10.1046/j.1365-2958.2001.02495.x. [DOI] [PubMed] [Google Scholar]
- 11.Kaadige MR, Lopes JM. Analysis of Opi1p repressor mutants. Curr. Genet. 2006;49:30–38. doi: 10.1007/s00294-005-0021-6. [DOI] [PubMed] [Google Scholar]
- 12.Heyken WT, Repenning A, Kumme J, Schuller HJ. Constitutive expression of yeast phospholipid biosynthetic genes by variants of Ino2 activator defective for interaction with Opi1 repressor. Mol. Microbiol. 2005;56:696–707. doi: 10.1111/j.1365-2958.2004.04499.x. [DOI] [PubMed] [Google Scholar]
- 13.Suka N, Suka Y, Carmen AA, Wu J, Grunstein M. Highly specific antibodies determine histone acetylation site usage in yeast heterochromatin and euchromatin. Mol. Cell. 2001;8:473–479. doi: 10.1016/s1097-2765(01)00301-x. [DOI] [PubMed] [Google Scholar]
- 14.Lo WS, Trievel RC, Rojas JR, Duggan L, Hsu J, Allis CD, Marmorstein R, Berger SL. Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol. Cell. 2000;5:917–926. doi: 10.1016/s1097-2765(00)80257-9. [DOI] [PubMed] [Google Scholar]
- 15.Lo WS, Duggan L, Emre NC, Belotserkovskya R, Lane WS, Shiekhattar R, Berger SL. Snf1 – a histone kinase that works in concert with the histone acetyltransferase Gcn5 to regulate transcription. Science. 2001;293:1142–1146. doi: 10.1126/science.1062322. [DOI] [PubMed] [Google Scholar]
- 16.Lo WS, Gamache ER, Henry KW, Yang D, Pillus L, Berger SL. Histone H3 phosphorylation can promote TBP recruitment through distinct promoter-specific mechanisms. EMBO J. 2005;24:997–1008. doi: 10.1038/sj.emboj.7600577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elkhaimi M, Kaadige MR, Kamath D, Jackson JC, Biliran H, Lopes JM. Combinatorial regulation of phospholipid biosynthetic gene expressed by the UME6, SIN3 and RPD3 genes. Nucleic Acids Res. 2000;28:3160–3167. doi: 10.1093/nar/28.16.3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kadosh D, Struhl K. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deactylase to target promoters. Cell. 1997;89:365–371. doi: 10.1016/s0092-8674(00)80217-2. [DOI] [PubMed] [Google Scholar]
- 19.Kadosh D, Struhl K. Targeted recruitment of the Sin3-Rpd3 histone deactylase complex generates a highly localized domain of repressed chromatin in vivo. Mol. Cell Biol. 1998;18:5121–5127. doi: 10.1128/mcb.18.9.5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rundlett SE, Carmen AA, Kobayashi R, Bavykin S, Turner BM, Grunstein M. HDA1 and RPD3 are members of distinct yeast histone deactylase complexes that regulate silencing and transcription. Proc. Natl. Acad. Sci. U. S. A. 1996;93:14503–14508. doi: 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rundlett SE, Carmen AA, Suka N, Turner BM, Grunstein M. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature. 1998;392:831–835. doi: 10.1038/33952. [DOI] [PubMed] [Google Scholar]
- 22.Ebbert R, Birkmann A, Schuller HJ. The product of the SNF2/SWI2 paralogue INO80 of Saccharomyces cerevisiae required for efficient expression of various yeast structural genes is part of a high-molecular-weight protein complex. Mol. Microbiol. 1999;32:741–751. doi: 10.1046/j.1365-2958.1999.01390.x. [DOI] [PubMed] [Google Scholar]
- 23.Peterson CL, Herskowitz I. Characterization of the yeast SWI1, SWI2, and SWI3 genes, which encode a global activator of transcription. Cell. 1992;68:573–583. doi: 10.1016/0092-8674(92)90192-f. [DOI] [PubMed] [Google Scholar]
- 24.Shen X, Mizuguchi G, Hamiche A, Wu C. A chromatin remodeling complex involved in transcription and DNA processing. Nature. 2000;406:541–544. doi: 10.1038/35020123. [DOI] [PubMed] [Google Scholar]
- 25.Shen X, Xiao H, Ranallo R, Wu WH, Wu C. Modulation of ATP-dependent chromatin-remodeling complexes by inositol polyphosphates. Science. 2003;299:112–114. doi: 10.1126/science.1078068. [DOI] [PubMed] [Google Scholar]
- 26.Kent NA, Karabetsou N, Politis PK, Mellor J. In vivo chromatin remodeling by yeast ISWI homologs Isw1p and Isw2p. Genes Dev. 2001;15:619–626. doi: 10.1101/gad.190301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sugiyama M, Nikawa J. The Saccharomyces cerevisiae Isw2p-Itc1p complex represses INO1 expression and maintains cell morphology. J. Bacteriol. 2001;183:4985–4993. doi: 10.1128/JB.183.17.4985-4993.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen C-H, Leblanc BP, Alfieri JA, Clark DJ. Remodeling of yeast CUP1 chromatin involves activator-dependent repositioning of nucleosomes over the entire gene and flanking sequences. Mol. Cell. Biol. 2001;21:534–547. doi: 10.1128/MCB.21.2.534-547.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eriksson PR, Mendiratta G, McLaughlin N, Wolfsberg T, Marino-Ramirez L, Pompa T, Jainerin M, Landsman D, Shen C-H, Clark DJ. The putative histone acetyltransferse encoded by the yeast SPT10 gene binds specifically to upstream activating sequences in the major core histone promoters. Mol. Cell. Biol. 2005;25:9127–9137. doi: 10.1128/MCB.25.20.9127-9137.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Griac P, Swede MJ, Henry SA. The role of phosphatidylcholine biosynthesis in the regulation of the INO1 gene of yeast. J. Biol. Chem. 1996;271:25692–25698. doi: 10.1074/jbc.271.41.25692. [DOI] [PubMed] [Google Scholar]
- 31.Lamping E, Paltauf F, Henry SA, Kohlwein SD. Isolation and characterization of a mutant of Saccharomyces cerevisiae with pleiotropic deficiencies in transcriptional activation and repression. Genetics. 1995;137:55–65. doi: 10.1093/genetics/137.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dasgupta A, Juedes SA, Sprouse RO, Auble DT. Mot1-mediated control of transcription complex assembly and activity. EMBO J. 2005;24:1717–1729. doi: 10.1038/sj.emboj.7600646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Richard-Roy H, Hager GL. Sequence-specific positioning of nucleosomes over the steroid-inducible MMTV promoter. EMBO J. 1987;6:2321–2328. doi: 10.1002/j.1460-2075.1987.tb02507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Simpson RT. Nucleosome positioning – occurrence, mechanisms, and functional consequences. Prog. Nucl. Acid. Res. Mol. Biol. 1991;40:143–184. doi: 10.1016/s0079-6603(08)60841-7. [DOI] [PubMed] [Google Scholar]
- 35.Shen C-H, Clark DJ. DNA sequence plays a major role in determining nucleosome positions in yeast CUP1 chromatin. J. Biol. Chem. 2001;276:35209–35216. doi: 10.1074/jbc.M104733200. [DOI] [PubMed] [Google Scholar]
- 36.Kim Y, McLaughlin N, Lindstrom K, Tsukiyama T, Clark DJ. Activation of Saccharomyces cerevisiae HIS3 results in Gcn4p-dependent, SWI/SNF-dependent moblization of nucleosomes over the entire gene. Mol. Cell. Biol. 2006;26:8607–8622. doi: 10.1128/MCB.00678-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kadosh D, Struhl K. Repression by Ume6 involves recruitment of a complex containing Sin3 corepressor and Rpd3 histone deactylase to target promoters. Cell. 1997;89:365–371. doi: 10.1016/s0092-8674(00)80217-2. [DOI] [PubMed] [Google Scholar]


