Abstract
The DLX gene family is a family of divergent homeobox genes which are related to the Drosophila distal-less (Dll) gene and has been reported to be expressed primarily in the forebrain and craniofacial structures. We have previously identified a new member of this family, DLX-7. We now report that this gene is expressed in normal hematopoietic cells and leukemia cell lines with erythroid characteristics. We used an antisense oligonucleotide targeted against the translation start site of DLX-7 mRNA to inhibit its expression in a human erythroleukemia cell line K562, which expresses DLX-7 at a high level. The antisense oligonucleotide efficiently reduced the DLX-7 mRNA, while control oligonucleotides, including a mutant oligonucleotide identical to the antisense sequence except for four nucleotide mismatches, had no effect on DLX-7 mRNA level. Inhibition of DLX-7 expression decreased the plating efficiency by ≈70% compared with control. The antisense treatment caused apoptosis, as shown by the terminal deoxynucleotidyl transferase-mediated deoxyuridine triphosphate-digoxigenin nick end labeling (TUNEL) method. Down-regulation of DLX-7 expression by antisense treatment was associated with a reduction in GATA-1 and c-myc mRNA levels. Thus, we conclude that DLX-7 is expressed in hematopoietic cells and that the inhibition of its expression results in the decreased levels of GATA-1 and c-myc genes, with an accompanying induction of apoptosis.
Keywords: divergent homeobox, TUNEL method, antisense oligonucleotide
The homeobox gene family encodes for a group of transcription factors characterized by a highly conserved 60-amino acid DNA binding motif (1, 2). The homeobox genes, located outside the four HOX clusters, are designated “divergent” homeobox genes. Some divergent homeobox genes have implicated in leukemogenesis. In acute lymphoblastic leukemia with t(1;19) translocation, a fusion protein created between the transcription factor E2A gene and a homeobox gene PBX (3, 4). In common childhood T cell acute leukemia with t(10;14) translocation, Hox-11 is transcriptionally activated (5, 6). TRX, also called MLL, located at 11q23 is rearranged in some biphenotypic leukemias (7).
DLX genes constitute a subfamily of divergent homeobox genes related to the Drosophila distal-less (Dll) gene (8–10). Six members in this family are found in the mouse and human genome, designated DLX-1, -2, -3, -5, -6, and -7 (9–14). These genes are expressed in the forebrain, craniofacial, and limb structures (12–19). We have recently described the identification of DLX-7, a new member of this family and have mapped it to human chromosome 17q23 near DLX-3 (14).
Antisense oligonucleotides have been employed as a powerful tool to modify gene expression (20, 21). This approach to the study of specific gene function has been successfully used to study c-myb (22), c-myc (23) and some homeobox genes such as HOXC6 (24) and HOXB7 (25) in hematopoietic progenitor assays.
In this report, we demonstrate that DLX-7 is expressed in normal bone marrow (BM) cells and at a particularly high levels in cell lines with the erythroid phenotype. Inhibition of DLX-7 gene expression by an antisense oligonucleotide directed against DLX-7 in K562 cells reduced the plating efficiency and induced apoptosis. The antisense treatment was accompanied by a reduction in GATA-1 and c-myc mRNA levels. These results suggest that the function of the DLX-7 gene may be to regulate events important in hematopoietic cell survival and/or proliferation.
MATERIALS AND METHODS
Cells and Cell Lines.
K562 and HEL cells were obtained from the American Type Culture Collection and maintained in RPMI 1640 culture medium supplemented with 10% fetal calf serum. For antisense experiments, K562 cells were grown in QBSF-58 serum free medium (Quality Biological, Gaithersburg, MD). Differentiation experiments using chemical inducers were performed by culturing cells in the presence of 1 × 10−7 M phorbol 12-O-tetradecanoyl phorbol-13-acetate (Sigma) (26, 27) or 4 × 10−6 M hemin (Sigma) (28, 29). Normal BM and peripheral blood (PB) samples were obtained from healthy volunteers or as discarded materials after consent. The mononuclear cells were purified by the Ficoll/Hypaque density gradient method from heparinized PB and BM aspirates.
Oligonucleotides Treatment.
Phosphodiester oligonucleotides were used (Oligos Etc., Wilsonville, OR). Tests were initially performed to assess the toxicity of the oligonucleotides in culture. No obvious toxic effects were observed in K562 cells up to a concentration of 80 μM. The oligonucleotide sequences used here were as follows: DLX-7-antisense, GACGGACAGTTTCATAAG; DLX-7-sense, CTTATGAAACTGTCCGTC; mutant-antisense, GACTGAAAGTGTCATACG (underlined bases indicate differences compared with the antisense sequence).
For oligonucleotide treatment, K562 cells were treated with streptolysin O essentially according to Spiller and Tidd (30) with some modifications. Cells (5 × 105) were washed in permeabilization buffer (137 mM NaCl/Pipes, pH 7.4/5.6 mM glucose/2.7 mM KCl/2.7 mM EGTA/1 mM Na-ATP/0.1% bovine serum albumin), and resuspended in 0.5 units/ml of streptolysin O (Sigma) in permeabilization buffer, preactivated with 5 mM dithiothreitol for 2 h, containing 20 μM oligonucleotide. After 20-min incubation at 37°C, 5 ml of QBSF-58 serum-free medium was added to stop the permeabilization reaction.
Clonogenic Assay.
The clonogenic assay for K562 cells were performed as described (31). Briefly, 500 cells were treated with oligonucleotides and plated in quadruplicate in a culture medium consisting of Iscove’s medium supplemented with 30% fetal bovine serum, 1% bovine serum albumin, 10−4 M 2-mercaptoethanol, 2 mM l-glutamine, and 0.8% methylcellulose (Stem Cell Technologies, Vancouver, BC, Canada). The colonies (>50 cells) were scored after 7 days of incubation at 37°C in 5% CO2.
Reverse Transcriptase–PCR (RT-PCR).
Expression of DLX-7 gene and other genes was determined by RT-PCR (32). Total RNA was extracted using the acid guanidium thiocyanate-phenol-chloroform method (33). For oligonucleotide treated cells, the RNA extraction was performed 3 h after termination of the oligonucleotide treatment, using 10 μg of glycogen as a carrier. One microgram of total RNA was reverse transcribed using 1 μg of oligo(dT) and 200 units of murine leukemia virus reverse transcriptase (BRL/GIBCO) under recommended conditions. One-tenth of this cDNA was used as the template in a PCR using the following upstream and downstream primers: DLX-7 (14), CTCCAGCCTGCAGCTGCA (base pairs 402–419 of the published sequence) and CCAAAGCTGTTGCCATAGCCAC (base pairs 698–677) (expected PCR product, 297 bp); HPRT (34), GGTGGAGATGATCTCTCAAC (base pairs 439–458) and TCCAGTTTCACTAATGACAC (base pairs 723–704) (285 bp); GATA-1 (35), CCATTGCTCAACTGTATGGAGGG (base pairs 235–257) and ACTATTGGGGACAGGGAGTGATG (base pairs 483–461) (249 bp); GATA-2 (36), AGCGTCTCCAGCCTCATCTTCCGCG (base pairs 745–769) and CGAGTCTTGCTGCGCCTGCTT (base pairs 1035–1016 (291 bp); SCL (37), CTCGGCAGCGGGTTCTTT (base pairs 168–185) and AGCAGCTTGGCCAAGAAGTT (base pairs 457–438) (290 bp); c-myc (38), TCTCCGTCCTCGGATTCTCT (base pairs 649–668) and GTCTCAGGACTCTGACACTG (base pairs 1003–984) (355 bp); c-myb (39), ATTAGGTAATGAATTGTAGCCAG (base pairs 2256–2278) and ACTTAGAGTAATGCTTTTACTGA (base pairs 2483–2461) (228 bp); γ-globin (40), AGATGCTGGAGGAGAAACCCTG (base pairs 119–140) and CGAAATGGATTGCCAAAACG (base pairs 411–392); c-abl (41), GCTGGACCCAGTGAAAATGACC (base pairs 530–551) and CAAGAAGCTGCCATTGATCCC (base pairs 814–794) (285 bp); bcl-2 (42), TTCTTTGAGTTCGGTGGGGTC (base pairs 1906–1926) and CTTCAGAGACAGCCAGGAGAAATC (base pairs 2112–2089) (207 bp); bcl-xL (43) CATGGCAGCAGTAAAGCAAGC (base pairs 380–400) and CTGCGATCCGACTCACCAATAC (base pairs 636–615). All primers used flank at least one intron.
The amplification conditions were denaturation at 94°C for 30 sec, annealing for 30 sec at 50°C for DLX-7, at 55°C for HPRT, c-myb, and γ-globin, at 58°C for GATA-1, at 60°C for SCL, c-myc, c-abl, bcl-2 and bcl-xL, and at 63°C for GATA-2, and extension at 72°C for 1 min for 25 cycles for DLX-7 and 20 cycles for other genes. Solution hybridization using 10% of the PCR product was performed in a thermal cycler with the oligonucleotide probe 5′-end labeled with [γ-32P]ATP (44). The probes used were as follows: DLX-7, CTGGCAGCGCAGCTCGGC (base pairs 478–495 of the published sequence); HPRT, CTTGCTGGTGAAAAGGACC (base pairs 573–591); GATA-1, TGGATGGAAAAGGCAGCACC (base pairs 371–390); GATA-2, ACGACTACAGCAGCGGACTC (base pairs 948–967); SCL, TCGAGTGAAGAGGAGACCTT (base pairs 224–243); c-myc, GAGCCCCTGGTGCTCCATGA )base pairs 706–725); c-myb, CTGGTATTTTAAAGGATCC (base pairs 2331–2349); γ-globin, CAGCTTTGGCAACCTGTCCTC (base pairs 184–205); c-abl, CACAATGGGGAATGGTGTGAAG (base pairs 647–668); bcl-2, ACAACATCGCCCTGTGGATGAC (base pairs 1970–1991); bcl-xL, ATGGGGTAAACTGGGGTCGCATTG (base pairs 532–555). Each hybridization sample was run on an 8% polyacrylamide gel, and the gel was autoradiographed at room temperature for 4 h.
Northern Blot Analysis.
Northern membrane with 2 μg of poly(A)+ RNA per lane (CLONTECH) was hybridized for 24 h at 42°C to [α-32P]dCTP-labeled probe prepared by the random priming labeling method. After hybridization, the membrane was washed under standard conditions and autoradiographed.
In Situ Detection of Apoptosis.
Cytospin preparations of K562 cells were prepared and fixed in 4% paraformaldehyde for 30 min, washed in PBS for 5 min, and incubated in 0.1% Triton X-100 for 2 min on ice and washed in PBS twice, after which the TUNEL (terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling) (45) assay was performed using the In Situ Cell Death Detection Kit (Boehringer Mannheim) according to the manufacturer’s instructions.
Statistical Analysis.
Statistical tests were performed using the statview (Abacus Concepts, Berkeley, CA) software. The number of each colony or the percentage of positive cells was shown as the mean ± SD. Comparisons of groups were analyzed using the Student’s t test or ANOVA test. Values of P < 0.05 were considered significant.
RESULTS
DLX-7 mRNA Expression in Normal and Leukemic Hematopoietic Cells.
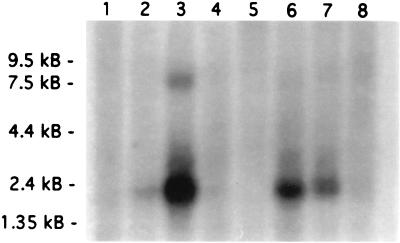
In a preliminary study, we used RT-PCR to survey the expression of DLX-7 gene in 50 leukemia cell lines and found that K562 erythroleukemia cell line expresses DLX-7 at a high level. The expression was confirmed by Northern blot analysis, which indicated a major transcript at 2.3 kb and a minor transcript at a 7.3 kb in K562 cells. Two additional nonhematopoietic cell lines, SW480 (colon adenocarcinoma) and A549 (lung adenocarcinoma), also expressed the DLX-7 gene (Fig. 1).
Figure 1.
Northern blot analysis demonstrating DLX-7 expression in leukemia cell lines and other cancer cell lines. Lanes: 1, HL-60; 2, HeLa; 3, K562; 4, MOLT-4; 5, Raji; 6, SW480; 7, A549; 8, G361.
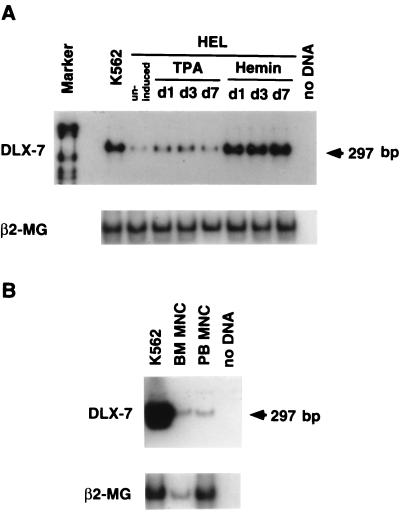
To determine whether DLX-7 gene is associated with the erythroid phenotype, we also examined HEL cells, which can be induced to undergo erythroid maturation when cultured in hemin (29). As indicated in Fig. 2A, the hemin treatment resulted in an increase in the DLX-7 mRNA to a level equivalent to K562 cells.
Figure 2.
(A) Expression of DLX-7 gene in HEL cells treated with 12-O-tetradecanoyl phorbol-13-acetate (TPA; 1 × 10−7 M) or hemin (4 × 10−6 M). RNA was reverse transcribed into cDNA, amplified by PCR using DLX-7-specific primers, and hybridized with a DLX-7 gene-specific internal oligonucleotide probe. Hemin treatment caused a marked increase in DLX-7 mRNA levels by day 1. (B) Expression of DLX-7 gene in normal BM cells and PB mononuclear cells.
DLX-7 is also expressed at detectable levels not only in BM cells but also PB mononuclear cells, indicating that the expression of this gene is not specific to leukemia cells (Fig. 2B).
Inhibition of DLX-7 mRNA Expression Level by an Antisense Oligonucleotide.
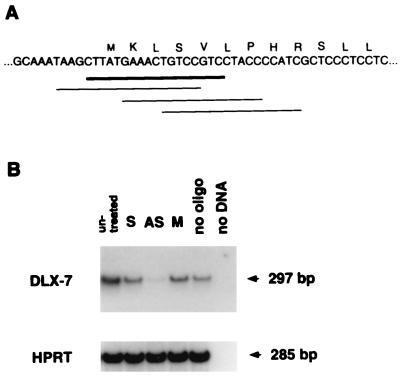
Antisense oligonucleotides inhibit gene expression by binding to its complementary site on the mRNA, resulting in an RNase H-mediated mRNA degradation (46). We tested the activity of several antisense oligonucleotides and selected one that was the most effective in reducing DLX-7 mRNA (Fig. 3A). The specificity of the antisense oligonucleotide effect was demonstrated by the lack of any effect of sense and mutant oligonucleotides (Fig. 3B). A negative control, HPRT gene, was not affected by antisense treatment.
Figure 3.
(A) Location and sequence of oligonucleotides assayed for their ability to interfere with the expression of DLX-7. The oligonucleotide exhibiting the maximal effect is shown as thick line. (B) Effect of DLX-7 antisense oligonucleotide on DLX-7 mRNA expression. RNA was extracted from K562 cells 3 h after treatment with sense (lane S), antisense (lane AS), mutant (lane M), and no (lane no) oligonucleotides and analyzed by RT-PCR. Amplification of hypothanthine phosphoribosyltransferase (HPRT) as a control is shown at the bottom. The final concentration of antisense and negative control oligonucleotides used was 20 μM.
Decreased DLX-7 Expression Results in Reduced Plating Efficiency.
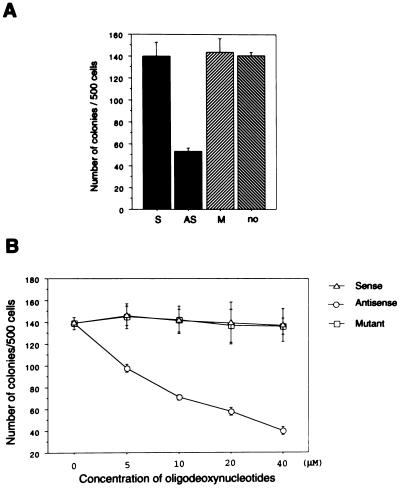
To investigate whether DLX-7 is required for K562 cell growth, we determined the plating efficiency of K562 cells treated with oligonucleotides. The DLX-7 antisense treatment at 20 μM suppressed the plating efficiency by ≈60%, while negative control oligonucleotides had no effect (Fig. 4A). Furthermore, the inhibitory effect of the DLX-7 antisense oligonucleotide was dose-dependent, ranging from 30% inhibition at 5 μM to 70% inhibition at 40 μM (Fig. 4B).
Figure 4.
(A) Effect of DLX-7 antisense treatment on the plating efficiency of K562 cells. After exposure to the oligonucleotides as indicated, 500 cells were plated per well in quadruplicate and cultured for 7 days in methylcellulose, after which colonies were counted. The concentration of the oligonucleotides was 20 μM. (B) Dose-response curve of oligonucleotides on the plating efficiency of K562 cells. Studies were carried out as in A, with the oligonucleotide concentration varied as indicated.
Reduced DLX-7 Expression Results in Apoptosis.
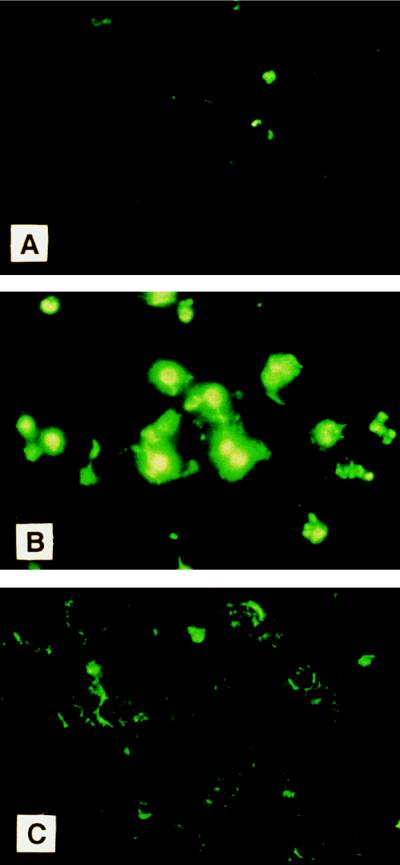
To determine whether the reduced plating efficiency is due to induction of apoptosis, K562 cells treated with antisense oligonucleotides were stained by the TUNEL method to detect in situ endonucleolytic cleavage characteristic of apoptosis. There was no detectable apoptosis at 3 h (data not shown), but after 24 h of exposure of oligonucleotides, significant apoptosis was detected in antisense-treated K562 cells compared with control cultures (Fig. 5). Thus, the reduced plating efficiency of K562 cells by DLX-7 antisense treatment is likely to be due to induction of apoptosis.
Figure 5.
In situ analysis for DNA fragmentation by the TUNEL method. At 24 h after oligonucleotide treatment, cytocentrifugation preparation of K562 cells were stained by TUNEL method. (A) No treatment. (B) Treatment with antisense. (C) Treatment with sense. Apoptotic cells were identified using fluorescent d-UTP.
Analysis of Gene Regulation by DLX-7.
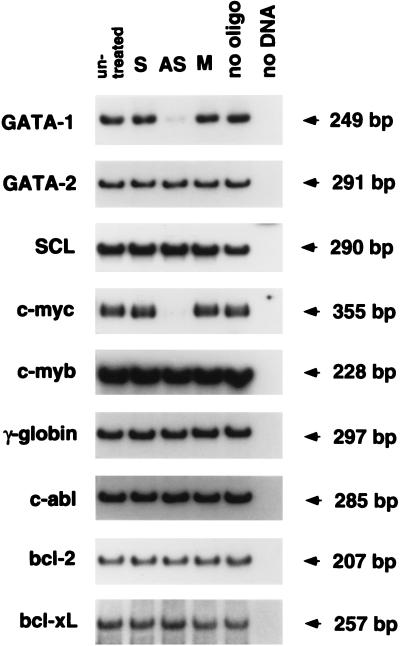
Because homeobox genes are likely to control the expression of other genes, we surveyed the expression of several genes known to be important in hematopoiesis, cell proliferation, or apoptosis. Three hours after the DLX-7 antisense treatment, K562 cells had decreased levels of GATA-1 and c-myc mRNA compared with various negative controls (Fig. 6). In contrast, GATA-2, SCL, c-myb, γ-globin, c-abl, bcl-2, and bcl-xL mRNA were unchanged (Fig. 6). We have not been able to locate any homology between the antisense oligonucleotide sequence and either GATA-1 or c-myc cDNA sequence, indicating that the effect of the antisense oligonucleotide is likely to be secondary to the effect on DLX-7 expression.
Figure 6.
Analysis of gene expression in K562 cells treated with DLX-7 antisense oligonucleotide. Total RNA was extracted 3 h after oligonucleotide treatment and analyzed by RT-PCR using the primers and probe for each gene.
DISCUSSION
Dlx genes such as Dlx-1, -2, -5, and -6 have previously been reported to be expressed primarily in the brain and craniofacial structures (9–19). We have found that DLX-7 is expressed at high levels in cell lines with erythroid characteristics such as K562 and HEL, as well as at low levels in normal BM cells. Hemin-induced erythroid differentiation of HEL cells resulted in an increase in DLX-7 mRNA levels after 24 h of exposure to hemin. We have also found that other members of the DLX gene family are expressed in hematopoietic cells (K.T., unpublished data).
We took advantage of its expression in well studied leukemia cell lines to study the possible function of DLX-7. Several authors have overexpressed homeobox genes to investigate their role in hematopoiesis (29, 30, 47, 48). The inhibition of gene expression by antisense oligonucleotides has also shown to be useful in elucidating homeobox gene function (24, 25). In this study, we have used an antisense oligonucleotide directed against DLX-7.
The antisense oligonucleotide treatment reduced GATA-1 and c-myc mRNAs at 3 h after the treatment, at a time when no apoptosis is detectable by the TUNEL assay. Apoptosis is seen at 12 h after antisense treatment and is accompanied by a reduction in the plating efficiency. The apoptosis might be mediated directly by DLX-7 or through downstream genes. At least one homeobox gene has been shown to prevent apoptosis (49). Since induction of cell death is preceded by down regulation of c-myc and GATA-1, down-regulation of one or both of these genes may be sufficient to account for the observed apoptosis (50–52). c-myc is well known to be involved in cellular decisions regarding apoptosis or proliferation (53–55), and decreasing the c-myc expression is known to cause apoptosis in leukemia cells (51, 52). GATA-1 has also recently been shown to control apoptosis. GATA-1 deficient embryonic stem cells undergo apoptosis at the proerythroblast stage (52) and overexpression of GATA-1 prevents estrogen-induced apoptosis (56). Therefore, apoptosis induced by DLX-7 gene inhibition might be due to inhibition of GATA-1 and/or c-myc gene expression, and the role of DLX-7 gene might simply be to maintain expression of GATA-1 and c-myc.
The relationship between the c-myc and GATA-1 gene expression and DLX-7 gene expression may either be direct or indirect. Homeobox-containing proteins are known to act at both transcriptional and posttranscriptional levels (57). Preliminary evidence suggests that the loss of GATA-1 mRNA in antisense-treated cells is due to increased mRNA instability (T.S., unpublished data). On the other hand, c-myc mRNA has a short half-life (58), which is not altered in antisense oligonucleotide treated K562 cells (T.S., unpublished data), suggesting that the loss of c-myc mRNA occurs at the transcriptional level.
Because DLX-7 is expressed at high levels in erythroleukemia cell lines, mediated by hemin in the case of HEL cells, and at low levels in the normal BM, it is tempting to speculate that the DLX-7 gene may be involved in some aspect of erythropoiesis, possibly in the regulation of apoptosis that occurs during normal erythropoiesis (59). Further studies are needed to address the function of DLX-7 and other DLX genes in normal hematopoiesis and to determine whether other DLX genes play a similar role in hematopoietic cells.
Acknowledgments
We are grateful for the continued support and encouragement of the Marcia Slater Society for Research in Leukemia. T.S. is a Florence A. Carter Fellow in Leukemia Research of the American Medical Association–Educational Research Fund. K.T. is supported by a Junior Faculty Research Award from the American Cancer Society. This work was supported by National Institutes of Health Grants R01DK45118 and RO1GM09966, a grant-in-aid from the New York City chapter of the American Heart Association, and the Kaplan Cancer Center (PO30CA16087) at New York University.
ABBREVIATIONS
- RT-PCR
reverse transcriptase–PCR
- PB
peripheral blood
- BM
bone marrow
- TUNEL
terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling
References
- 1.Scott M P, Tamkun J W, Hartzell G W. Biochem Biophys Acta. 1989;989:25–48. doi: 10.1016/0304-419x(89)90033-4. [DOI] [PubMed] [Google Scholar]
- 2.McGinnis W, Krumlauf R. Cell. 1992;68:283–302. doi: 10.1016/0092-8674(92)90471-n. [DOI] [PubMed] [Google Scholar]
- 3.Nourse J, Mellentin J D, Galili N, Wilkinson J, Stanbridge E, Smith S D, Cleary M L. Cell. 1990;60:535–545. doi: 10.1016/0092-8674(90)90657-z. [DOI] [PubMed] [Google Scholar]
- 4.Kamps M P, Murre C, Sun H, Baltimore D. Cell. 1990;60:547–555. doi: 10.1016/0092-8674(90)90658-2. [DOI] [PubMed] [Google Scholar]
- 5.Hatano M, Roberts C W, Minden M, Crist W M, Korsmeyer S J. Science. 1991;253:79–82. doi: 10.1126/science.1676542. [DOI] [PubMed] [Google Scholar]
- 6.Lu M, Gong Z Y, Shen W F, Ho A D. EMBO J. 1991;10:2905–2910. doi: 10.1002/j.1460-2075.1991.tb07840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Djabali M, Selleri L, Parry P, Bower M, Young B D, Evans G A. Nat Genet. 1992;2:113–118. doi: 10.1038/ng1092-113. [DOI] [PubMed] [Google Scholar]
- 8.Cohen S M, Bronner G, Kuttner F, Jurgens G, Jackle H. Nature (London) 1989;338:432–434. doi: 10.1038/338432a0. [DOI] [PubMed] [Google Scholar]
- 9.Price M, Lemaistre M, Pischetola M, DiLauro R, Duboule D. Nature (London) 1991;351:748–751. doi: 10.1038/351748a0. [DOI] [PubMed] [Google Scholar]
- 10.Porteus M H, Bulfone A, Ciaranello R D, Rubenstein J L R. Neuron. 1991;7:221–229. doi: 10.1016/0896-6273(91)90260-7. [DOI] [PubMed] [Google Scholar]
- 11.Simeone A, Acampora D, Pannese M, D’Esposito M, Stornaiuolo A, Gulisano M, Mallamaci A, Kastury K, Druck T, Huebner K, Boncinelli E. Proc Natl Acad Sci USA. 1994;91:2250–2254. doi: 10.1073/pnas.91.6.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robinson G W, Wray S, Mahon K A. New Biol. 1991;3:1183–1194. [PubMed] [Google Scholar]
- 13.Selski D, J, Thomas N E, Coleman P D, Rogers K E. Gene. 1993;132:301–303. doi: 10.1016/0378-1119(93)90212-l. [DOI] [PubMed] [Google Scholar]
- 14.Nakamura S, Stock D W, Wydner K L, Bollekens J A, Takeshita K, Nagai B M, Chiba S, Kitamura T, Freeland T M, Zhao Z, Minowada J, Lawrence J B, Weiss K M, Ruddle F H. Genomics. 1996;38:314–324. doi: 10.1006/geno.1996.0634. [DOI] [PubMed] [Google Scholar]
- 15.Price M. J Neurobiol. 1993;24:1385–1399. doi: 10.1002/neu.480241010. [DOI] [PubMed] [Google Scholar]
- 16.Ozcelik T, Porteus M H, Rubenstein J L, Francke U. Genomics. 1992;13:1157–1161. doi: 10.1016/0888-7543(92)90031-m. [DOI] [PubMed] [Google Scholar]
- 17.Dolle P, Price M, Duboule D. Differentiation (Berlin) 1992;49:93–99. doi: 10.1111/j.1432-0436.1992.tb00773.x. [DOI] [PubMed] [Google Scholar]
- 18.Bulfone A, Kim H J, Puelles L, Porteus M H, Grippo J F, Rubenstein J L. Mech Dev. 1993;40:129–140. doi: 10.1016/0925-4773(93)90071-5. [DOI] [PubMed] [Google Scholar]
- 19.Bulfone A. J Neurosci. 1993;13:3155–3172. doi: 10.1523/JNEUROSCI.13-07-03155.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tidd D M. Anticancer Res. 1990;10:1169–1182. [PubMed] [Google Scholar]
- 21.Stein C A, Cheng Y-C. Science. 1993;261:1004–1012. doi: 10.1126/science.8351515. [DOI] [PubMed] [Google Scholar]
- 22.Gewirtz A M, Calabretta B. Science. 1988;242:1303–1306. doi: 10.1126/science.2461588. [DOI] [PubMed] [Google Scholar]
- 23.Wickstrom E L, Bacon T A, Gonzalez A, Freeman D L, Lyman G H, Wickstrom E. Proc Natl Acad Sci USA. 1988;85:1028–1032. doi: 10.1073/pnas.85.4.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takeshita K, Bollekens J A, Hijiya N, Ratajczak M, Ruddle F H, Gewirtz A M. Proc Natl Acad Sci USA. 1993;90:3535–3538. doi: 10.1073/pnas.90.8.3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lill M C, Fuller J F, Herzig R, Crooks G M, Gasson J C. Blood. 1995;85:692–697. [PubMed] [Google Scholar]
- 26.Tetteroo P A T, Massaro F, Mulder A, Gelder S-V, Von Dem Borne A E G K. Leukemia Res. 1984;8:197–206. doi: 10.1016/0145-2126(84)90143-7. [DOI] [PubMed] [Google Scholar]
- 27.Papayannopoulou T, Nakamoto B, Yokochi T, Chait A, Kannagi R. Blood. 1983;62:832–845. [PubMed] [Google Scholar]
- 28.Hoffman R, Ibrahim N, Murnane M J, Diamond A, Forget B G, Levere R D. Blood. 1980;56:567–570. [PubMed] [Google Scholar]
- 29.Martin P, Papayannopoulou T. Science. 1982;216:1233–1235. doi: 10.1126/science.6177045. [DOI] [PubMed] [Google Scholar]
- 30.Spiller D G, Tidd D M. Antisense Res Dev. 1995;5:13–21. doi: 10.1089/ard.1995.5.13. [DOI] [PubMed] [Google Scholar]
- 31.Lemoli R M, Fortuna A, Tafuri A, Fogli M, Amabile M, Grande A, Ricciardi M R, Petrcci M T, Bonsi L, Bagnara G-P, Visani G, Martinelli G, Ferrari S, Tura S. Blood. 1996;87:3852–3859. [PubMed] [Google Scholar]
- 32.Ehrlich G D, Greenberg S, Abbott M A. In: PCR Protocols. Innis M A, Gelfand D H, Sninski J J, White T J, editors. San Diego: Academic; 1990. p. 325. [Google Scholar]
- 33.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 34.Konecki D S, Brennand J, Fuscoe J C, Caskey C T, Chinault A C. Nucleic Acids Res. 1982;10:6763–6775. doi: 10.1093/nar/10.21.6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tsai S F, Martin D I K, Zon L I, D’Andrea A D, Wong G G, Orkin S H. Nature (London) 1989;339:446–451. doi: 10.1038/339446a0. [DOI] [PubMed] [Google Scholar]
- 36.Dorfman D M, Wilson D B, Bruns G A P, Orkin S H. J Biol Chem. 1992;267:1279–1285. [PubMed] [Google Scholar]
- 37.Begley C G, Aplan P D, Denning S M, Haynes B F, Waldmann T A, Kirsch I R. Proc Natl Acad Sci USA. 1989;86:10128–10132. doi: 10.1073/pnas.86.24.10128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watt R, Stanton L W, Marcu K B, Gallo R C, Croce C M, Rovera G. Nature (London) 1983;303:725–729. doi: 10.1038/303725a0. [DOI] [PubMed] [Google Scholar]
- 39.Majello B, Kenyon L C, Dalla-Favera R. Proc Natl Acad Sci USA. 1986;83:9636–9640. doi: 10.1073/pnas.83.24.9636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Slightom J L, Blechl A E, Smithies O. Cell. 1980;21:627–638. doi: 10.1016/0092-8674(80)90426-2. [DOI] [PubMed] [Google Scholar]
- 41.Shtivelman E, Lifshitz B, Gale R P, Roe B A, Canaani E. Cell. 1986;47:277–284. doi: 10.1016/0092-8674(86)90450-2. [DOI] [PubMed] [Google Scholar]
- 42.Tsujimoto Y, Croce C M. Proc Natl Acad Sci USA. 1986;83:5214–5218. doi: 10.1073/pnas.83.14.5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boise L H, Gonzalez-Garcia M, Postema C E, Ding L, Lindsten T, Turka L A, Mao X, Nunez G, Tompson C B. Cell. 1993;74:597–608. doi: 10.1016/0092-8674(93)90508-n. [DOI] [PubMed] [Google Scholar]
- 44.Shimamoto T, Ohyashiki K, Ohyashiki J H, Kawakubo K, Fujimura T, Iwama H, Nakazawa S, Toyama K. Blood. 1995;86:3173–3180. [PubMed] [Google Scholar]
- 45.Gavrieli Y, Sherman Y, Ben-Sasson S A. J Cell Biol. 1992;119:493–501. doi: 10.1083/jcb.119.3.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wagner R W. Nature (London) 1994;372:333–335. doi: 10.1038/372333a0. [DOI] [PubMed] [Google Scholar]
- 47.Shen W-F, Detmer K, Mathews C H E, Hack F M, Morgan D A, Largman C, Lawrence H J. EMBO J. 1992;11:983–989. doi: 10.1002/j.1460-2075.1992.tb05137.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sauvageau G, Thorsteinsdottir U, Eaves C J, Lawrence H J, Largman C, Lansdorp P M, Humphries R K. Genes Dev. 1995;9:1753–1765. doi: 10.1101/gad.9.14.1753. [DOI] [PubMed] [Google Scholar]
- 49.Dear T N, Colledge W H, Carlton M B L, Lavenir I, Larson T, Smith A J H, Warren A J, Evans M J, Sofroniew M V, Rabbitts T H. Development (Cambridge, UK) 1995;121:2909–2915. doi: 10.1242/dev.121.9.2909. [DOI] [PubMed] [Google Scholar]
- 50.Alnemri E S, Fernandes T F, Haldar S, Croce C M, Litwack G. Cancer Res. 1992;52:491–495. [PubMed] [Google Scholar]
- 51.Oritani K, Kaisho T, Nakajima K, Hirano T. Blood. 1992;80:2298–2305. [PubMed] [Google Scholar]
- 52.Weiss M J, Orkin S H. Proc Natl Acad Sci USA. 1995;92:9623–9627. doi: 10.1073/pnas.92.21.9623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Evan G I, Wyllie A H, Gilbert C S, Littlewood T D, Land H, Brooks M, Waters C M, Penn L Z, Hancock D C. Cell. 1992;69:119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- 54.Bissonnette R P, Echeverri F, Mahboubi A, Green D R. Nature (London) 1992;359:552–554. doi: 10.1038/359552a0. [DOI] [PubMed] [Google Scholar]
- 55.Harrington E A, Bennett M R, Fanidi A, Evan G I. EMBO J. 1994;13:3286–3295. doi: 10.1002/j.1460-2075.1994.tb06630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Blobel G A, Orkin S H. Mol Cell Biol. 1996;16:1687–1694. doi: 10.1128/mcb.16.4.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rivera-Pomer R, Niessing D, Schmidt-Ott U, Gehring W J, Jackle H. Nature (London) 1996;379:746–749. doi: 10.1038/379746a0. [DOI] [PubMed] [Google Scholar]
- 58.Stewart M J, Litz-Lackson S, Burgess G S, Williamson E A, Leibowitz D S, Boswell H S. Leukemia. 1995;9:1499–1507. [PubMed] [Google Scholar]
- 59.Kelley L, Green W, Hicks G, Bondurant M C, Koury M, Ruley H. Mol Cell Biol. 1994;14:4183–4192. doi: 10.1128/mcb.14.6.4183. [DOI] [PMC free article] [PubMed] [Google Scholar]