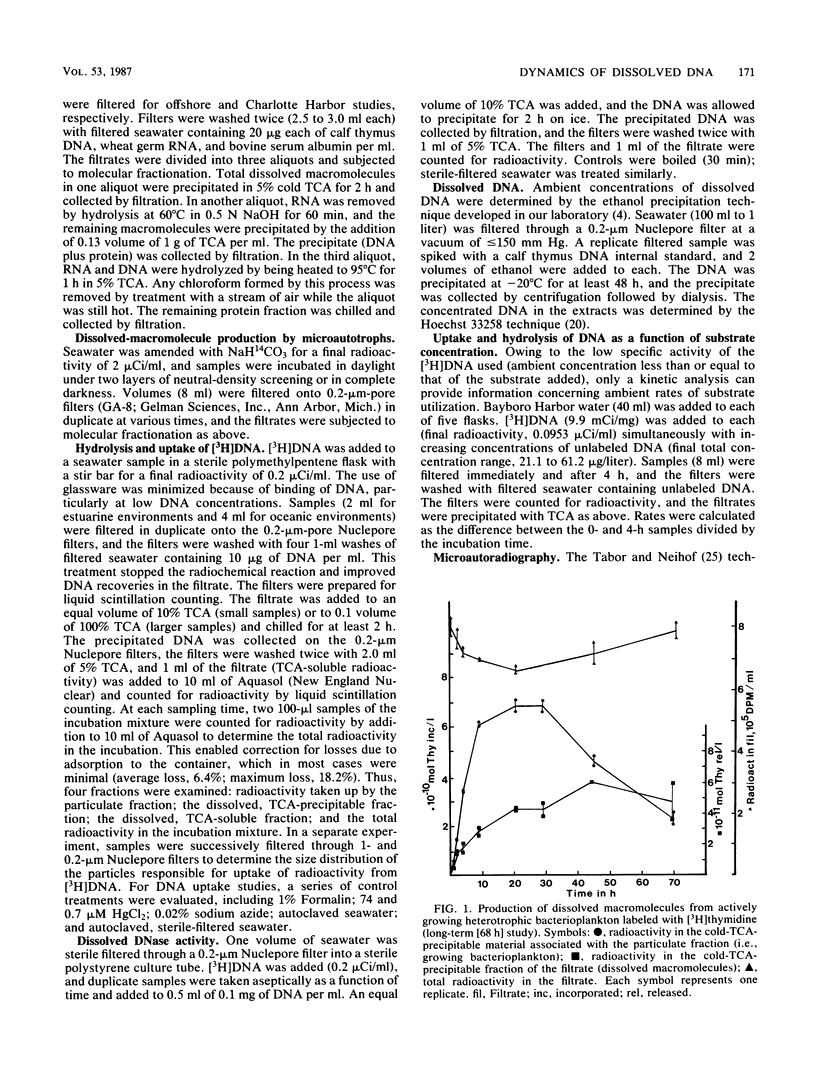
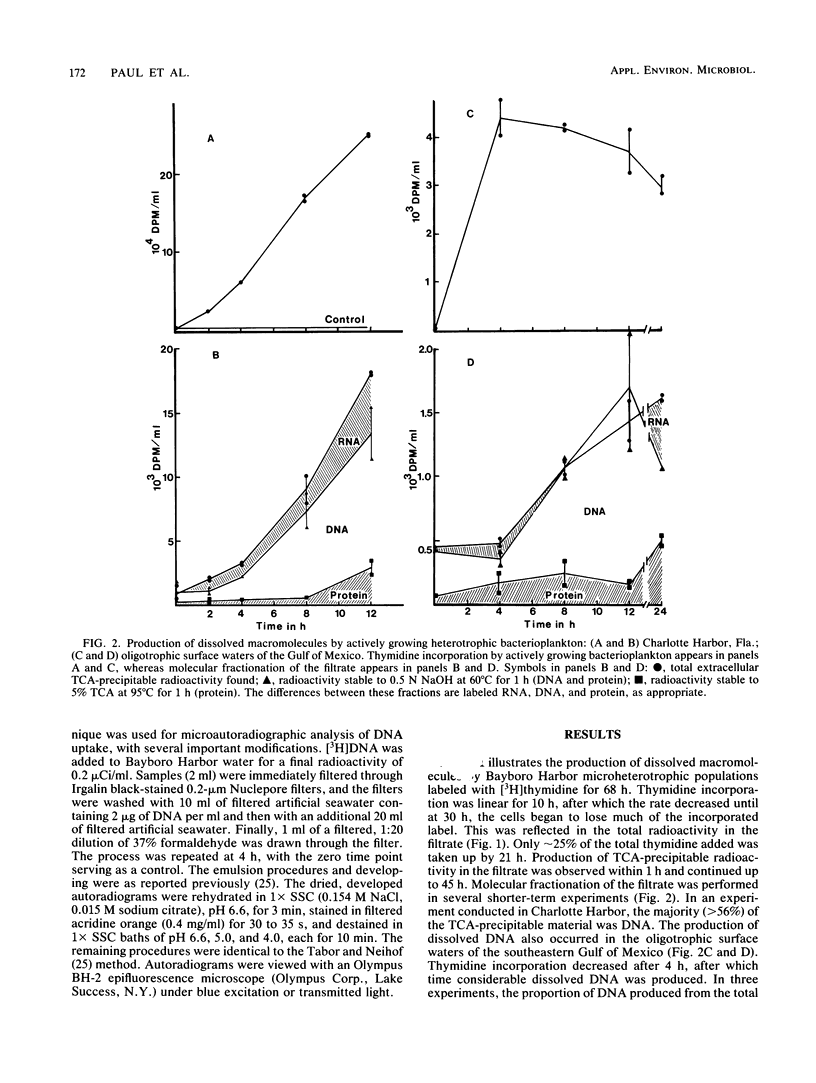
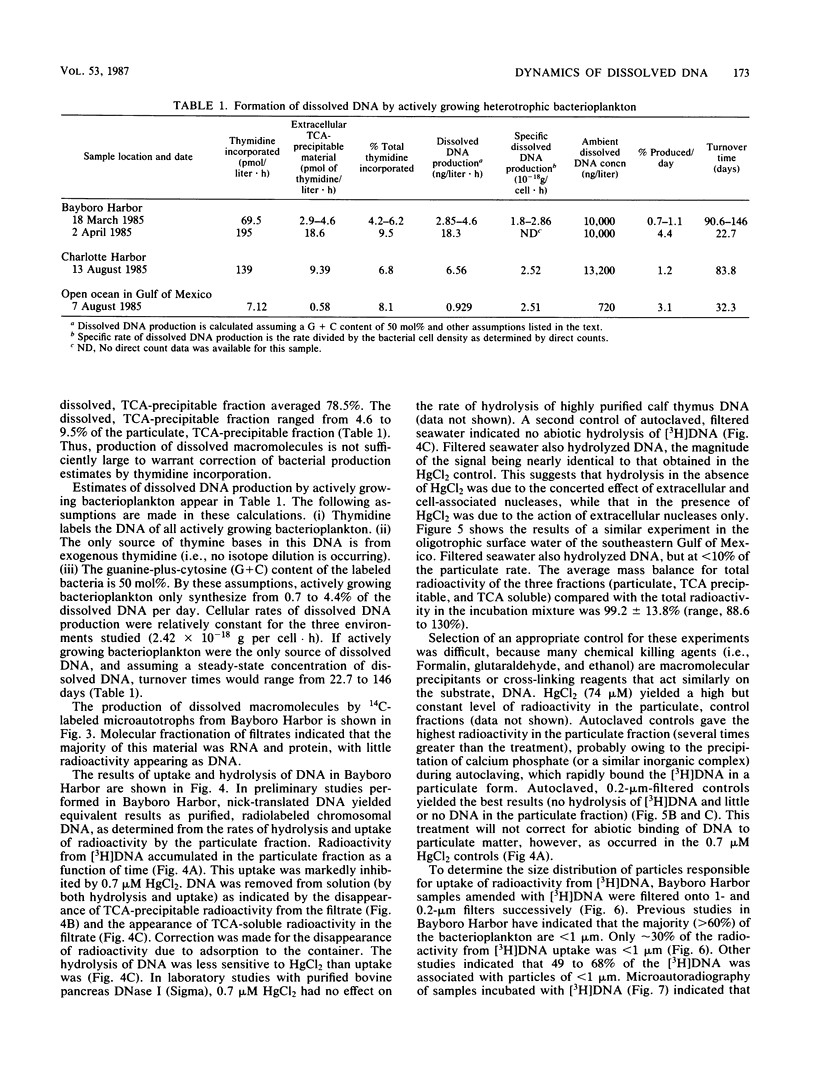
Abstract
The production and turnover of dissolved DNA in subtropical estuarine and oligotrophic oceanic environments were investigated. Actively growing heterotrophic bacterioplankton (i.e., those capable of [3H]thymidine incorporation) were found to produce dissolved DNA, presumably through the processes of death and lysis, grazing by bacteriovores, and excretion. Production of dissolved DNA as determined by [3H]thymidine incorporation was less than or equal to 4% of the ambient dissolved DNA concentration per day. In turnover studies, the addition of [3H]DNA (Escherichia coli chromosomal) to seawater resulted in rapid hydrolysis and uptake or radioactivity by microbial populations. DNA was hydrolyzed by both cell-associated and extracellular nucleases, in both estuarine and offshore environments. Kinetic analysis performed for a eutrophic estuary indicated a turnover time for dissolved DNA as short as 6.5 h. Microautoradiographic studies of bacterial populations in Tampa Bay indicated that filamentous and attached bacteria took up most of the radioactivity from [3H]DNA. Dissolved DNA is therefore a dynamic component of the dissolved organic matter in the marine environment, and bacterioplankton play a key role in the cycling of this material.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aardema B. W., Lorenz M. G., Krumbein W. E. Protection of sediment-adsorbed transforming DNA against enzymatic inactivation. Appl Environ Microbiol. 1983 Aug;46(2):417–420. doi: 10.1128/aem.46.2.417-420.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DOWD J. E., RIGGS D. S. A COMPARISON OF ESTIMATES OF MICHAELIS-MENTEN KINETIC CONSTANTS FROM VARIOUS LINEAR TRANSFORMATIONS. J Biol Chem. 1965 Feb;240:863–869. [PubMed] [Google Scholar]
- Deflaun M. F., Paul J. H., Davis D. Simplified method for dissolved DNA determination in aquatic environments. Appl Environ Microbiol. 1986 Oct;52(4):654–659. doi: 10.1128/aem.52.4.654-659.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham J. B., Istock C. A. Genetic exchange in Bacillus subtilis in soil. Mol Gen Genet. 1978 Nov 9;166(3):287–290. doi: 10.1007/BF00267620. [DOI] [PubMed] [Google Scholar]
- Maeda M., Taga N. Extracellular nuclease produced by a marine bacterium. II. Purification and properties of extracellular nuclease from a marine Vibrio sp. Can J Microbiol. 1976 Oct;22(10):1443–1452. doi: 10.1139/m76-214. [DOI] [PubMed] [Google Scholar]
- Novitsky J. A. Degradation of dead microbial biomass in a marine sediment. Appl Environ Microbiol. 1986 Sep;52(3):504–509. doi: 10.1128/aem.52.3.504-509.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul J. H., Myers B. Fluorometric determination of DNA in aquatic microorganisms by use of hoechst 33258. Appl Environ Microbiol. 1982 Jun;43(6):1393–1399. doi: 10.1128/aem.43.6.1393-1399.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard P. C., Moriarty D. J. Validity of the tritiated thymidine method for estimating bacterial growth rates: measurement of isotope dilution during DNA synthesis. Appl Environ Microbiol. 1984 Dec;48(6):1076–1083. doi: 10.1128/aem.48.6.1076-1083.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servais P., Billen G., Rego J. V. Rate of bacterial mortality in aquatic environments. Appl Environ Microbiol. 1985 Jun;49(6):1448–1454. doi: 10.1128/aem.49.6.1448-1454.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjostrom D. A., Forsdyke D. R. Isotope-dilution analysis of rate-limiting steps and pools affecting the incorporation of thymidine and deoxycytidine into cultured thymus cells. Biochem J. 1974 Feb;138(2):253–262. doi: 10.1042/bj1380253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith H. O., Danner D. B., Deich R. A. Genetic transformation. Annu Rev Biochem. 1981;50:41–68. doi: 10.1146/annurev.bi.50.070181.000353. [DOI] [PubMed] [Google Scholar]
- Stewart G. J., Carlson C. A. The biology of natural transformation. Annu Rev Microbiol. 1986;40:211–235. doi: 10.1146/annurev.mi.40.100186.001235. [DOI] [PubMed] [Google Scholar]
- Tabor P. S., Neihof R. A. Improved microautoradiographic method to determine individual microorganisms active in substrate uptake in natural waters. Appl Environ Microbiol. 1982 Oct;44(4):945–953. doi: 10.1128/aem.44.4.945-953.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]