Abstract
c-Myb, the cellular homologue of the transforming gene of the avian myeloblastosis virus, is preferentially expressed in all hematopoietic lineages, including T and B lymphocyte lineages. In T lymphocytes, c-Myb expression appears to be required for cell cycle progression and proliferation. To further investigate the role of c-Myb in T cell proliferation and survival, interleukin (IL) 2-dependent CTLL-2 cells were transfected with a constitutively active c-myb or with a c-myb antisense construct able to down-regulate endogenous Myb levels, and the transfectants were assessed for proliferation and survival in low concentrations of IL-2 and for susceptibility to dexamethasone-induced apoptosis. Compared with control cells, CTLL-2 cells constitutively expressing c-Myb proliferate in low concentrations of IL-2 and are less susceptible to apoptosis induced by IL-2 deprivation or treatment with dexamethasone. In contrast, cells transfected with an antisense c-myb construct do not proliferate in low concentrations of IL-2 and undergo apoptosis upon IL-2 deprivation or dexamethasone treatment more rapidly than parental cells. Overexpression of c-Myb was accompanied by up-regulation of BCL-2 expression. In transient transfection assays, the murine bcl-2 promoter was efficiently transactivated by c-Myb, but such effect was observed also in cells transfected with a DNA binding-deficient c-myb construct. Moreover, in gel retardation assays, a 38-bp oligomer in the shortest bcl-2 promoter segment regulated by c-Myb formed a specific complex with nuclear extracts from c-Myb-transfected CTLL-2 cells. Thus, these results strongly suggest that c-Myb, in addition to regulating T cell proliferation, protects T lymphocytes from apoptosis by induction of BCL-2 expression, which involves a c-Myb-dependent mechanism of promoter regulation.
The protooncogene c-myb is the normal cellular homolog of the transforming gene of the avian myeloblastosis virus (1, 2). Expression of c-Myb is predominant in, but not restricted to, cells of the hematopoietic system, including T lymphocytes. Many studies have shown that c-Myb is involved in the control of T cell proliferation (3–8), perhaps acting as a regulator of entry into S-phase and of DNA synthesis (9, 10). c-Myb is expressed at high levels in immature thymocytes and may be a regulator of T cell differentiation, as suggested by partial block of thymopoiesis in transgenic mice expressing a c-myb dominant–negative construct (11). Recently, it also has been shown that oligodeoxynucleotides complementary to c-myb mRNA inhibit growth and induce apoptosis in human Burkitt lymphoma cells (12), which is consistent with the possibility that, in normal T lymphocytes, c-Myb also might be involved in the regulation of the process of apoptosis. To address this possibility, a constitutively active c-myb was transfected into the interleukin (IL) 2-dependent cytotoxic T cell line CTLL-2 (13), which is susceptible to apoptosis in response to several death-inducing signals (14–18). CTLL-2 cells constitutively expressing c-Myb were protected from apoptosis induced by IL-2 deprivation or dexamethasone (DEX) treatment. Moreover, such reduced propensity toward apoptosis of c-myb-transfected CTLL-2 cells was associated with overexpression of BCL-2, a well characterized apoptosis inhibitor (19) previously shown to protect these cells from apoptosis (20).
MATERIALS AND METHODS
Cell Lines.
CTLL-2 cells were purchased from American Type Culture Collection. Cells were cultured in RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum and human recombinant (hr) IL-2 (50 units/ml). Tk−ts13 hamster fibroblasts (21) were maintained in culture as described (22).
DNA Constructs and Electroporation.
Expression plasmids were prepared using standard recombinant DNA methods and PCR techniques. The human c-myb full length cDNA (LXSNc-myb) and an internally deleted mutant that encodes a protein deficient in DNA binding [LXSN(R1-c-myb)] have been described (23).
To obtain a c-myb construct in the antisense orientation, a segment of the murine c-myb cDNA corresponding to the transactivation domain (nucleotides 701-1099 relative to the transcription initiation site) was amplified by reverse transcriptase PCR using murine c-myb-specific primers (5′ primer, GGTTTGGGCATGCCTCACCT; 3′ primer, GGATCTGCAGGCAGAGATGG) and total RNA from myeloid precursor 32Dcl3 cells. The amplified c-myb cDNA fragment was cloned into pCRII (TA cloning kit, Invitrogen), digested with EcoRI, and subcloned in the antisense orientation in pLXSN (pLXSN-TAS-c-myb).
The bcl-2–CAT constructs were prepared as follows: (i) pBcl-2(6)CAT was made by digesting plasmid pUCBcl-2 (24) with EcoRI, blunt-ending with Klenow enzyme, and then digesting with Pstl; the resulting 6.0-Kb fragment containing ≈4.6 Kb of 5′ flanking region and the entire 5′ untranslated region of the bcl-2 gene was subcloned into the HindIII-blunted/Pstl sites of pCAT basic vector (Promega); (ii) pBcl-2(3.2)CAT was made by digesting pBcl-2(6)CAT with Nhel, blunt-ending with Klenow enzyme, and then digesting with Pstl; the resulting 3.2-Kb fragment, which contains ≈1.8 Kb of the 5′ flanking region and the entire 5′ untranslated region, was subcloned into the HindIII-blunted/Pstl sites of pCAT basic vector; (iii) pBcl-2(2.8)CAT was made by digesting pBcl-2(6)CAT with EcoRV, blunt-ending with Klenow, and then digesting with Pstl; the resulting 2.8-Kb fragment was subcloned into the HindIII-blunted/Pstl sites of pCAT basic vector; (iv) pBcl-2(2.6)CAT was made by digesting pBcl-2(6)CAT with BstEII, blunt-ending with Klenow, and then digesting with Pstl; the resulting 2.6 Kb fragment was subcloned into the HindIII-blunted/Pstl sites of pCAT basic vector; (v) pBcl-2(2.4)CAT was made by digesting pBcl-2(6)CAT with Scal/Pstl and subcloning the resulting 2.4-Kb fragment into the HindIII-blunted/Pstl sites of pCAT basic vector; (vi) pBcl-2(2.1)CAT was made by digesting pBcl-2(6)CAt with Accl, blunt-ending with Klenow, and then digesting with Pstl; the resulting 2.1-Kb fragment was subcloned into the HindIII-blunted/Pstl sites of pCAT basic vector; (vii) pBcl-2(1.7)CAT was made by digesting pBcl-2(6)CAT with Apal, blunt-ending with T4 polymerase, and then digesting with Pstl; the resulting 1.7-Kb fragment was subcloned into the HindIII-blunted/Pstl sites of pCAT basic vector; and (viii) pBCL-2(1.6)CAT was made by digesting pBcl-2(6)CAT with Smal/Pstl. The resulting 1.6-Kb fragment containing 200 nucleotides of the 5′ flanking region, and the untranslated region (≈1.4 Kb) from the transcription initiation site to the Pstl site, 20 bp upstream of the ATG start codon, was subcloned into HindIII/Klenow-blunted/Pstl-digested pCAT basic vector.
CTLL-2 cells (5 × 106) were resuspended in 0.5 ml of PBS and electroporated (Gene Pulser; Bio-Rad) as described (25). Then, cells were resuspended in 20 ml of RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum and 50 units/ml hrIL-2. After 48 h, cells were seeded in 96-well plates in G418-containing medium (0.5 mg/ml) to obtain individual clones. Two weeks later, clones were expanded and screened for c-Myb expression by Western blot analysis.
Chloramphenicol Acetyltransgerase (CAT) Analysis.
CAT assays were performed as described (22). Tk−ts13 cells (21) were transfected by the calcium phosphate precipitation method (26) with 5 μg of reporter plasmids (pBcl-2-CAT constructs) and 1 μg of pSV β-galactosidase, which contains the bacterial β-galactosidase gene driven by the simian virus 40 promoter, with or without effector plasmids (pLXSNc-myb and pLXSΔR1c-myb) at a 1:1 molar ratio with the reporter plasmids.
After 48 h, cells were harvested, and proteins were extracted by freeze-thawing and normalized for transfection efficiency by β-galactosidase assay as described by the manufacturer (Promega). Cellular lysates were incubated with 14C-labeled chloramphenicol (NEN) and acetyl coenzyme A (Sigma), and CAT activity was measured by thin layer chromatography followed by autoradiography and densitometry, as described (22).
Northern Analysis.
Total RNA was extracted from 5 × 106 CTLL-2 cells using RNAzol (Biotec, Galveston, TX) according to the manufacturer’s instructions. RNA was electrophoresed (10 μg/lane) through 1% agarose gels, blotted onto a nylon membrane (Amersham), and hybridized to a 32P-labeled, 0.9-Kb EcoRI fragment of the human bcl-2 cDNA according to standard procedures (27). After stripping residual radioactivity, filters were hybridized with a glyceraldehyde-3-phosphate dehydrogenase cDNA probe (28) used for normalization of RNA loading and hybridization efficiency.
Western Analysis.
Equal numbers of cells were washed twice with ice-cold PBS and were lysed in 0.1 ml of Hepes buffer [10 mM Hepes, pH 7.5/150 mM NaCl/10% glycerol (vol/vol)/1 mM EDTA/1 mM DTT) containing 0.5% (vol/vol) Nonidet P-40 in the presence of protease inhibitors at the indicated concentration (1 mM phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin, 25 μg/ml aprotinin, 100 μg/ml pepstatin, and 1 mM benzamidine). Lysate preparation, SDS/PAGE, transfer to nitrocellulose membranes (Schleicher & Schuell), membrane blocking, and incubation with an antibody [murine monoclonal anti-c-Myb antibody (Upstate Biotechnology, Lake Placid, NY), rabbit polyclonal anti-BCL-2 antibody (Calbiochem), rabbit polyclonal anti-BCL-XL (Santa Cruz Biotechnology), or anti-β-actin (Oncogene Sciences)] were performed according to standard procedures (27). After incubation with sheep anti-mouse or anti-rabbit IgG conjugated to horseradish peroxidase (Amersham), bound proteins were detected using chemiluminescence substrates according to the manufacturer’s instructions (Amersham).
Gel Retardation Assay.
Nuclear extracts were obtained from parental and c-myb-transfected CTLL-2 cells as described (29). Nuclear extracts (15 μg) were used for gel retardation assays. Lysates in binding buffer (25 mM Hepes·KOH, pH 7.5/50 mM KCl/10 μM ZnSO4/10% glycerol/0.1% Nonidet P-40/1 mM DTT) were incubated with 0.12 μg of poly(dI·dC) per microgram for 10 min on ice. A γ-32P-end labeled, double-stranded oligonucleotide probe (5 × 104 cpm) corresponding to nucleotides −293 to −255 (CCAGCGTACGCCGCGGGTGGCCGCCACCCCAGGCCACG) of the bcl-2 promoter was added to the binding reaction mixes and incubated for 15 min at room temperature. Binding reaction mixes were electrophoresed in native 5% PAGE gels at low ionic stringency (0.25 × Tris–borate–EDTA). Gels were dried and exposed to x-ray films for autoradiography.
[3H]-Thymidine Incorporation Assay.
Cells (1 × 105 in 0.1 ml) were plated in 24-well plates, and 24 or 48 h later, [3H]-thymidine was added at 1 μCi per well. Cells were collected 6 h later, deposited on Millipore glass filters, and washed three times with 10% (vol/vol) trichloroacetic acid. Filters were dissolved in vials containing scintillation fluid (Fisher) and the amount of incorporated [3H]-thymidine was measured using a β-scintillation counter.
Detection of Apoptosis by Flow Cytometry.
Apoptosis, induced by DEX or by IL-2 deprivation, was detected by evaluating the reduction of the propidium iodide fluorescence in the apoptotic nuclei. The cell pellet (0.5 × 106) was fixed in ice-cold 70% ethanol for 10 min on ice and then resuspended in 0.5 ml of lysis buffer (0.1% Nonidet P-40/0.5 μg of DNAse-free RNase in PBS); after 10 min at room temperature, 2.5 μg of propidium iodide was added, and the samples were incubated at 4°C for 15 min. The fluorescence of propidium iodide-stained DNA was quantitated by an Epson Coulter cytometer equipped with a single 488-argon laser.
In Situ Apoptosis Detection.
The TUNEL (terminal deoxynucleotidyltransferase-mediated UTP end labeling) assay was performed using the TACS kit purchased from Trevigen (Gaithersburg, MD). In brief, 106 cells were washed with ice-cold PBS, fixed in 3.7% formaldehyde, and stored in 80% ethanol. Fixed cells were immobilized onto a clean slide and left at room temperature until dried. Slides were washed in 70% ethanol and then air dried. Then, cells were treated with proteinase K, and the endogenous peroxidase was removed by 2% (vol/vol) hydrogen peroxide treatment. Then, DNA was labeled with biotinylated nucleotides using terminal transferase. Samples were incubated with the streptavidine–horseradish peroxidase conjugate and then with the Blue Label (Trevigen) substrate. After counterstaining and fixation, 200 cells/slide were counted.
RESULTS
IL-2 Dependence in CTLL-2 Cells Overexpressing c-Myb or Transfected with a c-myb Antisense Construct.
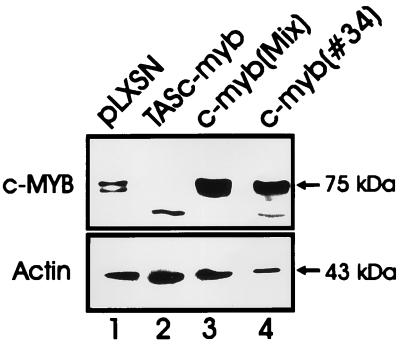
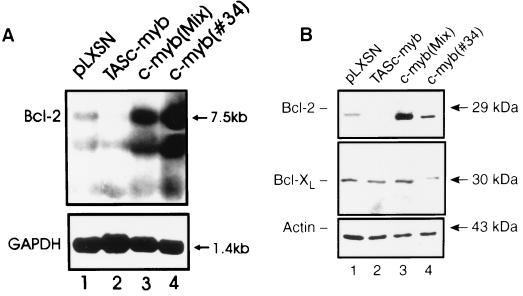
CTLL-2 cells were transfected with the pLXSN empty vector, with pLXSN-c-myb, and with pLXSN-TAS-c-myb. Several G418-resistant clones were isolated. Western blot analysis revealed increased levels of Myb protein in c-myb-transfected cells (Fig. 1, lanes 3 and 4) but almost complete absence of the endogenous Myb protein in cells transfected with the antisense construct (Fig. 1, lane 2).
Figure 1.
Expression of Myb protein in transfected CTLL-2 cells. Lysates were obtained from CTLL-2 cells (2 × 106 in 50 units/ml IL-2) transfected with the empty vector (LXSN) or with the full length c-myb cDNA (c-myb MIX and c-myb 34) or with a c-myb antisense construct (TAS–c-myb). Myb protein levels were detected using a commercial anti-Myb mAb as described (23).
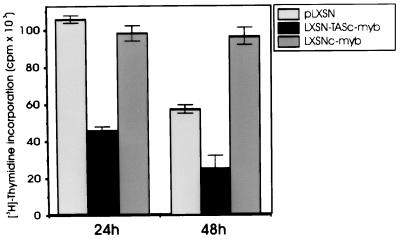
Parental cells and cells transfected with the antisense construct were maintained in culture for more than 48 h only in the presence of 50 units/ml of hrIL-2 whereas the IL-2 requirement of c-myb-transfected cells was much lower because these cells were cultured and were all alive in the presence of 15 units/ml of hrIL-2. To assess the effect of a low dose of IL-2 (15 units/ml) on the proliferation rate of parental and transfected cells, a thymidine incorporation assay with two different pulses (at 24 and 48 h) was performed. At 48 h, CTLL-2 cells constitutively expressing c-Myb exhibited an incorporation rate higher than that of vector-transfected cells (Fig. 2). Instead, cells transfected with the antisense construct incorporated 2- to 3-fold less thymidine than CTLL-2 cells transfected with the empty vector or with c-myb (Fig. 2).
Figure 2.
Thymidine incorporation in c-myb-transfected CTLL-2 cells. [3H]-thymidine counts from triplicate wells containing 105 cells cultured in the presence of 15 units/ml IL-2. [3H]-thymidine was added at 24 or 48 h of culture and assessed as described.
c-Myb Protects CTLL-2 Cells from Apoptosis Induced by IL-2 Deprivation and DEX Treatment.
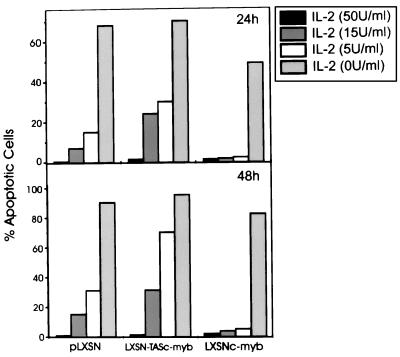
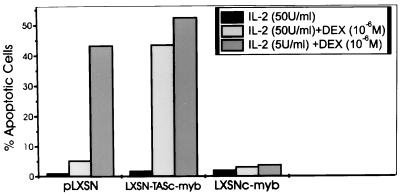
IL-2 deprivation can induce apoptosis in CTLL-2 cells (15–18). To determine the effect of c-Myb overexpression on the propensity of these cells to undergo apoptosis, transfected cells were grown in different hrIL-2 doses (from 0 to 50 units/ml), and the percentage of apoptotic cells was evaluated cytometrically. c-Myb protects CTLL-2 cells from apoptosis at IL-2 concentrations as low as 5 units/ml, but it does not confer IL-2 independence (Fig. 3). Cells transfected with the antisense c-myb construct were more susceptible (2- to 4-fold) than the parental cells to apoptosis induced by IL-2 deprivation (Fig. 3). Because CTLL-2 cells are known to be sensitive to DEX treatment at low doses of IL-2, transfected cells were cultured at 50 and 5 units/ml of hrIL-2 in the presence of DEX used at a concentration of 10−6 M. Cells transfected with the empty vector were very sensitive to DEX when cultured in 5 units/ml of hrIL-2 for 24 h; in marked contrast, c-myb transfected cells were completely resistant to DEX-induced apoptosis (Fig. 4). Conversely, cells transfected with the antisense construct were more sensitive to DEX-induced apoptosis than control cells and were not resistant to DEX even if cultured in 50 units/ml of hrIL-2 (≈8-fold more sensitive than control cells). The fraction of apoptotic cells in cultures deprived of IL-2 or treated with DEX was independently evaluated using the TUNEL method. The results of such analyses were virtually identical to those obtained by flow cytometry (data not shown).
Figure 3.
Apoptosis in cultures of c-myb-transfected cells seeded at different IL-2 concentrations. Apoptotic cells were evaluated cytometrically (propidium iodide staining) in cells cultured at the indicated IL-2 concentrations for 24 or 48 hours. Representative of three independent experiments with similar results.
Figure 4.
Apoptosis in c-myb-transfected CTLL-2 cultures upon DEX treatment in the presence of decreasing IL-2 concentrations. Apoptotic cells were evaluated cytometrically (propidium iodide staining) in cultures maintained for 24 h in 50 or 5 units/ml IL-2 and 106 M/liter DEX. Representative of three different experiments with similar results.
To assess if c-myb-transfected cells can survive IL-2 deprivation or DEX treatment via an autocrine mechanism, ELISA assays were performed to detect murine IL-2 secretion in the supernatants of cells transfected with the empty vector or the full length c-myb cDNA and cultured at different concentrations of hrIL-2. No autocrine production of IL-2 was detected in cultures of c-Myb-transfected CTLL-2 cells at any of the various hrIL-2 concentrations (data not shown).
Expression of BCL-2 in CTLL-2 Cells Overexpressing c-Myb.
To assess potential mechanisms associated with the reduced susceptibility to apoptosis of c-myb-transfected CTLL-2 cells, levels of BCL-2 and BCL-XL, two apoptosis inhibitors, were measured in control CTLL-2 cells and in cells overexpressing c-Myb. BCL-XL protein levels were essentially identical in vector- and c-myb-transfected CTLL-2 cells (Fig. 5B). By contrast, compared with vector-transfected CTLL-2 cells, Northern and Western blot analyses revealed that bcl-2 mRNA and protein levels were up-regulated in c-myb-transfected cells and down-regulated in cells transfected with the antisense construct (Fig. 5, A and B).
Figure 5.
BCL-2 and BCL-XL expression in c-myb-transfected CTLL-2 cells. Bcl-2 mRNA (A) and BCL-2 and BCL-XL proteins (B) levels were analyzed by Northern and Western blot analyses, respectively, in c-myb-transfected CTLL-2 cells. Expression of glyceraldehyde-3-phosphate dehydrogenase and β-actin was measured as a control of equal RNA and protein loading, respectively. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
c-Myb Transactivation of CAT Gene Expression Driven by the Murine bcl-2 5′ Flanking Region.
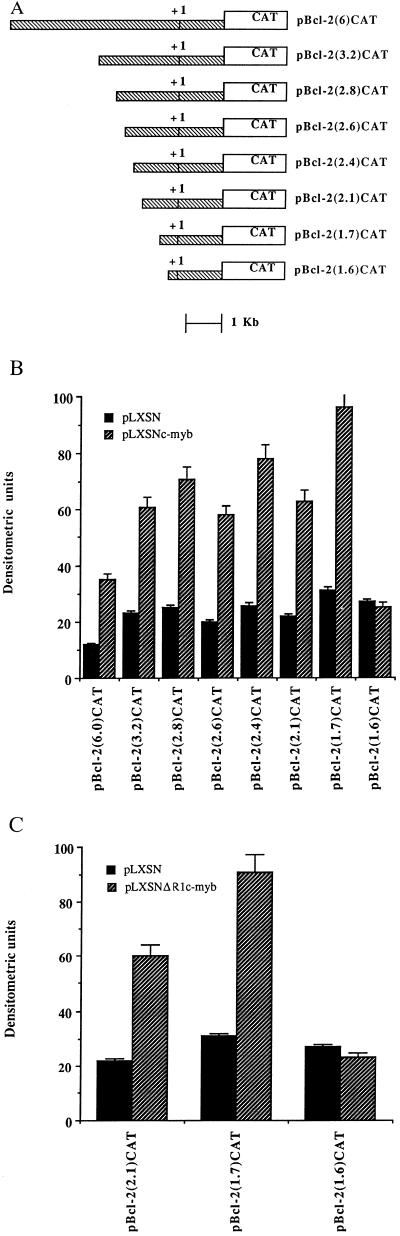
To determine whether the up-regulation of BCL-2 expression in c-myb-transfected cells was due to a c-Myb-induced enhancement of bcl-2 promoter activity, we assessed the ability of c-Myb to transactivate CAT reporter constructs containing segments of decreasing length of the murine bcl-2 5′ flanking region (24). CAT assays were performed in transiently transfected Tk−ts13 hamster fibroblasts, which do not express endogenous c-Myb at detectable levels (30). The LXSN-c-myb effector had no effect on the 1.6-Kb CAT reporter construct, which contained the entire 5′ untranslated region and 0.2 Kb of the 5′ flanking sequence upstream of the transcription initiation site (24); it induced, however, a 3- to 4-fold increase in CAT expression driven by all other bcl-2 5′ flanking sequence segments (Fig. 6), including pBcl-2(1.7)CAT, which contains only 293 bp upstream of the transcription initiation site. A c-myb construct encoding an internally deleted Myb protein unable to interact with canonical Myb binding sites (23) was still able to transactivate (3- to 4-fold induction) the 5′ flanking region bcl-2-CAT reporter plasmids (Fig. 6), consistent with a DNA binding-independent mechanism of bcl-2 promoter regulation.
Figure 6.
c-Myb transactivation of the murine bcl-2 promoter. (A) Diagram of bcl-2 promoter–CAT constructs. (B) Transactivation of bcl-2/CAT plasmids by full length c-Myb. Tk−ts13 cells were transfected with bcl-2–CAT constructs in the presence of the empty vector (▪) or the full length c-myb cDNA (▨) at a 1:1 molar ratio. CAT assays were performed as described (22). (C) Transactivation of bcl-2/CAT plasmids by a DNA binding-deficient c-myb construct. Tk−ts 13 cells were transfected with the indicated bcl-2–CAT constructs in the presence of the empty vector (▪) or the DNA binding-deficient c-Myb (▨) at a 1:1 molar ratio. Histograms (B and C) represent the results of three different experiments ± SD.
Detection of a c-Myb-Dependent Complex at Nucleotides −293 to −255 of the bcl-2 Promoter.
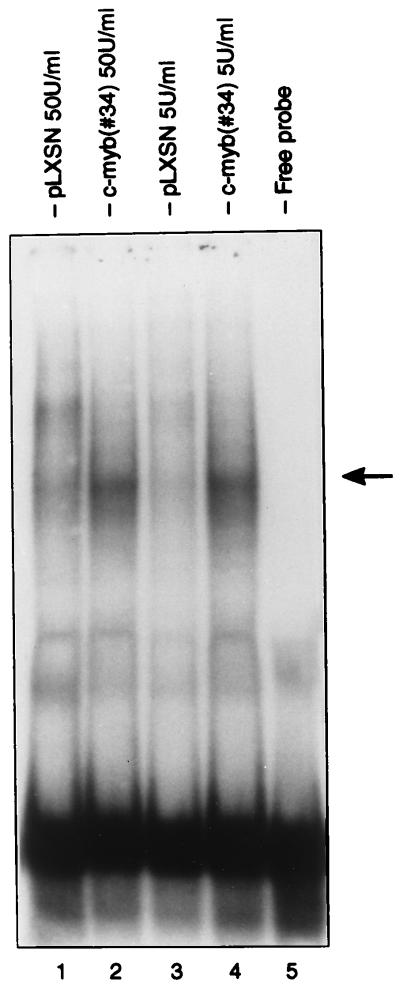
To determine whether c-Myb transactivation of the bcl-2 promoter from nucleotides −293 to −200 correlated with specific protein(s) interaction in that fragment, gel shift assays were performed with nuclear extracts from parental and c-myb-transfected CTLL-2 cells. Cells cultured in the presence of 50 units/ml IL-2 or shifted for 16 h in 5 units/ml IL-2 were used in the experiments. No differences in complexes formation were detected using a 32P-labeled oligoprobe corresponding to nucleotides −255 to −200 (not shown); by contrast, a specific complex was detected in nuclear extracts from c-myb-transfected CTLL-2 cells using a 32P-labeled oligoprobe corresponding to nucleotides −293 to −255 (Fig. 7).
Figure 7.
Gel retardation assay of a 38-bp oligomer from nucleotides −293 to −255 of the bcl-2 5′ flanking region with nuclear extracts from c-Myb-transfected CTLL-2 cells. Representative of three independent experiments. The arrow indicates the c-Myb-dependent specific complex.
DISCUSSION
In the present investigation, we assessed whether c-Myb is involved in the regulation of apoptosis by studying the effect of constitutive c-Myb expression or by assessing the consequences of inhibiting endogenous Myb expression in CTLL-2 cells, an IL-2-dependent cytotoxic T cell line. Parental cells and cells transfected with the antisense construct were maintained in culture for more than 48 h only in the presence of 50 units/ml IL-2; in contrast, c-myb-transfected cells were able to survive at suboptimal concentrations of IL-2 (5–15 units/ml). This correlated with a proliferative advantage of c-myb-transfected cells, as indicated by their higher thymidine incorporation rate, compared with parental and antisense c-myb-transfected CTLL-2 cells. CTLL-2 cells readily undergo apoptosis if cultured in the absence of IL-2 or if treated with DEX (14–18). In marked contrast, c-myb-transfected CTLL-2 cells were resistant to apoptosis induced by IL-2 deprivation or by DEX treatment. Conversely, c-myb antisense-transfected cells were more susceptible to apoptosis induced by either method.
The importance of c-Myb in the proliferation of hematopoietic cells was initially demonstrated by use of antisense oligodeoxynucleotides; such compounds greatly reduced proliferation of several myeloid leukemia cell lines (31) and of normal human T lymphocytes (9). Consistent with these observations, transgenic mice carrying a c-myb dominant–negative construct showed a partial impairment of thymopoiesis and inhibition of mature T cell proliferation (11). Moreover, treatment of Burkitt lymphoma cell lines with c-myb antisense oligodeoxynucleotides leads to inhibition of cell growth and induces apoptosis (12). Together, these findings and the results reported here indicate that c-Myb is important not only in the regulation of cell proliferation but also in the control of the process of apoptotic cell death. An autocrine mechanism was not responsible for the c-Myb-induced protection from apoptosis; there was no evidence of IL-2 secretion in cultures of c-myb-transfected cells. To assess other mechanisms by which c-Myb protects CTLL-2 cells from apoptosis, we undertook experiments to evaluate expression levels of intracellular mediators of the apoptosis-resistant phenotype. CTLL-2 cells and T cell hybridomas overexpressing BCL-2 are resistant to apoptosis induced by IL-2 deprivation and glucocorticoid treatment, respectively (20, 32), so BCL-2 expression levels were evaluated in c-myb-transfected cells. Constitutive expression of c-Myb was associated with up-regulation of bcl-2 mRNA and protein, consistent with the possibility that c-Myb acts as an apoptotic repressor by inducing BCL-2 expression. Indeed, in T lymphocytes, BCL-2 expression is induced by mitogens and by IL-2 with kinetics similar to that of c-Myb (33). Moreover, such an increase in Bcl-2 levels is transcriptionally regulated (33). Thus, it is conceivable that c-Myb is involved in the regulation of bcl-2 transcription. In support of this hypothesis, CAT assays indicate that c-Myb directly modulates the activity of the bcl-2 promoter. A Bcl-2–CAT construct containing the 5′ untranslated region and 200 nucleotides of 5′ flanking region (Bcl-2–1.6) was not responsive to c-Myb. In contrast, several bcl-2 promoter constructs containing segments of increasing length of the bcl-2 5′ flanking region were responsive to c-Myb. The shortest promoter regulated by c-myb includes a 38-bp segment that formed a specific complex with nuclear extracts from c-myb-transfected CTLL-2 cells. Canonical Myb binding sites are not present in this region, and a DNA binding-deficient c-myb construct also was capable of enhancing Bcl-2-driven CAT activity (Fig. 6a), so the c-Myb transactivation effect seems due to a DNA binding-independent mechanism.
A DNA binding-independent mechanism of promoter regulation by genes of the Myb family has been reported for human HSP70 (34). Myb inducibility of this promoter was dependent on the presence of an intact TATA box and upstream sequences (35). However, the bcl-2 promoter lacks a canonical TATA box (24), suggesting that the ability of c-Myb to regulate the bcl-2 promoter in a DNA binding-independent manner involves other mechanisms. More recently, protein–protein interactions involving c-Myb and cAMP response element binding protein have been reported (35, 36). Such interaction enhances the transcription-activating function of c-Myb, perhaps providing a bridge to the basal transcription machinery. Whether these interactions are involved in the regulation of the bcl-2 promoter is now under investigation. The 38-bp oligomer, which is the site of a Myb-dependent complex (Fig. 7), includes two copies of the CACCC motif. This motif has been reported to mediate the transactivation of the murine GATA-1 promoter induced by a myb–ets-containing retrovirus (37), raising the possibility that the bcl-2 promoter also is regulated by c-Myb through this site.
In summary, our studies support an essential role of c-Myb in the survival of growth factor-dependent T lymphocytes and, perhaps, of other hematopoietic cell types. Such effect is, in part, dependent on transcriptional activation of the expression of BCL-2, the prototype of a protein with anti-apoptosis properties. These data are consistent with the results of two recent studies indicating that c-Myb and v-Myb protect T lymphocytes and myeloid cells from apoptosis by regulating BCL-2 expression (38, 39). However, at variance with the results of those studies, we show here that c-Myb regulation of bcl-2 promoter activity is independent from specific interactions with Myb binding sites. Whether the mechanism of bcl-2 promoter regulation is due to protein–protein interactions involving c-Myb or to Myb induction of a bcl-2 transcription regulator is presently under investigation.
Acknowledgments
We thank Massimo Negrini for the generous gift of plasmid PUC Bcl-2 and Andrew Engelhard for critical reading of this manuscript. This work was supported in part by National Institutes of Health Grant RO1 CA46782 (B.C.) and by a grant of the Associazione Italiana Ricerca sul Cancro (C.F.). P.S. was supported by an Associazione Italiana Ricerca sul Cancro fellowship; and R.M. was supported by a minority student fellowship from Thomas Jefferson University.
ABBREVIATIONS
- IL
interleukin
- hrIL-2
human recombinant IL-2
- DEX
dexamethasone
References
- 1.Gonda T J, Bishop J M. J Virol. 1983;46:212–219. doi: 10.1128/jvi.46.1.212-220.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klempnauer K H, Ramsay G, Bishop J M, Moscovici J P, Moscovici C, McGrath J P, Levinson A P. Proc Natl Acad Sci USA. 1983;33:345–350. doi: 10.1016/0092-8674(83)90416-6. [DOI] [PubMed] [Google Scholar]
- 3.Torelli G, Selleri L, Donelli A, Ferrari S, Emilia D, Venturelli D, Moretti L, Torelli U. Mol Cell Biol. 1985;5:2847–2887. doi: 10.1128/mcb.5.10.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stern J B, Smith K A. Science. 1986;233:203–206. doi: 10.1126/science.3523754. [DOI] [PubMed] [Google Scholar]
- 5.Reed J C, Alpers J D, Nowell P C, Hoover R C. Proc Natl Acad Sci USA. 1986;83:3982–3986. doi: 10.1073/pnas.83.11.3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thompson C B, Challoner P B, Neiman P E, Groudine M. Nature (London) 1986;319:374–380. doi: 10.1038/319374a0. [DOI] [PubMed] [Google Scholar]
- 7.Lipsick J S, Boyle W J. Mol Cell Biol. 1987;7:3358–3360. doi: 10.1128/mcb.7.9.3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pouza D C. Mol Cell Biol. 1989;9:342–348. [Google Scholar]
- 9.Gewirtz A, Anfossi G, Venturelli D, Valpreda S, Sims R, Calabretta B. Science. 1989;245:180–183. doi: 10.1126/science.2665077. [DOI] [PubMed] [Google Scholar]
- 10.Churilla A M, Braciale T, Braciale V. J Exp Med. 1989;170:105–121. doi: 10.1084/jem.170.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Badiani P, Corbella P, Kioussis D, Marvel J, Weston K. Genes Dev. 1994;8:770–782. doi: 10.1101/gad.8.7.770. [DOI] [PubMed] [Google Scholar]
- 12.Joshi S S, Wu A G, Verbik D J, Algarra S M, Bishop M R, Pirruccelo S J, Iversen P L, Jackson J D, Kessinger M A, Sharp J G. Int J Oncol. 1996;8:815–820. doi: 10.3892/ijo.8.4.815. [DOI] [PubMed] [Google Scholar]
- 13.Gillis S, Smith K A. Nature (London) 1977;268:154–156. doi: 10.1038/268154a0. [DOI] [PubMed] [Google Scholar]
- 14.Perrin-Wolff M, Bertoglio J, Bressac B, Bohuon C, Pallardy M. Biochem Pharmacol. 1995;50:103–110. doi: 10.1016/0006-2952(94)00527-s. [DOI] [PubMed] [Google Scholar]
- 15.Thiele D L, Lipsky P E. J Immunol. 1992;148:3950–3957. [PubMed] [Google Scholar]
- 16.Perillo N L, Pace K E, Seilhamer J J, Baum L. Nature (London) 1995;378:736–739. doi: 10.1038/378736a0. [DOI] [PubMed] [Google Scholar]
- 17.Gruber J, Sgone R, Hu Y H, Beug H, Wick G. Eur J Immunol. 1994;24:1115–1121. doi: 10.1002/eji.1830240516. [DOI] [PubMed] [Google Scholar]
- 18.Azuma Y, Onishi Y, Sato Y, Kizaki H. J Biochem. 1995;118:312–318. doi: 10.1093/oxfordjournals.jbchem.a124908. [DOI] [PubMed] [Google Scholar]
- 19.Cory S. Annu Rev Immunol. 1995;13:513–543. doi: 10.1146/annurev.iy.13.040195.002501. [DOI] [PubMed] [Google Scholar]
- 20.Deng G, Podack E R. Proc Natl Acad Sci USA. 1993;90:2189–2193. doi: 10.1073/pnas.90.6.2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Talavera A, Basilico C. J Cell Physiol. 1977;92:425–436. doi: 10.1002/jcp.1040920310. [DOI] [PubMed] [Google Scholar]
- 22.Melotti P, Ku D-H, Calabretta B. J Exp Med. 1994;179:1023–1028. doi: 10.1084/jem.179.3.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sala A, Bellon T, Melotti P, Peschle C, Calabretta B. Blood. 1995;86:3404–3412. [PubMed] [Google Scholar]
- 24.Negrini M, Silini E, Kozak C, Tsujimoto Y, Croce C M. Cell. 1987;49:455–463. doi: 10.1016/0092-8674(87)90448-x. [DOI] [PubMed] [Google Scholar]
- 25.Ku D-H, Wen S-C, Engelhard A, Nicolaides N C, Lipson K E, Marino T A, Calabretta B. J Biol Chem. 1993;268:2255–2259. [PubMed] [Google Scholar]
- 26.Huttner K M, Barbosa J A, Scangos G A, Pratcheva D D, Ruddle F H J. J Cell Biol. 1981;91:153–156. doi: 10.1083/jcb.91.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 28.Fort P, Piechaczyk M, Sabrouty S, Dani C, Jeaniteur P, Blanchard J. Nucleic Acids Res. 1985;13:1431–1437. doi: 10.1093/nar/13.5.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perrotti D, Melotti P, Skorski T, Casella I, Peschle C, Calabretta B. Mol Cell Biol. 1995;15:6075–6087. doi: 10.1128/mcb.15.11.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Travali S, Reiss K, Ferber A, Petralia S, Mercer W E, Calabretta B, Baserga R. Mol Cell Biol. 1991;11:731–736. doi: 10.1128/mcb.11.2.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anfossi G, Gewirtz A, Calabretta B. Proc Natl Acad Sci USA. 1989;86:3379–3383. doi: 10.1073/pnas.86.9.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Merrion S A, Moreno M B, Petrak D, Zacharchuck C M. J Immunol. 1995;155:4644–4652. [PubMed] [Google Scholar]
- 33.Reed J C, Tsujimoto T, Alpers J D, Croce C M, Nowell P C. Science. 1987;236:1295–1298. doi: 10.1126/science.3495884. [DOI] [PubMed] [Google Scholar]
- 34.Foos G, Natour S, Klempnauer K-H. Oncogene. 1993;8:1775–1783. [PubMed] [Google Scholar]
- 35.Dai P, Akimaru I, Tanaka Y, Hou D-X, Yasukawa T, Kanei-Ishii C, Takahashi T, Ishii S. Genes Dev. 1996;18:528–540. doi: 10.1101/gad.10.5.528. [DOI] [PubMed] [Google Scholar]
- 36.Oelgeschlager M, Janknecht R, Krieg J, Schreck S, Lüscher B. EMBO J. 1996;15:2771–2780. [PMC free article] [PubMed] [Google Scholar]
- 37.Hoffman L-S, Aurigemma R E, Sun B, Ruscetti S K. Oncogene. 1996;13:1939–1042. [PubMed] [Google Scholar]
- 38.Taylor D, Badiani P, Weston K. Genes Dev. 1996;10:2732–2744. doi: 10.1101/gad.10.21.2732. [DOI] [PubMed] [Google Scholar]
- 39.Frampton J, Ramquist T, Graf T. Genes Dev. 1996;10:2720–2731. doi: 10.1101/gad.10.21.2720. [DOI] [PubMed] [Google Scholar]