Abstract
Ligation of the cell surface receptor Fas/APO-1 (CD95) by its specific ligand or by anti-Fas antibodies rapidly induces apoptosis in susceptible cells. To characterize the molecular events involved in Fas-induced apoptosis, we examined the contribution of two subgroups of the mitogen-activated protein (MAP) kinase family, the Jun kinases or stress-activated protein kinases (JNKs/SAPKs) and the extracellular signal-regulated kinases (ERKs), in a Fas-sensitive neuroblastoma cell line. Here we show that both JNK and ERK protein kinases were activated upon Fas crosslinking through a Ras-dependent mechanism. Interference with either the JNK or ERK pathway by ectopic expression of dominant-interfering mutant proteins blocked Fas-mediated apoptosis. ERK activation was transient and associated with induced expression of the Fas receptor. In contrast, JNK activation was sustained and correlated with the onset of apoptosis. These data indicate that the ERK and the JNK groups of MAP kinases cooperate in the induction of cell death by Fas. Inhibition of Fas killing by an interleukin 1β-converting enzyme (ICE)-like protease inhibitor peptide did not modify Fas-induced JNK activation upon Fas ligation. In contrast, changes in Bcl-2 level due to expression of sense and antisense vectors influenced the sensitivity to Fas killing and Fas-induced JNK activation.
The Fas/APO-1 protein is a 45-kDa glycosylated transmembrane protein belonging to the tumor necrosis factor (TNF) receptor family (1, 2). The Fas ligand (FasL) is a 40-kDa protein displaying significant homology to the members of the TNF family and is primarily expressed on activated T cells (3, 4). Activation of Fas by its ligand or by crosslinking with anti-Fas antibodies induces apoptosis (2, 3, 5, 6). Apoptosis induced by Fas has been shown to be involved in the regulation of lymphocyte death in the peripheral immune system (7–11) and T cell-mediated cytotoxicity (12–16).
Several gene products involved in Fas-mediated apoptosis have been characterized. The interleukin 1β-converting enzyme (ICE)-like family of cysteine proteases has been implicated in Fas killing, since ICE inhibitors and transient expression of the pox-virus-derived serpin inhibitor CrmA or an antisense ICE construct block Fas-induced apoptosis (17–19). Another enzyme implicated in Fas-induced cell death is sphingomyelinase, which hydrolyzes sphingomyelin to generate second-messenger ceramides (20–22).
Recently, Jun kinases (JNKs) have been implicated in two different models of cell death, apoptosis induced by nerve growth factor (NGF) deprivation in differentiated rat PC12 pheochromocytoma cells (23) and by environmental stresses (24, 25). JNKs can be activated by subjecting cells to environmental stresses or by exposure to pro-inflammatory cytokines, such as TNF and interleukin 1 (26–30).
The Bcl-2 oncogene, originally identified in a subset of B cell lymphomas (31–33), has been associated with resistance to apoptosis in a variety of mammalian systems, including growth factor withdrawal from hematopoietic and neuronal cells, treatment with glucocorticoids, calcium ionophores or cytotoxic drugs, and γ irradiation (34). Bcl-2 is a member of a multigene family that includes Bcl-x (35), Mcl-1 (36), A1 (37), Bax (38), Bad (39), Bak (40) in mammals, and ced-9 in the nematode Caenorhabditis elegans (41). Similarly to Bcl-2, Bcl-xL promotes cell survival. In contrast, Bax, Bad, Bak, and Bcl-xS, interact with Bcl-2 and/or Bcl-xL and antagonize their protective effect. The mode of action of Bcl-2 in the inhibition of cell death is unknown.
Similar to Fas, TNFα induces cell death through interaction with its receptor. Several potent activators of JNKs, such as TNFα, cycloheximide, or exposure to heat shock, can also potentiate Fas-mediated apoptosis (5), suggesting that activation of these kinases may activate or contribute to the cell death pathway triggered by Fas. We investigated the contribution of the mitogen-activated protein (MAP) kinases to Fas-mediated killing in a Fas-sensitive neuroblastoma cell line. We found that Fas crosslinking with an anti-Fas antibody activated JNK and extracellular signal-regulated kinase (ERK) MAP kinases. Interference with either the JNK or the ERK pathway by ectopic expression of dominant-interfering proteins blocked Fas-mediated apoptosis. Inhibition of Fas killing by an ICE inhibitor did not modify Fas-induced JNK activation. In contrast, changes in Bcl-2 expression level influenced specifically Fas-stimulated JNK protein activity.
MATERIALS AND METHODS
Materials.
The pCDNA3 plasmids encoding M2-tagged JNK1 wild-type or JNK1 APF mutant or hemagglutinin (HA)-tagged JNK1 wild type were described previously (27). The glucocorticoid-inducible Ha-Ras(Asn17):pMMTV Ha-Ras(Asn-17) plasmid was provided by G. Cooper (Harvard University). A murine MEK1 K→R cDNA, provided by R. L. Erikson (Harvard University), was cloned in a cytomegalovirus (CMV) vector containing tet operator sequences, a tetracycline-repressible trans-activator, and the neomycin-resistance gene, provided by A. Fattaey (Onyx Pharmaceuticals). The activated MEK1 kinase (CMVΔN3-MEK) plasmid was provided by N. G. Ahn (University of Colorado). A 1.9-kb human Bcl-2 cDNA, obtained from BK-KS-H Bcl-2, provided by S. Korsmeyer (Washington University School of Medicine), was subcloned into the expression plasmid pCEP9. The pCEP9 vector is identical to the Epstein–Barr virus (EBV)-based episomal expression vector pCEP4 (Invitrogen) except for the substitution of the hygromycin-resistance gene for the neomycin-resistance gene.
Cell Death Assay.
Cells (1.5 × 105 per well) were plated in 35-mm multiwells plates. Anti-Fas IgM (clone CH-11, Upstate Biotechnology) was added to the cells for the indicated times. The number of apoptotic cells was assessed by nuclear staining with Hoechst 33258 (0.1 μg/ml) in PBS after fixation in methanol/acetic acid (3:1, vol/vol) and visualization on a fluorescence microscope. Six to seven hundred cells were counted for each time point.
For 24-hr cell death assay, most of the dead cells are fragmented into debris and cannot be stained with Hoechst 33258. Therefore, the total percentage of cell death was estimated by crystal violet staining of the remaining viable cells as follows. Cells were plated in either 96-well (2 × 104 per well) or in 35-mm multiwell (1.5 × 105 per well) plates. After 18 hr, the anti-Fas antibody was added for the indicated times. The wells were washed twice with PBS to eliminate dead cells. Viable cells were then stained and fixed by addition of 200 μl of 0.05% crystal violet in 20% ethanol/0.37% formaldehyde/80% water solution for 10 min. The wells were rinsed with water and dried. The optical density (OD) was measured at 595 nm after lysing the cells with 100 μl of methanol. A background value of crystal violet staining for wells containing only the culture medium was subtracted from each OD measurement. The percentage of cell death is calculated as follows: 100 − [(OD for assay/OD for control well) × 100].
Flow Cytometric Analysis.
Cells were incubated 1 hr on ice with 20 μg/ml mouse anti-human Fas antibody or a control mouse IgM antibody (PharMingen) in PBS containing 1% fetal bovine serum and 0.02% Tween 20. The cells were washed and incubated 30 min on ice with fluorescein isothiocyanate (FITC)-labeled goat anti-mouse IgM. After washing twice in PBS, the cells were fixed in PBS containing 1% formaldehyde (pH 7.0) prior to analysis. Flow cytometric analysis was carried out using a FACScan (Becton Dickinson).
Transfections.
Prior to electroporation, cells were washed twice in ice-cold PBS and then resuspended on ice at 5 × 106 cells per 0.5 ml of PBS with plasmid DNA (20 μg). The cells were electroporated (180 V, 960 μF) in a Bio-Rad Gene Pulser. After 10 min on ice, the cells were transferred to fresh complete medium and cultured for 36 hr before addition of G418 (Geneticin; GIBCO) at a concentration of 0.4–0.8 mg/ml. To avoid problems with clonal variation, the transfected cells were selected for 2–3 weeks for neomycin resistance and all the clones (50–100 per transfection) were pooled.
Kinase Assay.
Cells (1.5 × 105 per well) were plated in 35-mm multiwell plates. Anti-Fas IgM was added at a concentration of 1 μg/ml to the cells for the indicated times. Cell lysis and immune complex kinase assays were done as described (26). JNK and ERK proteins were immunoprecipitated with polyclonal antibodies to JNK1 (N19 and C17) and ERK1 (C16) (Santa Cruz Biotechnology). After SDS/PAGE, the extent of phosphorylation was quantitated by measuring the radioactivity in the gel bands and background regions with a liquid scintillation counter. The kinase activities are expressed relative to cells incubated without anti-Fas (1.0) (time 0).
Immunoblot Analysis.
Cells were extracted with ELB buffer (50 mM Hepes, pH 7.0/250 mM NaCl/0.5 mM EDTA/0.1% Nonidet P-40) containing protease inhibitors (0.2 mM phenylmethanesulfonyl fluoride, 1 μg/ml aprotinin). SDS/PAGE was performed using 100 μg of total protein per sample, followed by transfer to nitrocellulose filters and sequential incubation with the primary antibodies and a horseradish peroxidase-linked antibody to mouse or rabbit immunoglobulin (Amersham International, Buckinghamshire, England). Immunoblots were developed according to the ECL procedure (Amersham).
RESULTS
Fas-Induced Apoptosis in SHEP Cells.
For our analysis we selected SHEP, a human neuroblastoma cell line, from among a panel of Fas-positive and -sensitive human tumor cell lines of diverse histological origin, including multiple carcinomas, neuroglial tumors, and sarcomas. Eighty-five to 90% of SHEP cells expressed Fas with a mean intensity greater than 100-fold over background levels (Fig. 1A). Addition of anti-Fas antibody to SHEP cells caused morphological changes characteristic of apoptosis, such as loss of cell-to-cell contact, membrane blebbing, cell fragmentation, nuclear condensation, and formation of nuclear bodies (Fig. 1 B and C).
Figure 1.
Induction of cell death by Fas in SHEP cells. (A) Fas expression in SHEP cells. Fas was detected by surface immunofluorescence staining and flow cytometry, using the anti-human Fas IgM (clone CH11). The black lines indicate background staining with an isotype-matched control antibody. (B) Phase-contrast photomicrographs of a control SHEP cell culture (−) and 12 hr after anti-Fas antibody addition (1 μg/ml) (+), showing loss of cell contact and adherence (one arrow) and membrane blebbing (two arrows). (×200.) (C) Hoechst 33258 staining of control SHEP cells (−) and anti-Fas-treated cells (+) (1 μg/ml for 4 hr) showing condensed nuclei (one arrow), and nuclear bodies (two arrows) in Fas-treated cells. (×200.)
JNK Protein Kinase Activation in Fas-Induced Apoptosis.
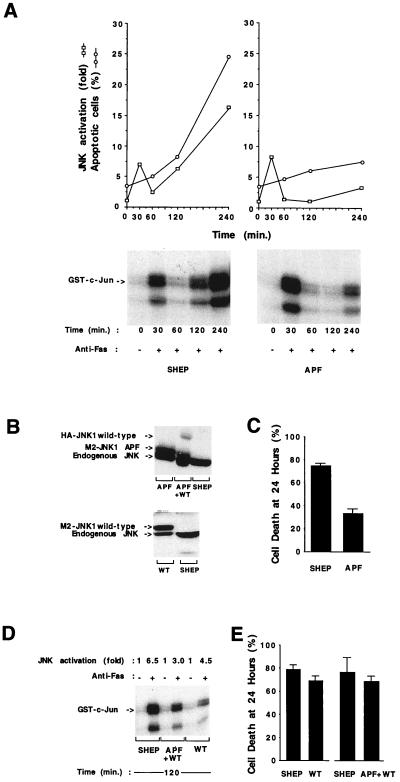
A marked increase in JNK activity was detected after anti-Fas antibody addition (Fig. 2A). JNK activity increased within 30 min after anti-Fas addition to SHEP cells and then decreased to background levels. In our hands, the initial peak of activity was not detected by other polyclonal antibodies raised against JNK1, suggesting that a cross-reacting kinase, possibly another Jun kinase, may account for the early peak of activation. A secondary sustained increase in JNK protein kinase activity occurred at 120 min. This increase in JNK activity coincided with the onset of cell death (Fig. 2A), raising the possibility that this kinase might contribute to the Fas-apoptosis signaling pathway. To examine the role of JNK in Fas-mediated apoptosis, an expression construct of a kinase-inactive JNK was transfected into SHEP cells (Fig. 2B). The kinase-inactive JNK mutant (APF) is the double point mutation that changes the phosphorylation sites Thr-183 and Tyr-185 to Ala and Phe, respectively (27). This mutation blocks JNK activation. To distinguish between endogenous and transfected proteins, the expression constructs were engineered to encode epitope-tagged versions of the kinases. Transfected cells were selected for neomycin resistance in G418 for approximately 2 weeks and pooled. The population of transfected cells represented between 50 and 100 individual clones. The use of this mixed population eliminates clonal variation as a possible explanation for differences observed between control SHEP cells and SHEP expressing dominant-negative JNK (APF). The expression of exogenous JNK was similar to the endogenous level of JNK (Fig. 2B).
Figure 2.
JNK protein kinase and Fas-induced apoptosis. (A) Time course of endogenous JNK activation and apoptosis in SHEP cells and SHEP cells expressing dominant-negative JNK1 APF (APF). □, JNK activity relative to control cells without anti-Fas. ○, Percentage of cells undergoing apoptosis, assessed by nuclear staining with Hoechst 33258. Six to seven hundred cells were counted for each time point. Autoradiography showing the phosphorylated c-Jun after SDS/PAGE. Transfections were done by electroporation with 20 μg of the pCDNA3 plasmid encoding M2-tagged JNK1 APF mutant (APF). Data shown are representative of three different experiments. (B) Levels of expression of the endogenous and transfected JNK1 proteins examined by Western blot analysis using two polyclonal anti-JNK1 antibodies (C17 and N19). (C) Percentage of cell death in SHEP and APF 24 hr after addition of anti-Fas antibody (100 ng/ml). The percentage of cell death compared with the control (no anti-Fas antibody) is shown. Fas killing assays were performed in triplicate and data are expressed as the mean ± SD of nine experiments. (D and E) JNK1 activation (D) and cell death (E) in SHEP cells, JNK1 wild-type (WT), or JNK1 APF together with JNK1 wild-type (APF/WT) transfected cells. Cells were transfected by electroporation with 20 μg of the pCDNA3 plasmid encoding M2-tagged JNK1 wild-type (WT) in association with the empty pCDNA3 vector or JNK1 APF in association with hemagglutinin (HA)-tagged JNK1 wild type (APF/WT). Fas killing assays were performed in triplicate and data are expressed as the mean of four experiments for SHEP/WT and two experiments for SHEP/APF+WT.
The immediate increase in Fas-stimulated JNK activity was observed in APF cells. As mentioned earlier, this may represent the activation of a structurally related kinase, which cannot be inhibited by the JNK1 mutant protein APF. However the subsequent sustained activation of JNK caused by treatment of SHEP cells with anti-Fas was blocked by transfection of the dominant-negative JNK1 (APF) mutant (Fig. 2A). Expression of APF effectively blocked the apoptosis induced by Fas in the first 4 hr after crosslinking (Fig. 2A). Even after 24 hr of incubation with anti-Fas (100 ng/ml), only 33% of the APF-transfected cells were killed, as compared with 74% of the parental cells (Fig. 2C). In these cells, the activation of JNK by anisomycin, another potent activator of JNK/SAPK (26), was also inhibited to the same extent. No changes in Fas expression in APF cells were detected (data not shown). The cells transfected with wild-type JNK1 (WT) (Fig. 2B) did not show any significant difference (compared with parental SHEP cells) in both JNK activation (Fig. 2D) and sensitivity to Fas killing (Fig. 2E). The inhibitory effect on JNK activity and Fas-mediated apoptosis caused by the dominant-negative APF mutant could be reversed by cotransfection with wild-type JNK1 (APF/WT) (Fig. 2 D and E).
Dependence of JNK Activation by Anti-Fas on Ras.
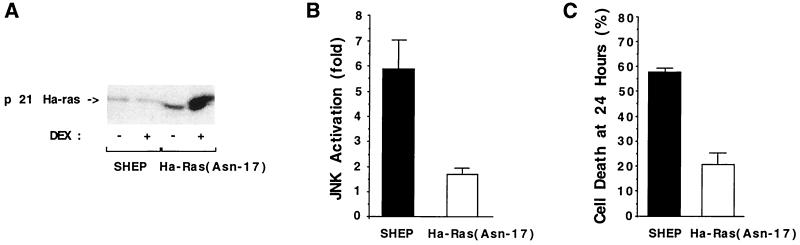
To test whether Ras is required for Fas signaling, we investigated the effect of expressing a dominant-negative Ras on Fas-induced JNK activation. SHEP cells were transfected with a plasmid expressing a dexamethasone-inducible dominant-interfering Ha-Ras(Asn-17) mutant (Fig. 3A) (42). Pools of transfected cells, isolated by short-term drug selection, were used in this study. A marked inhibition of Fas-induced JNK activation was observed in Ha-Ras(Asn-17)-expressing cells, suggesting that Fas-induced JNK activation was Ras dependent (Fig. 3B). Moreover, the mutated Ras protein also blocked Fas-mediated killing (Fig. 3C). The levels of JNK activation or Fas-mediated apoptosis were slightly reduced by the presence of dexamethasone in SHEP cells.
Figure 3.
JNK activation by Fas is Ras dependent. (A) Level of expression of the Ha-Ras(Asn-17) protein after induction by dexamethasone. Cells were transfected with 20 μg of the glucocorticoid-inducible Ha-Ras(Asn17):pMMTV Ha-Ras(Asn-17) plasmid. Cells were incubated with 5 × 10−7 M dexamethasone (DEX+) or vehicle alone (DEX−) for 18 hr and lysed in ELB. The level of expression of the Ras (endogenous and mutant) proteins was examined by Western blot analysis using an anti-human Ras polyclonal antibody (Upstate Biotechnology). (B) Inhibition of Fas-induced JNK activation by expression of Ha-Ras(Asn-17). Transfected and control SHEP cells were incubated with 5 × 10−7 M dexamethasone for 18 hr and then treated with anti-Fas (1 μg/ml) for 2 hr. Cells were collected and lysates were assayed for JNK activity in an immune complex kinase assay. The kinase activities are expressed relative to cells incubated without anti-Fas. Data represent the mean ± SD of two independent assays. (C) Inhibition of Fas killing by expression of Ha-Ras(Asn-17). Transfected Ha-Ras(Asn-17) and control SHEP cells were cultured with 5 × 10−7 M dexamethasone for 18 hr. The cells were then treated with anti-Fas for 24 hr (100 ng/ml). The percentage of dead cells was determined by a crystal violet staining/spectrophotometric assay. Data represent mean ± SD of two independent assays performed in triplicate.
ERK Protein Kinase Activation in Fas-Mediated Apoptosis.
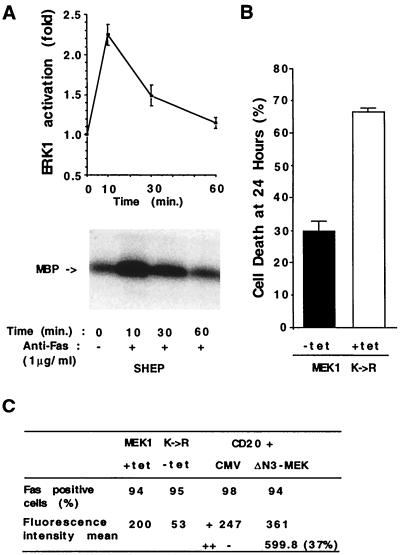
Activation of Ras triggers at least two different signaling cascades that activate different MAP kinases: the JNKs (27, 43) and the ERKs (44–48). We examined ERK activity after Fas ligation in an immune-complex kinase assay using myelin basic protein as a substrate. A small and transient activation of ERK1 (approximately 2-fold) was observed in Fas-treated cells approximately 10 to 30 min after anti-Fas antibody addition (Fig. 4A). This activation was also blocked in Ha-Ras(Asn-17)-expressing cells but not in APF cells (data not shown).
Figure 4.
ERK protein kinase and Fas-induced apoptosis. (A) Time course of ERK1 activity in SHEP cells after treatment with anti-Fas antibody (1 μg/ml). Data represent mean ± SD of three independent assays. A representative autoradiography showing the phosphorylated myelin basic protein (MBP) after SDS/PAGE is shown. (B) Inhibition of Fas killing by expression of MEK1 K→R. Transfected MEK1 K→R cells were cultured in the presence (+tet) or after removal (−tet) of tetracycline for 9 hr. The cells were then treated with anti-Fas for 24 hr (100 ng/ml). The percentage of dead cells was determined by a crystal violet staining/spectrophotometric assay. Data represent mean ± SD of three independent assays performed in triplicate. (C) Fas expression in parental, MEK1 K→R (±tet), and activated ΔN3-MEK-transfected cells. Fas was detected by surface immunofluorescence staining and flow cytometry. Cells expressing the mutated MEK1 K→R protein were cultured in the presence (+tet) or after removal (−tet) of tetracycline for 24 hr and stained with anti-Fas antibody. Cells expressing activated MEK1 (ΔN3-MEK) were obtained in a transient cotransfection assay as described (49). SHEP cells were transiently transfected with 2 μg of CMV CD20 in combination with CMV ΔN3-MEK (5 μg) (ΔN3-MEK) (50), provided by N. G. Ahn, or the CMV vector (CMV) (5 μg). CD20-positive cells were selected by flow cytometry. Three thousand CD20 cells were analyzed for Fas expression. In ΔN3-MEK-transfected cells, two distinct populations of CD20 cells, exhibiting different amount of Fas staining, could be distinguished. The mean fluorescence intensity for the ΔN3-MEK and CMV total cell populations are indicated by +. The mean fluorescence intensity of the subpopulation of ΔN3-MEK cells displaying stronger Fas staining is indicated as ++.
To evaluate the relevance of ERK1 activation in the Fas-apoptotic pathway, we tranfected SHEP cells with dominant-negative MEK1 (MEK1 K→R) cloned in a tetracycline-regulatable expression vector. Pools of transfected cells, isolated by short-term drug selection, were used in this study. ERK activation, but not JNK’s, was inhibited in anti-Fas-treated MEK1 K→R-transfected cells after induction of the dominant-negative MEK1 by tetracycline removal (data not shown). When assayed for Fas-induced apoptosis, MEK1 K→R cells (−tet) were resistant to Fas killing, compared with cells maintained in the presence of tetracycline (+tet) (Fig. 4B).
To investigate the role of the ERK signaling pathway in Fas killing, we examined the expression of Fas by flow cytometry. We found that dominant-negative MEK1 caused a decrease in the expression of Fas on SHEP cells (Fig. 4C). This decreased expression of Fas may account, in part, for the resistance of the MEK1 K→R (−tet) cells to Fas-mediated apoptosis. Moreover, transient transfection of SHEP cells with an activated MEK1 kinase (ΔN3-MEK) (50) up-regulated Fas expression (Fig. 4C). Thirty-seven percent of the ΔN3-MEK1-transfected cells displayed a mean intensity of fluorescence double that of the control CMV vector-transfected cells.
Effect of Fas-Mediated Apoptosis Inhibitors on JNK Activity.
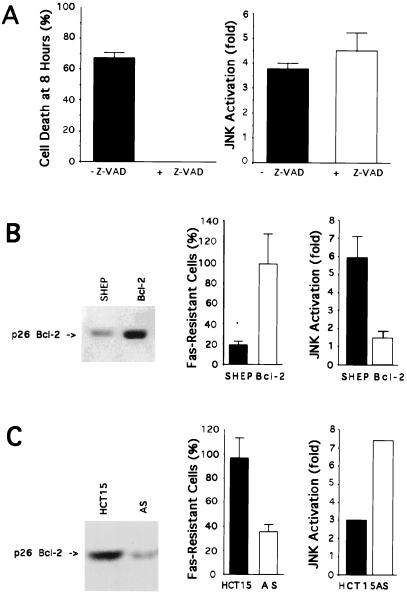
The ICE-related protease cascade is required for induction of apoptosis through the Fas signaling pathway (51). To investigate whether inhibition of ICE activity influences Fas-induced JNK activation, we treated SHEP cells with anti-Fas antibody (1 μg/ml) in the presence of the ICE-like protease inhibitor peptide Z-VAD-fmk (50 μM) and determined JNK activity. As expected, we found that preincubation with Z-VAD-fmk completely prevented Fas killing (Fig. 5A). No change in Fas-stimulated JNK activity was observed (Fig. 5A).
Figure 5.
Effect of an ICE-inhibitor and Bcl-2 on Fas-induced cell death and JNK activation. (A) SHEP cells were incubated for 1 hr with or without the ICE-like protease inhibitor peptide Z-VAD-fmk (50 μM) (Enzyme Systems Products). Anti-Fas (1 μg/ml) was added to the wells for 8 hr. Cell death was determined after 8 hr by a crystal violet staining/spectrophotometric assay. The mean percentage of cell death is shown. Data represent the average of two different experiments performed in triplicate. Error bars indicate the SD. JNK protein kinase activities were measured 4 hr after addition of the anti-Fas. The kinase activities are expressed relative to cells incubated without anti-Fas. Data represent the average of two different experiments. Error bars indicate the SD. (B) Cells were transfected by electroporation using the vector pCEP9 containing the mouse Bcl-2 cDNA in the sense orientation: (Bcl-2). Bcl-2 (p26) protein levels in control SHEP and Bcl-2 cells were determined by immunoblotting. Cells were plated in 96-well plates and the anti-Fas antibody (100 ng/ml) was added for 24 hr. Cell survival was determined after 24 hr by a crystal violet staining/spectrophotometric assay. The mean percentage of cells resistant to cell death after 24 hr is shown. Data represent the average of triplicate cultures from 10 independent experiments. Error bars indicate the SD. SHEP and Bcl-2 cells were plated in six-well plates. Anti-Fas antibody (1 μg/ml) was added to the cells for 4 hr and JNK protein kinase activity was measured. The kinase activities are expressed relative to cells incubated without anti-Fas. Data represent the average from two different experiments performed in duplicate. Error bars indicate the SD. (C) Cell survival and JNK protein kinase assays in parental and Bcl-2 antisense-transfected HCT-15 cells. HCT-15 cells were transfected by electroporation using the vector pCEP9 alone (HCT-15) or containing the mouse Bcl-2 cDNA in the antisense orientation (AS). Data represent the average of triplicate cultures from three independent experiments. Error bars indicate the SD. One kinase assay is shown. The experiment was repeated twice and despite different JNK activity levels, the increase in JNK activation in AS as compared with HCT-15 was reproducibly observed.
Overexpression of Bcl-2 prevents apoptosis in response to a number of stimuli, including Fas activation (34). To explore the effect of Bcl-2 on JNK activity, SHEP neuroblastoma cells were transfected with a plasmid vector expressing Bcl-2. After selection in G418, the obtained mass population (Bcl-2) was tested for the presence of the p26 Bcl-2 protein and sensitivity to Fas killing. Immunoblot analysis demonstrated that the transfected cells express Bcl-2 at a higher level than the untransfected SHEP cells (Fig. 5B). As expected, the mass population expressing Bcl-2 was resistant to Fas killing. At a dose of 100 ng/ml of anti-Fas, more than 95% of the Bcl-2 cells were resistant to Fas-mediated apoptosis, compared with 20% of the control SHEP cells (Fig. 5B). The resistance of the mass population was not a result of loss of Fas-receptor expression, as confirmed by flow cytometric analysis. No change in Fas receptor expression was detected (data not shown). When we compared JNK protein kinase activity in SHEP and Bcl-2 cells, we observed that the second phase of JNK activation caused by treatment with anti-Fas was reduced (Fig. 5B). Typically, a reduction of 65–75% in JNK activation was observed in Bcl-2 compared with SHEP. No change in JNK protein level was observed in Bcl-2 compared with SHEP (data not shown). The reduction in JNK activity inversely correlated with the degree of resistance to Fas killing observed in Bcl-2 cells. When we measured ERK activation in Bcl-2-transfected cells, we did not observe any changes (data not shown).
To demonstrate more definitively that endogenous Bcl-2 expression can influence Fas-mediated apoptosis and JNK activation, we designed experiments employing antisense inhibition of Bcl-2 expression in the HCT-15 colon cell line. HCT-15 was chosen because it expresses Fas, has high levels of Bcl-2, and shows a low sensitivity to Fas-mediated killing, suggesting that Fas is functional in this cell line. In HCT-15 cells transfected with an antisense Bcl-2 vector (AS) (Fig. 5C), Fas engagement resulted in the killing of 65% of the cell population (representing over 50 individual transfected clones) as compared with 5% of the cells transfected with the control vector (HCT-15) (Fig. 5C). In these cells, reduced Bcl-2 expression was associated with an enhancement of apoptotic cell death. Cells transfected with the control vector (HCT-15) retained the resistant phenotype of the parental cell line. No change in Fas receptor expression was detected by flow cytometric analysis (data not shown). By decreasing the level of Bcl-2 expression with an antisense strategy (AS), we could also restore JNK activation upon Fas ligation (Fig. 5C).
DISCUSSION
The data presented here show that JNK, ERK, and Ras are involved in Fas-mediated apoptosis. JNKs have recently been implicated in apoptosis induced by environmental stresses (24, 25) and by NGF deprivation in differentiated rat PC12 pheochromocytoma cells (23). In NGF withdrawal-induced apoptosis, cell death can be prevented by activation of a survival pathway, such as ERK. In contrast, during Fas-mediated apoptosis, the JNK and ERK groups of MAP kinases cooperate in the induction of cell death. Here, we show that the ERK signaling pathway increases Fas expression. Whether the only function of ERK MAP kinase is the regulation of Fas expression remains to be established. However, it is likely that additional targets of the ERK pathway (46), such as the transcription factors c-Myc (52) and c-Fos (53), which are involved in the control of apoptosis, may contribute to signaling by Fas. JNK activation by Fas is Ras dependent. As previously described (20), a dominant-interfering Ras protein also blocked Fas-mediated killing, confirming that the signaling cascade leading from Ras to JNK activation is implicated in the Fas death pathway. Additional small GTP-binding proteins, including members of the Rho group, may also contribute to Fas-induced JNK activation (54).
Two proteins binding to the intracellular part of the Fas receptor at the level of one region required for death induction (55) (the death domain), have been identified. These are MORT1/FADD (Fas-associated protein with death domain) (56, 57) and RIP (receptor-interacting protein) (58). When expressed ectopically, these proteins can induce cell death. Recently, the protein FLICE/MACH (59, 60), which interacts with FADD/MORT1 and possesses a region homologous to the ICE-related protease family, was identified. This protein may represent the link between Fas and the ICE-like protease cascade that has been implicated in Fas-induced apoptosis (51). This cascade includes members of the Ced-3-like and the ICE-like subfamilies (51). We did not observe any change in Fas-induced JNK activation in cells rendered Fas-resistant by blocking the ICE-like protease activity with the inhibitor peptide Z-VAD-fmk, suggesting that JNK activation is not dependent on the protease cascade activation. These data also confirm that JNK activation is not a secondary effect of the death induced by Fas.
We showed that changes in Bcl-2 expression level influence specifically Fas-stimulated JNK protein activity. No difference in the time course or extent of ERK activation was observed, consistent with the observation that no changes in Fas expression were detected between parental and Bcl-2 sense and antisense transfected cells. These data implicate Bcl-2 in the control of Fas-mediated apoptosis and possibly other cell death programs, in part, by regulating the pathway leading to JNK activation. The mechanism that accounts for Bcl-2 inhibition of JNK activation remains to be elucidated. Bcl-2 may activate a JNK-specific phosphatase or, alternatively, inhibit the activation of another component of the signaling cascade leading from the Fas receptor to JNK activation. Bcl-2 can bind to R-Ras, a member of the Ras superfamily of GTPases (61), and antagonizes the ability of activated R-Ras to accelerate cell death upon interleukin 3 withdrawal from FL5.12 cells (62). Bcl-2 could regulate the function of other members of the Ras family, such as Ha-Ras, whose function is required for Fas-induced JNK activation. Alternatively, other kinases downstream from Ras in the signaling pathway regulating JNK activation may also be regulated by Bcl-2. Bcl-2 family members interact with each other to regulate apoptosis. Bax heterodimerizes with Bcl-2 and antagonizes its anti-apoptotic activity (38). Whether Bax blocks a Bcl-2-mediated survival pathway or Bcl-2 inhibits a Bax-mediated apoptotic pathway is unclear (34). It is possible that Bcl-2 inhibits the activation of JNK indirectly by binding to Bax, which in this model would be an activator of this pathway. In some cases, Fas has been reported to promote cell proliferation (63). Similarly, JNK activation has been associated with nonapoptotic signals (27). Whether this Fas-stimulatory effect on cell growth is mediated by the same cascades and depends on the expression of genes susceptible to interfere with the apoptotic program, such as Bcl-2, remains to be elucidated.
The JNK-signaling pathway and the ICE-related proteases cascade are both activated and required for Fas-induced apoptosis. Moreover, apoptosis induced by overexpression of Ced-3 or ICE can also be blocked by Bcl-2 (64). The cowpox crmA gene product, which binds to and inhibits ICE (65, 66), blocks apoptosis induced by Fas (17–19) and also prevents apoptosis induced by NGF deprivation of neuronal cells (67), a model in which the JNK cascade has been implicated (23). Identification of the respective targets of the ICE-like protease and the JNK protein kinase cascades, the relationship between these two apoptotic signaling pathways, and the mechanism of action of Bcl-2 represent important questions for future research. The relative contribution of these protease and kinase cascades to the apoptotic process remains to be established.
Acknowledgments
We thank G. M. Cooper for the pMMTV Ha-Ras(Asn-17) expression plasmid, R. L. Erickson for the MEK1 K→R cDNA and the anti-MEK1 antibody, A. Fattaey for the tetracycline-regulatable vector, N. G. Ahn for the ΔN3MEK expression plasmid, S. Korsmeyer for the Bcl-2 cDNA, and P. Alonso for his help with the flow cytometry analysis. L. Yamasaki, J. LaBaer, J. Koh, and E. A. Harrington are acknowledged for their careful reading of the manuscript. I.S. thanks B. Dynlacht and L. Berg for interesting discussion and for their support through the course of this endeavor. E.H. is an American Cancer Society Research Professor. R.J.D. is an investigator of the Howard Hughes Medical Institute. This work was supported by grants from the National Institutes of Health to R.I.T., E.H., and R.D.
ABBREVIATIONS
- MAP
mitogen-activated protein
- JNK
Jun kinase
- ERK
extracellular signal-regulated kinase
- ICE
interleukin 1β-converting enzyme
- TNF
tumor necrosis factor
- NGF
nerve growth factor
- CMV
cytomegalovirus
- Z-VAD-fmk
N-benzyloxycarbonyl-Val-Ala-Asp-fluoromethylketone
References
- 1.Oehm A, Behrmann I, Falk W, Pawlita M, Maier G, et al. J Biol Chem. 1992;267:10709–10715. [PubMed] [Google Scholar]
- 2.Itoh N, Yonehara S, Ishii A, Yonehara M, Mizushima S, Sameshima M, Hase A, Seto Y, Nagata S. Cell. 1991;66:233–243. doi: 10.1016/0092-8674(91)90614-5. [DOI] [PubMed] [Google Scholar]
- 3.Suda T, Takahashi T, Golstein P, Nagata S. Cell. 1993;75:1169–1178. doi: 10.1016/0092-8674(93)90326-l. [DOI] [PubMed] [Google Scholar]
- 4.Suda T, Nagata S. J Exp Med. 1994;179:873–879. doi: 10.1084/jem.179.3.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yonehara S, Ishii A, Yonehara M. J Exp Med. 1989;169:1747–1756. doi: 10.1084/jem.169.5.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trauth B C, Klas C, Peters A M, Matzku S, Moller P, Falk W, Debatin K M, Krammer P H. Science. 1989;245:301–305. doi: 10.1126/science.2787530. [DOI] [PubMed] [Google Scholar]
- 7.Dhein J, Walczak H, Baumler C, Debatin K M, Krammer P H. Nature (London) 1995;373:438–441. doi: 10.1038/373438a0. [DOI] [PubMed] [Google Scholar]
- 8.Ju S T, Panka D J, Cui H, Ettinger R, el-Khatib M, Sherr D H, Stanger B Z, Marshak-Rothstein A. Nature (London) 1995;373:444–448. doi: 10.1038/373444a0. [DOI] [PubMed] [Google Scholar]
- 9.Rathmell J C, Cooke M P, Ho W Y, Grein J, Townsend S E, Davis M M, Goodnow C C. Nature (London) 1995;376:181–184. doi: 10.1038/376181a0. [DOI] [PubMed] [Google Scholar]
- 10.Brunner T, Mogil R J, LaFace D, Yoo N J, Mahboubi A, Echeverri F, Martin S J, Force W R, Lynch D H, Ware C F, et al. Nature (London) 1995;373:441–444. doi: 10.1038/373441a0. [DOI] [PubMed] [Google Scholar]
- 11.Rothstein T L, Wang J K, Panka D J, Foote L C, Wang Z, Stanger B, Cui H, Ju S T, Marshak-Rothstein A. Nature (London) 1995;374:163–165. doi: 10.1038/374163a0. [DOI] [PubMed] [Google Scholar]
- 12.Walsh C M, Glass A A, Chiu V, Clark W R. J Immunol. 1994;153:2506–2514. [PubMed] [Google Scholar]
- 13.Stalder T, Hahn S, Erb P. J Immunol. 1994;152:1127–1133. [PubMed] [Google Scholar]
- 14.Rouvier E, Luciani M F, Golstein P. J Exp Med. 1993;177:195–200. doi: 10.1084/jem.177.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lowin B, Hahne M, Mattmann C, Tschopp J. Nature (London) 1994;370:650–652. doi: 10.1038/370650a0. [DOI] [PubMed] [Google Scholar]
- 16.Kagi D, Vignaux F, Ledermann B, Burki K, Depraetere V, Nagata S, Hengartner H, Golstein P. Science. 1994;265:528–530. doi: 10.1126/science.7518614. [DOI] [PubMed] [Google Scholar]
- 17.Los M, Van de Craen M, Penning L C, Schenk H, Westendorp M, Baeuerle P A, Droge W, Krammer P H, Fiers W, Schulze-Osthoff K. Nature (London) 1995;375:81–83. doi: 10.1038/375081a0. [DOI] [PubMed] [Google Scholar]
- 18.Enari M, Hug H, Nagata S. Nature (London) 1995;375:78–81. doi: 10.1038/375078a0. [DOI] [PubMed] [Google Scholar]
- 19.Tewari M, Dixit V M. J Biol Chem. 1995;270:3255–3260. doi: 10.1074/jbc.270.7.3255. [DOI] [PubMed] [Google Scholar]
- 20.Gulbins E, Bissonnette R, Mahboubi A, Martin S, Nishioka W, Brunner T, Baier G, Baier-Bitterlich G, Byrd C, Lang F, Kolesnick A, Altman A, Green D. Immunity. 1995;2:341–351. doi: 10.1016/1074-7613(95)90142-6. [DOI] [PubMed] [Google Scholar]
- 21.Cifone M G, De Maria R, Roncaioli P, Rippo M R, Azuma M, Lanier L L, Santoni A, Testi R. J Exp Med. 1994;180:1547–1552. doi: 10.1084/jem.180.4.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tepper C G, Jayadev S, Liu B, Bielawska A, Wolff R, Yonehara S, Hannun Y A, Seldin M F. Proc Natl Acad Sci USA. 1995;92:8443–8447. doi: 10.1073/pnas.92.18.8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xia Z, Dickens M, Raingeaud J, Davis R J, Greenberg M E. Science. 1995;270:1326–1331. doi: 10.1126/science.270.5240.1326. [DOI] [PubMed] [Google Scholar]
- 24.Verheij M, Bose R, Lin X L, Yao B, Jarvis W D, Grant S, Birrer M J, Szabo E, Zon L I, Kyriakis J M, Friedman A H, Fuks Z, Kolesnick R N. Nature (London) 1996;380:75–79. doi: 10.1038/380075a0. [DOI] [PubMed] [Google Scholar]
- 25.Chen Y R, Meyer C F, Tan T H. J Biol Chem. 1996;272:631–634. doi: 10.1074/jbc.271.2.631. [DOI] [PubMed] [Google Scholar]
- 26.Kyriakis J M, Banerjee P, Nikolakaki E, Dai T, Rubie E A, Ahmad M F, Avruch J, Woodgett J R. Nature (London) 1994;369:156–160. doi: 10.1038/369156a0. [DOI] [PubMed] [Google Scholar]
- 27.Derijard B, Hibi M, Wu I H, Barrett T, Su B, Deng T, Karin M, Davis R J. Cell. 1994;76:1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- 28.Hibi M, Lin A, Smeal T, Minden A, Karin M. Genes Dev. 1993;7:2135–2148. doi: 10.1101/gad.7.11.2135. [DOI] [PubMed] [Google Scholar]
- 29.Galcheva-Gargova Z, Derijard B, Wu I H, Davis R J. Science. 1994;265:806–808. doi: 10.1126/science.8047888. [DOI] [PubMed] [Google Scholar]
- 30.Sluss H K, Barrett T, Derijard B, Davis R J. Mol Cell Biol. 1994;14:8376–8384. doi: 10.1128/mcb.14.12.8376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bakhshi A, Jensen J P, Goldman P, Wright J J, McBride O W, Epstein A L, Korsmeyer S J. Cell. 1985;41:899–906. doi: 10.1016/s0092-8674(85)80070-2. [DOI] [PubMed] [Google Scholar]
- 32.Cleary M L, Smith S D, Sklar J. Cell. 1986;47:19–28. doi: 10.1016/0092-8674(86)90362-4. [DOI] [PubMed] [Google Scholar]
- 33.Tsujimoto Y, Gorham J, Cossman J, Jaffe E, Croce C M. Science. 1985;229:1390–1393. doi: 10.1126/science.3929382. [DOI] [PubMed] [Google Scholar]
- 34.White E. Genes Dev. 1996;10:1–15. doi: 10.1101/gad.10.1.1. [DOI] [PubMed] [Google Scholar]
- 35.Boise L H, Gonzalez-Garcia M, Postema C E, Ding L, Lindsten T, Turka L A, Mao X, Nunez G, Thompson C B. Cell. 1993;74:597–608. doi: 10.1016/0092-8674(93)90508-n. [DOI] [PubMed] [Google Scholar]
- 36.Kozopas K M, Yang T, Buchan H L, Zhou P, Craig R W. Proc Natl Acad Sci USA. 1993;90:3516–3520. doi: 10.1073/pnas.90.8.3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin E Y, Orlofsky A, Berger M S, Prystowsky M B. J Immunol. 1993;151:1979–1988. [PubMed] [Google Scholar]
- 38.Oltvai Z N, Milliman C L, Korsmeyer S J. Cell. 1993;74:609–619. doi: 10.1016/0092-8674(93)90509-o. [DOI] [PubMed] [Google Scholar]
- 39.Yang E, Zha J, Jockel J, Boise L H, Thompson C B, Korsmeyer S J. Cell. 1995;80:285–291. doi: 10.1016/0092-8674(95)90411-5. [DOI] [PubMed] [Google Scholar]
- 40.Chittenden T, Harrington E A, O’Connor R, Flemington C, Lutz R J, Evan G I, Guild B C. Nature (London) 1995;374:733–736. doi: 10.1038/374733a0. [DOI] [PubMed] [Google Scholar]
- 41.Hengartner M O, Ellis R E, Horvitz H R. Nature (London) 1992;356:494–499. doi: 10.1038/356494a0. [DOI] [PubMed] [Google Scholar]
- 42.Cai H, Szeberenyi J, Cooper G M. Mol Cell Biol. 1990;10:5314–5323. doi: 10.1128/mcb.10.10.5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davis R J. Trends Biochem Sci. 1994;19:470–473. doi: 10.1016/0968-0004(94)90132-5. [DOI] [PubMed] [Google Scholar]
- 44.Cobb M H, Robbins D J, Boulton T G. Curr Opin Cell Biol. 1991;3:1025–1032. doi: 10.1016/0955-0674(91)90124-h. [DOI] [PubMed] [Google Scholar]
- 45.Blenis J. Proc Natl Acad Sci USA. 1993;90:5889–5892. doi: 10.1073/pnas.90.13.5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davis R J. J Biol Chem. 1993;268:14553–14556. [PubMed] [Google Scholar]
- 47.Hunter T. Cell. 1995;80:225–236. doi: 10.1016/0092-8674(95)90405-0. [DOI] [PubMed] [Google Scholar]
- 48.Marshall C J. Cell. 1995;80:179–185. doi: 10.1016/0092-8674(95)90401-8. [DOI] [PubMed] [Google Scholar]
- 49.van den Heuvel S, Harlow E. Science. 1993;262:2050–2054. doi: 10.1126/science.8266103. [DOI] [PubMed] [Google Scholar]
- 50.Mansour S J, Matten W T, Hermann A S, Candia J M, Rong S, Fukasawa K, Vande Woude G F, Ahn N G. Science. 1994;265:966–970. doi: 10.1126/science.8052857. [DOI] [PubMed] [Google Scholar]
- 51.Fraser A, Evan G. Cell. 1996;85:781–784. doi: 10.1016/s0092-8674(00)81005-3. [DOI] [PubMed] [Google Scholar]
- 52.Harrington E A, Fanidi A, Evan G I. Curr Opin Genet Dev. 1994;4:120–129. doi: 10.1016/0959-437x(94)90100-7. [DOI] [PubMed] [Google Scholar]
- 53.Smeyne R J, Vendrell M, Hayward M, Baker S J, Miao G G, Schilling K, Robertson L M, Curran T, Morgan J I. Nature (London) 1993;363:166–169. doi: 10.1038/363166a0. [DOI] [PubMed] [Google Scholar]
- 54.Whitmarsh, A. J. & Davis, R. J. (1996) J. Mol. Med., in press. [DOI] [PubMed]
- 55.Tartaglia L, Ayres T, Wong G, Goeddel D. Cell. 1993;74:845–853. doi: 10.1016/0092-8674(93)90464-2. [DOI] [PubMed] [Google Scholar]
- 56.Chinnaiyan A M, O’Rourke K, Tewari M, Dixit V M. Cell. 1995;81:505–512. doi: 10.1016/0092-8674(95)90071-3. [DOI] [PubMed] [Google Scholar]
- 57.Boldin M P, Varfolomeev E E, Pancer Z, Mett I L, Camonis J H, Wallach D. J Biol Chem. 1995;270:7795–7798. doi: 10.1074/jbc.270.14.7795. [DOI] [PubMed] [Google Scholar]
- 58.Stanger B Z, Leder P, Lee T H, Kim E, Seed B. Cell. 1995;81:513–523. doi: 10.1016/0092-8674(95)90072-1. [DOI] [PubMed] [Google Scholar]
- 59.Boldin M P, Goncharov T M, Goltsev Y V, Wallach D. Cell. 1996;85:803–815. doi: 10.1016/s0092-8674(00)81265-9. [DOI] [PubMed] [Google Scholar]
- 60.Muzio M, Chinnaiyan A M, Kischkel F C, O’Rourke K, Shevchenko A, Ni J, Scaffidi C, Bretz J D, Zhang M, Gentz R, Mann M, Krammer P H, Peter M E, Dixit V M. Cell. 1996;85:817–827. doi: 10.1016/s0092-8674(00)81266-0. [DOI] [PubMed] [Google Scholar]
- 61.Fernandez-Sarabia M J, Bischoff J R. Nature (London) 1993;366:274–275. doi: 10.1038/366274a0. [DOI] [PubMed] [Google Scholar]
- 62.Wang H G, Millan J A, Cox A D, Der C J, Rapp U R, Beck T, Zha H, Reed J C. J Cell Biol. 1995;129:1103–1114. doi: 10.1083/jcb.129.4.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Owen-Schaub L B, Radinsky R, Kruzel E, Berry K, Yonehara S. Cancer Res. 1994;54:1580–1586. [PubMed] [Google Scholar]
- 64.Miura M, Zhu H, Rotello R, Hartwieg E A, Yuan J. Cell. 1993;75:653–660. doi: 10.1016/0092-8674(93)90486-a. [DOI] [PubMed] [Google Scholar]
- 65.Ray C A, Black R A, Kronheim S R, Greenstreet T A, Sleath P R, Salvesen G S, Pickup D J. Cell. 1992;69:597–604. doi: 10.1016/0092-8674(92)90223-y. [DOI] [PubMed] [Google Scholar]
- 66.Komiyama T, Ray C A, Pickup D J, Howard A D, Thornberry N A, Peterson E P, Salvesen G. J Biol Chem. 1994;269:19331–19337. [PubMed] [Google Scholar]
- 67.Gagliardini V, Fernandez P A, Lee R K, Drexler H C, Rotello R J, Fishman M C, Yuan J. Science. 1994;263:826–828. doi: 10.1126/science.8303301. [DOI] [PubMed] [Google Scholar]