Abstract
Some epidemiological studies have suggested a possible link between human cytomegalovirus (HCMV) infection and various malignancies, and HCMV has been shown to transform cultured cells. However, viral DNA is not detected in most transformants, and the mechanism by which HCMV might contribute to oncogenesis has remained obscure. Here we show that the HCMV immediate early 1 and 2 genes can cooperate with the adenovirus E1A gene to generate transformed foci of primary baby rat kidney cells. HCMV gene expression is transient and viral DNA is not present in clonal cell lines derived from the transformed foci. We find that the HCMV immediate early proteins are mutagenic, and we propose that HCMV has the potential to contribute to oncogenesis through a “hit-and-run” mechanism, by inducing mutations in cellular genes.
Serological and molecular studies have suggested a possible association of human cytomegalovirus (HCMV) with the development of human malignancies, including cervical carcinoma, colon carcinoma, and prostate cancer (reviewed in refs. 1–5). A variety of laboratory observations demonstrate that HCMV has oncogenic potential, supporting the idea that HCMV might contribute to the induction of human tumors. HCMV has been shown to induce cervical neoplasia in mice (6), it can morphologically transform human (7, 8) as well as other mammalian (9–11) cells in culture, and HCMV-transformed cells can form tumors in experimental animals (6, 8). In vitro assays have identified three regions on the HCMV genome with transforming activity (reviewed in ref. 5). However, if HCMV infection does contribute to tumor induction in humans, the mechanism underlying HCMV-induced oncogenesis is very likely different from that of DNA viruses that are known to play a role in human malignancies. Whereas specific viral genes are present and expressed in tumors induced by agents such as Epstein–Barr virus and the human papillomaviruses, no specific HCMV DNA sequence element is reliably present and often no viral DNA can be detected in cells transformed by HCMV (reviewed in refs. 4 and 12).
We have previously reported (13) that the two most abundantly expressed immediate-early gene products of HCMV, IE1 and IE2 (reviewed in ref. 14), inhibit the induction of apoptosis by tumor necrosis factor α (TNF-α) or the adenovirus E1A oncoproteins. The fact that the IE1 and IE2 gene products are able to inhibit apoptosis raised the interesting possibility that they might cooperate with the adenovirus E1A oncoproteins to transform primary rodent cells, as do the adenovirus E1B 19-kDa (E1B) protein and the cellular Bcl-2 protein, each of which can block apoptosis (15, 16). In this report, we show that the IE1 and IE2 gene products can, indeed, cooperate with E1A to transform primary baby rat kidney (BRK) cells. Unexpectedly, however, we find that expression of the IE1 and IE2 proteins is transient; HCMV proteins and DNA are not present in transformed cell lines derived from the foci. The IE1 and IE2 gene products are mutagenic, and we propose that they contribute to transformation at least in part by a mutagenesis-based “hit-and-run” mechanism.
MATERIALS AND METHODS
Plasmids.
The mammalian expression plasmids pCMV-E1A (17), pCMV-19-kDa (18); pCGN-IE1 (IE1) and pCGN-IE2 (IE2) (13) and their parental vector pCGN (19) have all been described previously. pPuro-αIE1 and pPuro-αIE2 contain IE1 and IE2 cDNAs, respectively, in an antisense orientation in the pBabe-puro expression vector (20).
BRK Transformation Assay.
BRK cells, which have been used extensively to assay the transforming activity of adenovirus oncogenes (21, 22), were used to test the oncogenic potential of the HCMV immediate early genes. Kidneys from 6-day-old Fisher rats were treated with 0.125% trypsin to produce a uniform cell suspension. These cells were incubated in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal calf serum (FCS) for 4 days, then trypsinized and frozen in aliquots in 90% FCS plus 10% dimethyl sulfoxide. One day before transfection, aliquots of cells were thawed and counted, and 1.6 × 106 viable cells were seeded per 100-mm dish into medium containing 10% FCS. Transfection was by the calcium phosphate precipitation method (23), with 5 μg per dish of each of the expression plasmids. Parental vectors were used to equalize the amount of plasmid DNA among different transfections, and the total amount of DNA was adjusted to 30 μg per dish with salmon sperm DNA. After 16 hr, DNA precipitates were removed, and cells were allowed to recover in medium containing 10% FCS for 24 hr. Finally, cells from each dish were split into three 60-mm dishes and incubated in medium containing 5% FCS until foci appeared. Some foci were cloned and maintained in DMEM containing 10% FCS. Others were visualized with 4% Giemsa stain.
Assays for Gene Expression.
Immunoprecipitation of IE1 and IE2 proteins from 35S-labeled cell extracts was as described (16) with monoclonal antibody mAB810 (Chemicon), which interacts with the common amino-terminal domain shared by the two immediate early proteins. Expression of E1A was assayed by immunoprecipitation from unlabeled cell extracts by using the E1A-specific monoclonal antibody M73 (24). The immune complexes were resolved by electrophoresis on SDS/8% polyacrylamide gels, transferred to nitrocellulose, subjected to immunoblot analysis with antibody M73, and visualized by using ECL reagents (Amersham).
For analysis of protein expression by immunofluorescence, BRK cells seeded on coverslips were transfected by the calcium phosphate precipitation method (23) either with pCMV-E1A plus pCMV-19-kDa or with pCMV-E1A plus pCGN-IE1 and pCGN-IE2. At appropriate times after transfection, two coverslips were removed from each transfected culture, cells were fixed in phosphate-buffered saline containing 4% paraformaldehyde, and the preparations were stored in phosphate-buffered saline at 4°C. After coverslips were collected at all time points, the cells on them were allowed to react with the appropriate primary antibodies. For the two sets of coverslips collected from cells transfected with vectors expressing the E1A plus E1B-19-kDa proteins, one set was stained with anti-E1A antibody (M73), and the other was stained with monoclonal antibody to E1B-19-kDa (Oncogene Science). Coverslips with cells transfected with vectors expressing E1A, IE1, and IE2 proteins were allowed to react with either anti-E1A (M73) or anti-HCMV IE1/2 (mAB810) antibody. The samples were then stained with fluorescein isothiocyanate (FITC)-conjugated secondary antibodies and visualized with a confocal microscope (Bio-Rad model MRC600). Each image is from an independent coverslip.
Southern Hybridization.
Total cellular DNA was prepared from 1.5 × 107 cells of each cell line by using DNAzol (BRL). A 15-μg sample of each DNA was digested with EcoRI and BamHI (10 units/μg of DNA) for 12 hr and was resolved by electrophoresis on a 0.7% agarose gel. Southern blotting and hybridization were performed as described (16). The entire IE1 and IE2 cDNAs were used as templates to make probe by random-primed DNA synthesis.
Mutagenesis Assays.
The D422 CHO cell line (25) was a generous gift from L. A. Chasin (Columbia University). The cells were maintained in medium (α-MEM) containing 10% FCS. Subconfluent cultures of D422 were transfected by the calcium phosphate precipitation technique (23) with 5 μg of each expression plasmid per 100-mm dish. The total amount of DNA was adjusted to 30 μg per dish with parental vector and salmon sperm DNA. After transfection, cells were incubated in nonselective growth medium for 4 days. Mutation frequencies were determined by plating 2 × 105 treated cells into each 100-mm dish in medium containing 10% dialyzed FCS supplemented with either 200 μM 2,6-diaminopurine (selection for aprt mutants) or 20 μM 6-thioguanine (selection for hprt mutants). Drug-resistant colonies usually appeared within 7–8 days, and they were stained with 4% Giemsa solution. For each culture, 200 cells were also plated in nonselective medium to determine plating efficiency. As a control, D422 cells were treated with 0.2 mg/ml ethyl methanesulfonate (EMS) for 18 hr and then incubated in nonselective medium for 8 days before selection for drug-resistant colonies.
To assay for mutations in p53, immunoprecipitations were performed from 35S-labeled cell extracts as described (16), using monoclonal antibody pAb240, which specifically recognizes mutant p53. For DNA sequencing, the DNA-binding domain of p53 (amino acids 113–301) was obtained from selected BRK cell lines by reverse transcription followed by PCR amplification using the Pfu DNA polymerase (Stratagene). Two independent reverse transcription/PCRs were performed for each cell line. Amplified products were inserted in PCR-Script cloning vector (Stratagene) and sequenced.
RESULTS
HCMV IE1 and IE2 Can Cooperate with E1A for Transformation.
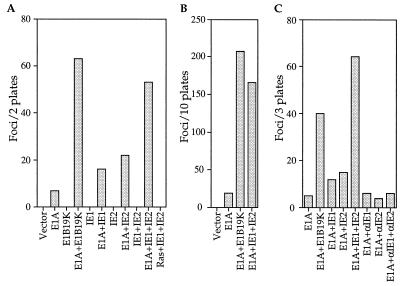
To evaluate the possibility that the HCMV IE1 and IE2 genes might have transforming potential, we transfected primary BRK cells with plasmids encoding the IE1 and IE2 proteins as well as the adenovirus E1A protein. IE1 or IE2 alone enhanced the focus-forming activity of E1A by a factor of 2–3 (Fig. 1A); while IE1 and IE2 together increased the transforming activity of E1A by a factor of about 8, a level comparable to the activity of E1A plus the adenovirus E1B protein (Fig. 1 A and B) or Bcl-2 (15). As reported previously (15), it was extremely difficult to clone the flat foci induced by E1A alone. Of the 17 E1A-induced foci that were tested, only one was recovered as a stable cell line (6%). In contrast, much higher cloning efficiencies were obtained for the multilayered foci induced by E1A plus E1B (9/12, 75%) or E1A plus IE1 and IE2 (12/18, 67%). These results clearly demonstrate that the IE1 and IE2 genes have transforming potential, and they show that there is cooperation between the two immediate early genes. In the absence of E1A, IE1 and IE2 did not produce any foci, nor did they complement activated Ras to transform BRK cells (Fig. 1A).
Figure 1.
The HCMV IE1 and IE2 genes cooperate with the adenovirus E1A gene to transform primary BRK cells. (A) Representative BRK transformation assay showing the number of foci that arise in response to transfection with different combinations of expression plasmids. (B) BRK transformation assays demonstrating that IE1 plus IE2 cooperate with E1A for transformation. The number of foci reported is the total from 10 plates assayed in four independent experiments. (C) Representative experiment showing that IE1 and IE2 cDNAs cloned in an antisense orientation (αIE1 and αIE2) have no transforming activity. The number of foci reported is the total from three 100-mm plates. In all experiments, the gene products expressed by transfected plasmids are listed below the bars. Cultures transfected with pCGN containing no insert are designated vector.
The morphological transforming regions of herpes simplex viruses do not specify any identifiable polypeptide products, and it was hypothesized that the DNA sequence itself is responsible for HSV-2-induced transformation (reviewed in refs. 12, 26, and 27). To determine whether this is also the case in our system, we reversed the direction of the IE1 and IE2 cDNAs in the expression vectors so that they resided in an antisense orientation relative to the promoter. These antisense plasmids failed to cooperate with E1A to transform BRK cells (Fig. 1C), suggesting that the IE1 and IE2 gene products are required for transformation.
IE1 and IE2 Expression Is Transient During Transformation.
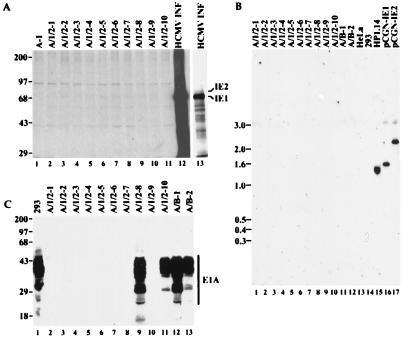
To gain insight to the mechanism by which IE1 and IE2 contribute to oncogenicity, we examined the expression of the transfected genes in a series of transformed BRK cell lines. In cell lines derived by transfection with E1A plus E1B, both E1A (Fig. 2C) and E1B (data not shown) proteins are readily detected in all cell lines tested. Unexpectedly, however, IE1 or IE2 protein was not detected in any of the E1A/IE1/IE2-derived cell lines, either by immunoblot analysis (not shown) or by immunoprecipitation of 35S-labeled cell extracts (Fig. 2A). IE1 and IE2 DNA sequences were completely absent from these cell lines, as judged by Southern blot analysis (Fig. 2B) or PCR amplification of DNA sequences located within the cDNAs (data not shown). Furthermore, E1A protein expression was detected only in a subset of such cell lines (Fig. 2C).
Figure 2.
HCMV IE1 and IE2 genes and their products are not present in transformed BRK cell lines. (A) Lack of expression of the IE1 or IE2 protein in transformed BRK cell lines when assayed by immunoprecipitation of 35S-labeled cell extracts using monoclonal antibody mAB810. A-1 is a cell line established by transfection with the E1A-expressing plasmid alone. A/1/2–1 to A/1/2–10 are cell lines transformed by E1A plus IE1 and IE2. Lanes designated HCMV INF display immunoprecipitates from HCMV-infected human foreskin fibroblasts; lane 13 is an exposure 1/8 as long as that presented for lanes 1–12. Numbers to the left of the gel mark the positions of marker proteins whose sizes are indicated in kDa. (B) Southern blot analysis showing the lack of IE1- or IE2-specific DNA in E1A/IE1/IE2-induced transformants (lanes 1–10). A/B-1 and A/B-2 are cell lines transformed with E1A and E1B-19-kDa. HeLa, 293, and HP1.14 (a derivative HeLa cells expressing IE1) are included as controls. pCGN-IE1 and pCGN-IE2 are IE1 and IE2 expression plasmids, respectively, in amounts equivalent to the rest of the samples, assuming they contain one copy of each of the HCMV genes per cell. Numbers to the left of the gel are marker DNA sizes in kb. (C) Expression of E1A protein as determined by immunoprecipitation followed by immunoblot analysis, both with monoclonal antibody M73 to E1A. Lanes are labeled as in A and B. Numbers to the left of the gel are marker protein sizes in kDa. Autoradiograms were scanned and cropped using Photoshop and figures were prepared using Freehand software.
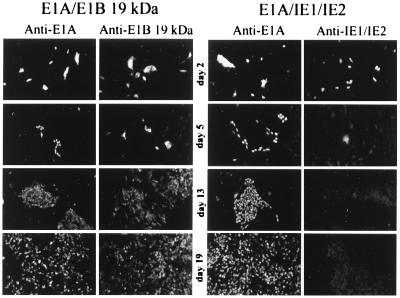
A time course experiment was performed to examine the expression of the transforming proteins after cotransfection into BRK cells. As shown in Fig. 3, the IE1 and IE2 proteins initially accumulated to high levels as judged by immunofluorescence, but their expression tapered off rapidly. By day 5 after transfection, virtually no IE1 or IE2 protein was detected by immunofluorescent staining, even in cells that had begun to form foci. The faint fluorescence evident in cells assayed for IE1 and IE2 expression on days 5, 13, and 19 is a nonspecific background signal. In contrast, the E1B protein was detected in cells transformed by the E1A plus E1B proteins throughout the entire 27-day time course, and the E1A protein was detected throughout the time course in cells transformed by E1A/IE1/IE2 or by E1A/E1B. Even though the IE1 and IE2 proteins disappeared rapidly, their transforming activity was evident: a similar number of foci formed whether the cells were transfected with E1A/E1B or with E1A/IE1/IE2 in the time course experiment (data not shown).
Figure 3.
The IE1 and IE2 gene products are expressed transiently during the transformation of BRK cells. BRK cells seeded on coverslips were transfected with expression plasmids for E1A and E1B-19-kDa (Left) or plasmids for E1A and IE1 and IE2 (Right). At 2, 5, 13, and 19 days after transfection, two coverslips from each transfection were fixed and kept in phosphate-buffered saline until coverslips for all time points were collected. Expression of the transfected genes was examined by indirect immunofluorescent staining with appropriate monoclonal antibodies. Anti-E1A, anti-E1B-19-kDa, and anti-IE1/2 identify the antibodies used for the immunofluorescent staining. Images are from independent coverslips. The images from days 13 and 19 display transformed foci whose cells were positive for E1A and E1B-19-kDa proteins, but completely negative for IE1 and IE2 protein expression. (×50).
IE1 and IE2 Exhibit Mutagenic Potential.
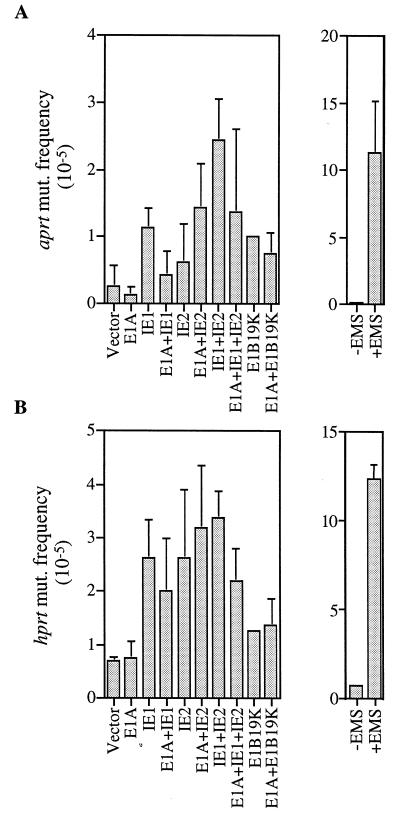
These results are difficult to fit into the conventional theory of viral oncogenesis, where the continuous expression of an oncogene is generally required to sustain a transformed phenotype. Rather, they are consistent with a “hit-and-run” model (reviewed in ref. 27) in which the HCMV genes act transiently to induce a transformed state. Several earlier reports indicating that HCMV infection results in cellular chromosome damage (28–31) prompted us to ask whether IE1 and IE2 might cause a “hit” by inducing mutations in cellular genes. To test this possibility, we examined whether the IE1 and IE2 proteins could alter the mutation rates of aprt and hprt, two cellular genes that are often used as indicators of mutation frequency. A CHO-derived cell line, D422, which is hemizygous for both aprt and hprt loci, was chosen as the indicator cell (25). As shown in Fig. 4, when IE1 and IE2 expression plasmids were introduced into D422, the frequency of mutations at the aprt and hprt loci increased by a factor of 4 to 10 over the background level, whereas transfection with the expression plasmid for E1A or the vector with no insert did not significantly change the mutation frequency. These results demonstrate that IE1 and IE2 have the capacity to induce mutations. In fact, considering that about 10% of the cells were transfected (data not shown), the actual mutation frequencies induced by the IE1 and IE2 proteins appear to be comparable to that induced by EMS, a chemical mutagen used in the experiments as a positive control (Fig. 4).
Figure 4.
The IE1 and IE2 gene products increase the frequency of mutation within the aprt (A) and hprt (B) genes. D422 cells were transfected with expression plasmids (identified below bars). After an incubation period of 4 days, aprt−/− and hprt−/− mutants were selected by the addition of 200 μM diaminopurine or 20 μM 6-thioguanine, respectively, to otherwise purine-free medium. The frequencies of mutation at the two loci for cells exposed to 0.2 mg/ml EMS for 18 hr are also shown.
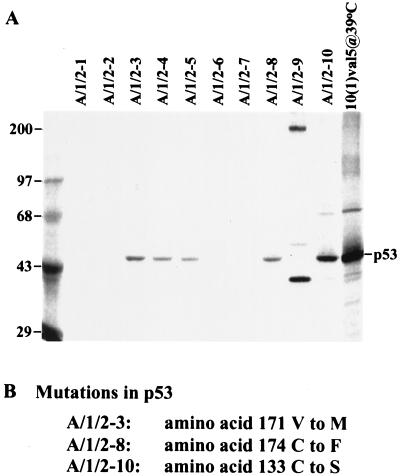
Since mutant p53 alleles are known to cooperate with the adenovirus E1A protein to transform rodent cells (32), we tested whether the p53 gene was altered in the E1A/IE1/IE2 transformants. Immunoprecipitation with an antibody (pAb240) that recognizes some mutant p53 alleles but not wild-type p53 revealed that at least 6 of 10 transformed cell lines contained mutant p53 (Fig. 5A). The p53 DNA-binding domain was amplified and sequenced in three of the transformants, and each was found to contain a missense mutation (Fig. 5B). It seems likely that the p53 mutations result from the temporary expression of the IE1 and IE2 gene products, since mutant p53 alleles arise only rarely in BRK cells receiving the E1A gene alone (33). As a result, the presence of mutant p53 alleles is consistent with a mutagenesis-based “hit-and-run” model for transformation. Further, since mutant p53 alone does not transform BRK cells (32), we anticipate that the cells lacking E1A protein (Fig. 2C) contain additional lesions in cellular genes that contribute to the transformed phenotype.
Figure 5.
BRK cells transformed by E1A, IE1, and IE2 often contain mutant p53 alleles. (A) Immunoprecipitation of 35S-labeled cell extracts with a mutant p53-specific antibody, pAb240. 10(1)val5@39°C indicates 10(1) cells harboring a temperature-sensitive form of p53 cultured at 39°C. The left-most lane displays marker proteins whose sizes in kDa are indicated to the left of the autoradiogram. (B) Identification of missense mutations in three E1A/IE1/IE2 transformants. Two independent PCR amplifications of the central DNA-binding domain of p53 were performed for each cell line, and two clones were isolated and sequenced from each amplification reaction. The A/1/2–3 and A/1/2–10 cell lines contain both mutant and wild-type p53 sequences, indicating that only one allele is mutated within the DNA-binding domain. All clones from A/1/2–8 contained a mutation, indicating that both alleles might be altered in these cells.
DISCUSSION
The two most abundant immediate-early products of HCMV, IE1 and IE2, can cooperate with the E1A oncoprotein to transform primary BRK cells (Fig. 1). The IE1 and IE2 products are present only transiently during the transformation process (Figs. 2 and 3), and they promote the accumulation of mutations (Figs. 4 and 5). The HCMV genes are lost after mutations arise that promote cell growth, presumably because continued mutagenesis would be deleterious to cell survival. Thus, the IE1 and IE2 proteins cooperate with E1A to sponsor “hit-and-run” transformation.
Earlier reports have documented the transforming potential of both herpes simplex virus and HCMV DNAs and have demonstrated that viral DNA is often not retained in transformed cells and tumor samples (4, 12, 26, 27, 34–38). Further, several studies have indicated that infection with herpesviruses or transfection with viral DNA induces mutations in cellular genes (12, 39–45). It has been hypothesized (12, 26) that herpesvirus DNAs might cause mutations by movement of insertion sequence-like elements present in the viral genomes. Our results demonstrate that the HCMV IE1 and IE2 coding regions do not have transforming potential when they are positioned in an antisense orientation relative to the promoter (Fig. 1C); transient expression of the immediate early gene products is required for transformation. As yet, we can only speculate on the mechanism by which the IE1 and IE2 products induce the accumulation of mutations. They might increase the error frequency of cellular DNA polymerases or interfere with cellular DNA repair processes. The latter possibility is intriguing because defects in mismatch repair genes play a role in some common human cancers (reviewed in ref. 46).
The finding that many of the transformed BRK cell lines contain mutated p53 alleles suggests that mutation of p53 might be one of the mechanisms by which IE1 and IE2 contribute to transformation. In a similar vein, Boldogh et al. (47) have shown that three different cell lines transformed by HCMV infection harbor an activating mutation in both alleles in H-Ras. However, in both of these cases it is unclear whether the mutations in H-Ras or p53 are a direct result of the mutagenic activity of HCMV gene products. The mutations could arise as the transformants are selected for growth in culture.
Given the mutagenic activity of IE1 and IE2, it is puzzling why they do not transform primary cells in the absence of E1A (Fig. 1A). A plausible explanation for this observation is that mutagenesis by IE1 and IE2 might require active DNA replication, which rarely happens in primary BRK cells unless E1A is present to stimulate DNA replication and cell cycle progression (48). In the established D422 cells used to assay mutation frequencies, however, active cell division might bypass the need for E1A.
Could the mutagenic activity of IE1 and IE2 contribute to cancer in humans? Cell cycle progression is inhibited in productively infected cells (49–52), and most infected cells are eventually killed as HCMV completes its replication cycle. However, defective virus particles arising spontaneously in an infected individual could infect cells in the absence of wild-type virus, and they could express a subset of the viral genes, including IE1 and IE2. Alternatively, the wild-type virus might express only subsets of its genes in certain cell types, as has been suggested for the interaction of HCMV with peripheral blood mononuclear cells (53, 54). Nonproductive infections could induce the accumulation of mutations in cellular growth regulatory genes, which ultimately leads to cell transformation. The induction of cervical neoplasia in mice by HCMV (4) is likely an example of an oncogenic event resulting from an abortive infection, because HCMV expresses a limited number of its genes in rodent cells (55, 56). If cell growth is induced by a nonproductive infection, then, as indicated above, the continued presence of mutagenic viral gene products would likely be detrimental. This would select for malignant cells that have lost the viral genome, and, as a result, it would be very difficult to implicate the widespread pathogen in tumor induction.
Acknowledgments
We thank Dr. L. Chasin (Columbia University) for cell lines and helpful advice on mutagenesis assays, J. Goodhouse for assistance with confocal microscopy, and C. Patterson for extracts from HCMV-infected cells. This work was supported by a grant from the National Cancer Institute (CA41086). Y.S. and H.Z. were Postdoctoral Fellows of the New Jersey Commission on Cancer Research. T.S. is an American Cancer Society Professor and an Investigator of the Howard Hughes Medical Institute.
ABBREVIATIONS
- HCMV
human cytomegalovirus
- BRK
baby rat kidney
- FCS
fetal calf serum
- EMS
ethyl methanesulfonate
References
- 1.Huang E-S, Boldogh I, Mar E C. In: Viruses Associated with Human Cancer. Fillips L, editor. NY: Dekker; 1983. pp. 161–194. [Google Scholar]
- 2.Huang E-S, Mar E-C, Boldogh I, Baskar J. Birth Defects. 1984;20:193–211. [PubMed] [Google Scholar]
- 3.Rapp F, Robbins D. Birth Defects. 1984;20:175–192. [PubMed] [Google Scholar]
- 4.Macnab J C M. J Gen Virol. 1987;68:2525–2550. doi: 10.1099/0022-1317-68-10-2525. [DOI] [PubMed] [Google Scholar]
- 5.Rosenthal L J, Choudhury S. In: Molecular Aspects of Human Cytomegalovirus Disease. Becker Y, Darai G, editors. NY: Springer; 1993. pp. 412–436. [Google Scholar]
- 6.Heggie A D, Wentz W B, Reagan J W, Anthony D D. Cancer Res. 1986;46:5211–5214. [PubMed] [Google Scholar]
- 7.Geder L, Lausch R, O’Neill F, Rapp F. Science. 1976;192:1134–1137. doi: 10.1126/science.179143. [DOI] [PubMed] [Google Scholar]
- 8.Smiley M L, Mar E-C, Huang E-S. J Med Virol. 1988;25:213–226. doi: 10.1002/jmv.1890250212. [DOI] [PubMed] [Google Scholar]
- 9.Albrecht T, Rapp F. Virology. 1973;55:53–61. doi: 10.1016/s0042-6822(73)81007-4. [DOI] [PubMed] [Google Scholar]
- 10.Boldogh I, Gonczol E, Vaczi L. Acta Microbiol Acad Sci Hung. 1978;25:269–275. [PubMed] [Google Scholar]
- 11.Yelle J, Hamelin C. Cell Biol Int Rep. 1986;10:493–499. doi: 10.1016/0309-1651(86)90023-8. [DOI] [PubMed] [Google Scholar]
- 12.Galloway D A, Buonaguro F M, Brandt C R, McDougall J K. Cancer Cells. 1986;4:355–361. [Google Scholar]
- 13.Zhu H, Shen Y, Shenk T. J Virol. 1995;69:7960–7970. doi: 10.1128/jvi.69.12.7960-7970.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mocarski E S. In: Fields Virology. Fields B N, Knipe D M, Howley P M, editors. Philadelphia: Lippincott–Raven; 1996. pp. 2447–2492. [Google Scholar]
- 15.Rao L M, Debbas M, Sabbatini P, Hockenberry D, Korsmeyer S, White E. Proc Natl Acad Sci USA. 1992;89:7742–7746. doi: 10.1073/pnas.89.16.7742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen Y, Shenk T. Proc Natl Acad Sci USA. 1994;91:8940–8944. doi: 10.1073/pnas.91.19.8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neil S D, Hemstrom C, Virtanen A, Nevins J R. Proc Natl Acad Sci USA. 1990;87:2008–2012. doi: 10.1073/pnas.87.5.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.White E, Cipriani R. Proc Natl Acad Sci USA. 1989;86:9886–9890. doi: 10.1073/pnas.86.24.9886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanaka M, Herr W. Cell. 1990;60:375–386. doi: 10.1016/0092-8674(90)90589-7. [DOI] [PubMed] [Google Scholar]
- 20.Morgenstern J P, Land H. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.White E, Cipriani R. Mol Cell Biol. 1990;10:120–130. doi: 10.1128/mcb.10.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moore M, Horikoshi N, Shenk T. Proc Natl Acad Sci USA. 1996;93:11295–11301. doi: 10.1073/pnas.93.21.11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wigler M, Pellicer A, Silverstein S, Axel R, Urlaub G, Chasin L. Proc Natl Acad Sci USA. 1979;76:1373–1376. doi: 10.1073/pnas.76.3.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harlow E, Franza B R, Schley C. J Virol. 1985;55:533–546. doi: 10.1128/jvi.55.3.533-546.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bradley W E, Letovanec D. Somatic Cell Genet. 1982;8:51–66. doi: 10.1007/BF01538650. [DOI] [PubMed] [Google Scholar]
- 26.Galloway D A, Nelson J A, McDougall J K. Proc Natl Acad Sci USA. 1984;81:4736–4740. doi: 10.1073/pnas.81.15.4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galloway D A, McDougall J K. Nature (London) 1983;302:21–24. doi: 10.1038/302021a0. [DOI] [PubMed] [Google Scholar]
- 28.Nachtigal M, Nachtigal S. Arch Roum Pathol Exp Microbiol. 1978;37:223–250. [PubMed] [Google Scholar]
- 29.Luleci G, Sakfzli M, Gunalp A. Acta Virol. 1980;24:341–345. [PubMed] [Google Scholar]
- 30.AbuBaker S, Au W W, Legator M S, Albrecht T. Environ Mol Mutagen. 1988;12:409–420. doi: 10.1002/em.2860120409. [DOI] [PubMed] [Google Scholar]
- 31.Albrecht T, Boldogh I, Fons M P, Lee CH, AbuBaker S, Russell J M, Au W W. Subcell Biochem. 1989;15:157–202. [PubMed] [Google Scholar]
- 32.Debbas M, White E. Genes Dev. 1993;7:546–554. doi: 10.1101/gad.7.4.546. [DOI] [PubMed] [Google Scholar]
- 33.White E. Genes Dev. 1993;7:2277–2284. doi: 10.1101/gad.7.12a.2277. [DOI] [PubMed] [Google Scholar]
- 34.Nelson J A, Fleckenstein B, Galloway D A, McDougall J K. J Virol. 1982;43:83–91. doi: 10.1128/jvi.43.1.83-91.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nelson J A, Fleckenstein B, Jahn G, Galloway D A, McDougall J K. J Virol. 1984;49:109–115. doi: 10.1128/jvi.49.1.109-115.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.El-Beik T, Razzaque A, Jariwalla R, Cihlar R L, Rosenthal L J. J Virol. 1986;60:645–652. doi: 10.1128/jvi.60.2.645-652.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jariwalla R J, Razzaque A, Lawson S, Rosenthal L J. J Virol. 1989;63:425–428. doi: 10.1128/jvi.63.1.425-428.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Razzaque A, Zhu F, Jones C. Virology. 1991;181:399–402. doi: 10.1016/0042-6822(91)90513-b. [DOI] [PubMed] [Google Scholar]
- 39.Schlehofer J R, zur Hausen H. Virology. 1982;122:471–475. doi: 10.1016/0042-6822(82)90247-1. [DOI] [PubMed] [Google Scholar]
- 40.Pilon L, Royal A, Langelier Y. J Gen Virol. 1985;66:259–265. doi: 10.1099/0022-1317-66-2-259. [DOI] [PubMed] [Google Scholar]
- 41.Pilon L, Langelier Y, Royal A. Mol Cell Biol. 1986;6:2977–2983. doi: 10.1128/mcb.6.8.2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brandt C R, Buonaguro F M, McDougall J K, Galloway D A. Nucleic Acids Res. 1987;15:561–573. doi: 10.1093/nar/15.2.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hwang C B C, Shillitoe E J. Virology. 1990;178:180–188. doi: 10.1016/0042-6822(90)90392-5. [DOI] [PubMed] [Google Scholar]
- 44.Clarke P, Clements J B. Virology. 1991;182:597–606. doi: 10.1016/0042-6822(91)90600-g. [DOI] [PubMed] [Google Scholar]
- 45.Hwang C B C, Shillitoe E J. Virology. 1991;181:620–629. doi: 10.1016/0042-6822(91)90895-i. [DOI] [PubMed] [Google Scholar]
- 46.Kolodner R D. Trends Biochem Sci. 1995;20:397–401. doi: 10.1016/s0968-0004(00)89087-8. [DOI] [PubMed] [Google Scholar]
- 47.Boldogh I, Huang E S, Rady P, Arany I, Tyring S, Albrecht T. Intervirology. 1994;37:321–329. doi: 10.1159/000150396. [DOI] [PubMed] [Google Scholar]
- 48.Shenk T E. In: Fields Virology. Fields B N, Knipe D M, Howley P M, editors. Philadelphia: Lippincott-Raven; 1996. pp. 2111–2148. [Google Scholar]
- 49.Jault F M, Jault J M, Ruchti F, Fortunato E A, Clark C, Corbeil J, Richman D D, Spector D H. J Virol. 1995;69:6697–6704. doi: 10.1128/jvi.69.11.6697-6704.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bresnahan W A, Boldogh I, Thompson E A, Albrecht T. Virology. 1996;224:150–160. doi: 10.1006/viro.1996.0516. [DOI] [PubMed] [Google Scholar]
- 51.Lu M, Shenk T. J Virol. 1996;70:8850–8857. doi: 10.1128/jvi.70.12.8850-8857.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dittmer, D. & Mocarski, E. S. (1997) J. Virol., in press. [DOI] [PMC free article] [PubMed]
- 53.Einhorn L, Ost A. J Infect Dis. 1984;149:207–214. doi: 10.1093/infdis/149.2.207. [DOI] [PubMed] [Google Scholar]
- 54.Rice G P A, Schrier R D, Oldstone M B A. Proc Natl Acad Sci USA. 1984;81:6134–6138. doi: 10.1073/pnas.81.19.6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Albrecht T, Nachtigal M, St. Jeor S C, Rapp F. J Gen Virol. 1976;30:167–174. doi: 10.1099/0022-1317-30-2-167. [DOI] [PubMed] [Google Scholar]
- 56.Kamiya S, Tanaka J, Ogura T, Ogura H, Sato H, Hatano M. Arch Virol. 1986;89:131–144. doi: 10.1007/BF01309884. [DOI] [PubMed] [Google Scholar]