Abstract
Effective antiviral agents will be of great value in controlling virus replication and delaying the onset of HIV-1-related disease symptoms. Current therapy involves the use of antiviral agents that target the enzymatic functions of the virus, resulting in the emergence of resistant viruses to these agents, thus lowering their effectiveness. To overcome this problem, we have considered the idea of developing novel agents from within HIV-1 as inhibitors of virus replication. The specificity of the Vpr protein for the HIV-1 virus particle makes it an attractive molecule for the development of antiviral agents targeting the events associated with virus maturation. We have generated chimeric Vpr proteins containing HIV-1-specific sequences added to the C terminus of Vpr. These sequences correspond to nine cleavage sites of the Gag and Gag–Pol precursors of HIV-1. The chimeric Vpr constructs were introduced into HIV-1 proviral DNA to assess their effect on virus infectivity using single- and multiple-round replication assays. The virus particles generated exhibited a variable replication pattern depending on the protease cleavage site used as a fusion partner. Interestingly, the chimeric Vpr containing the cleavage sequences from the junction of p24 and p2, 24/2, completely abolished virus infectivity. These results show that chimeric proteins generated from within HIV-1 have the ability to suppress HIV-1 replication and make ideal agents for gene therapy or intracellular immunization to treat HIV-1 infection.
Keywords: protease cleavage signal/virion maturation/precursor processing/colocalization
The HIV-1 life cycle shares several features common to all retroviruses. These features include virus attachment to a specific receptor, penetration, uncoating, reverse transcription, translocation of viral DNA from the cytoplasm to the nucleus, integration, expression of viral proteins, assembly, and maturation of virus particles (1). Virally encoded enzymatic activities, which are essential for the processes associated with virus infection, have all been used as targets for developing agents that interfere with virus replication (2–4). Unfortunately, the use of such antiviral agents has also resulted in the emergence of drug-resistant viruses (5–7). In comparison to the monotherapy, combination therapy involving multiple inhibitors has been shown to be effective (8–11). The emergence of drug-resistant viruses, however, will remain a problem with the continued use of antiviral agents to target viral enzymes. Hence, alternative strategies to contain HIV-1 replication are warranted. Toward this goal, an approach to generate a novel anti-HIV-1 agent from within the virus has been considered.
Among the auxiliary gene products of HIV-1, vpr, vif, and nef have been shown to be associated with virus particles to a varying extent (12–16). The virion-associated protein Vpr has been an intensive area of interest with respect to understanding the role of Vpr in virus infection. Vpr coding sequences (96 aa) are found to overlap Vif at the 5′ end and Tat at the 3′ end (17). Characteristic features of Vpr include virion incorporation, cell cycle arrest at the G2 stage, nuclear localization, participation in transport of the preintegration complex, demonstration of cation channel activity, and interaction with several candidate cellular proteins (18–28). Additionally, work from our laboratory and others has shown that Vpr is essential for optimum infection of macrophages (29, 30). Mutational analysis of Vpr has revealed the presence of critical domains needed for its virion incorporation and the importance of the predicted helical domain (amino acids 17–34) in such an event (22, 31–38). The virion specificity and abundance of Vpr in viral particles provide avenues for localizing antiviral agents to progeny virus, giving the ability to interfere with the assembly, maturation, and infectivity of HIV-1.
Upon initial synthesis as a polyprotein precursor, the HIV-1 aspartyl protease has the unique ability to autocatalyze its own cleavage from the Pr160 polyprotein precursor. After its release, the protease is then able to catalyze the cleavage at other sites generating the mature Gag proteins, p17, p24, p7, and p6 and the reverse transcriptase (RT) and integrase enzymes. The specific cleavage sites between the proteins in the polyprotein precursor recognized by HIV-1 protease are highly conserved among viral isolates (39).
To generate an effective anti-HIV-1 agent from within the virus, we have combined the protease cleavage site residues found in the Gag and Gag–Pol precursor proteins and the virion-specific feature of Vpr. The rationale for this approach is that an inappropriate presentation of chimeric Vpr (Vpr-C) proteins containing protease cleavage signal sequences might interfere with the processing of bona fide viral precursor polyproteins leading to the generation of incompletely processed noninfectious virus particles (2). The differential amount of Gag, Gag–Pol, and Vpr proteins present in the virus particle supports the feasibility of such an approach. This study has demonstrated that the above strategy is effective in interfering with HIV-1 virus replication.
MATERIALS AND METHODS
Plasmids.
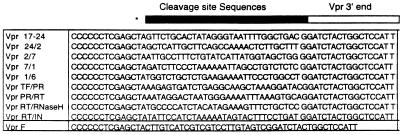
Cloning of wild-type and Vpr-C was carried out using the pCDNA3 expression vector as described (31–35). DNA fragments were amplified through PCR using the proviral clone pNL4–3 with primers containing HindIII and XhoI at the 5′ and 3′ end of the Vpr coding region, respectively. Sequences corresponding to the protease cleavage site residues were added in frame (10 aa) to the C terminus of Vpr coding sequences as part of the minus strand primer (see Figs. 1A and 2A). Sequences representing the Flag epitope (F) were also added to the C terminus of Vpr. All recombinant plasmids were verified by restriction enzyme cleavage and DNA sequence analysis.
Figure 1.
Minus strand primers (5′–3′) used to generate Vpr-C. Primers include restriction site, stop codon (∗), sequences specific for the cleavage site (solid rectangle), and sequences specific for the 3′ end of Vpr (open rectangle).
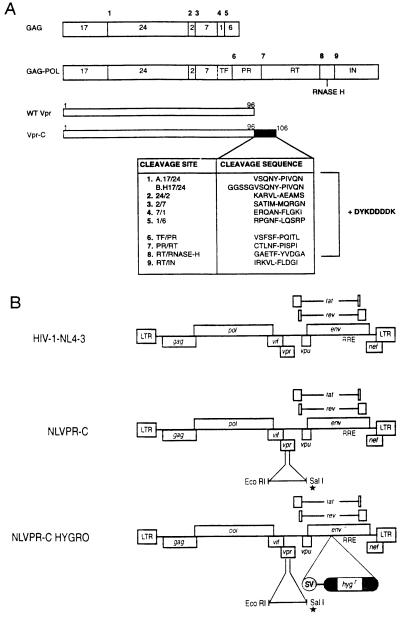
Figure 2.
(A) Schematic representation of the Gag and Gag–Pol precursors of HIV-1 indicating protease cleavage sites 1–9 (bold). The Vpr-C proteins contain the corresponding cleavage signal found in the Gag and Gag–Pol precursors indicated by number and abbreviation for the site, fused in-frame to the C terminus of Vpr resulting in a protein of 106 aa. Likewise, all Vpr-C constructs received a Flag epitope (DYKDDDDK) immediately following the cleavage site. The asterisk indicates the construct containing the Flag epitope alone. The dashed line indicates site of ribosomal frame shift. (B) Proviral clone pNL4–3 cleaved with EcoRI/SalI in Vpr coding region to allow insertion of EcoRI/XhoI generated fragment from Vpr-C (NLVPR-C). Vpr-C-derived fragments contain a stop codon (∗) so sequences downstream of SalI are out of frame. NLVPR-C HYGRO contains an SV-Hygr cassette in the env gene for selection of positive clones in single-round replication assay.
Vpr-C fragments were prepared by excision from the respective expression vector at the EcoRI and XhoI sites for insertion into the proviral DNA, pNL4–3, cleaved at the unique sites EcoRI and SalI (see Fig. 2B). This strategy does not interfere with the overlap region of vif and tat.
In Vitro Transcription/Translation and RIA Analysis of Vpr-C Proteins.
The coupled T7 transcription/translation system (Promega) was used for characterizing the expression of the Vpr-C clones. Incubation conditions were followed according to manufacturer’s instructions.
Radioimmunoprecipitation assay (RIPA) analysis of in vitro translated proteins was carried out using polyclonal antiserum to Vpr as described (35).
Infection/Transfection, Metabolic Labeling, Immunoprecipitation, and Western Blot Analysis.
For expression studies, the recombinant vaccinia virus vTF7-3 that expresses T7 RNA polymerase in infected cells was used. HeLa cells at 106 cells per 35-mm tissue culture dish were first infected with the virus at a multiplicity of infection of 10 for 1 hr and subsequently transfected via the Lipofectin method (Life Technologies, Gaithersburg, MD) with the Vpr-C expression vectors in conjunction with an HIV-1 Gag expression vector for virion incorporation analysis. Transfected cells were washed in PBS and starved in DMEM (without sera, Met and Cys) for a total of 1 hr, followed by labeling with 35S protein labeling mix (DuPont/NEN) at 200 μCi/ml (1.2 Ci/mmol; 1 Ci = 37 GBq) for a total of 5 hr. The culture medium, cleared of cellular debris by low speed centrifugation, was subsequently centrifuged at 25,000 rpm for 90 min, and virus-like-particles were suspended in RIPA buffer. Cells were washed twice in PBS and lysed in RIPA buffer as above. Immunoprecipitation analysis subsequently followed using polyclonal HIV-1 Vpr, Gag, and Flag epitope antibodies, respectively. Immunoprecipitated proteins were separated on 15% SDS/PAGE and immunoblot analysis was carried out as described (Santa Cruz Biotechnology).
Generation of Virus upon Transfection of HIV-1 Proviral DNA and Virus Infectivity Studies.
Both wild-type- and Vpr-C-containing proviral DNA were transfected into rhabdomayosarcoma cells as described (40). Virus particles released into the culture medium were harvested 72–120 hr posttransfection and quantitated by RT and p24 antigen assay (40). The virus infectivity studies were carried out using established CD4+ CEM cells as targets. Three million cells were incubated with virus innoculum, normalized on the basis of RT activity or p24 antigen levels, for 2 hr at 37°C. Infected cells were then washed and resuspended in RPMI 1640 medium. Aliquots from infected cultures were taken once a week and split to keep the cell concentration at one million cells/ml.
Single Cycle Replication Assay.
Using a previously published strategy, proviral DNAs containing Vpr-C were cleaved with the NheI restriction endonuclease, and a simian virus 40 (SV40) early promoter/enhancer hygromycin gene cassette (SV-Hygr) was inserted, leading to the disruption of the env gene (41–43). pED84, which contains the insertion of the SV-Hygr cassette in the env gene of pNL4-3 was used as the control plasmid for transfection experiments and contains a wild-type vpr gene (44). To generate virus particles capable of only a single round of replication, cotransfection of Cos cells was performed with the Vpr-C modified proviral clones (NLVpr-C-HYGRO) and an amphotropic murine leukemia virus (A-MLV) envelope glycoprotein (Env-gp) expression plasmid, pSV-A-MLV-env (see Fig. 2B) (45).
Virus particles released into the medium were harvested 72 hr posttransfection and cleared of cellular debris by low speed centrifugation. An aliquot was used to infect HeLa T4 cells. At 48 hr postinfection, selection of hygromycin resistance was initiated with media containing 200 μg/ml hygromycin B. After 9–11 days, hygromycin-resistant colonies were stained with 0.5% crystal violet in 50% methanol and counted.
RESULTS
Construction of Vpr-C Containing HIV-1 Protease Cleavage Signal Sequences.
The structural proteins Gag and Gag–Pol of HIV-1 are synthesized as precursor polyproteins and contain a total of 12 cleavage sites recognized by the virus-encoded protease allowing for precursor processing and virus maturation (46). The specificity of HIV-1 protease is found to lie in the detection and cleavage of a scissile bond (Met-Met, Leu-Ala/Phe, Tyr-Pro, Phe-Pro/Tyr/Leu) within the minimum context of four residues 5′ (P1–P4) and three residues 3′ (P1′–P3′) to the site of cleavage (2, 47–49). In this study, nine major sites for generating Vpr-C were selected. Primers that comprise sequences corresponding to the nine cleavage sites, followed by a termination codon, were synthesized to allow for the addition of sequences at the C terminus of Vpr by PCR (Fig. 1). In addition, following each of the cleavage sites, an 8 aa Flag epitope was added to aid in antibody detection of the chimeric proteins (23). As a control, we have generated a Vpr-C containing only a Flag epitope at the C terminus. The proviral clone pNL4-3 was used as the template for generating replication-competent proviral clones containing the Vpr-C. The details of cleavage site residues used for generating Vpr-C are presented in Fig. 2A. The designation of Vpr-C is indicated by the abbreviation for the respective protease cleavage site. The cleavage signals added onto our Vpr-C proteins include the P5 and P5′ residues specific to each of the respective sites, which includes the minimum protease recognition site. The Vpr-C clones were verified for sequence integrity.
Expression and Virion Incorporation of Vpr-C.
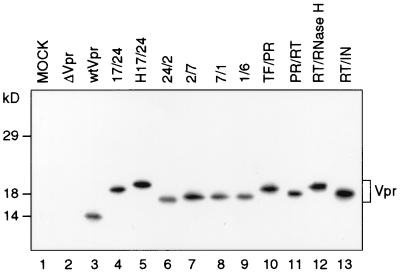
We used an in vitro T7 expression system to verify the expression of each of the Vpr-C proteins. In vitro translated Vpr-C proteins were immunoprecipitated with polyclonal Vpr antiserum. As expected, each Vpr-C protein was expressed at levels equal to the wild-type Vpr protein (Fig. 3). As reported earlier, the wild-type Vpr migrates to 14 kDa (32). The different Vpr-C proteins displayed altered mobility in comparison to wild-type Vpr. This may be due to the added signal sequences containing highly acidic and hydrophobic residues. The Vpr-C Flag protein (Vpr-F) with the Flag addition, made mostly of acidic residues, also migrated differently than wild-type Vpr (data not shown).
Figure 3.
RIPA analysis of in vitro transcribed and translated Vpr-C proteins. Antiserum to Vpr was used as described. Vpr-C proteins exhibit a shift in migration and appear around 18 kDa. Designation of the Vpr constructs are described in legend to Fig. 2.
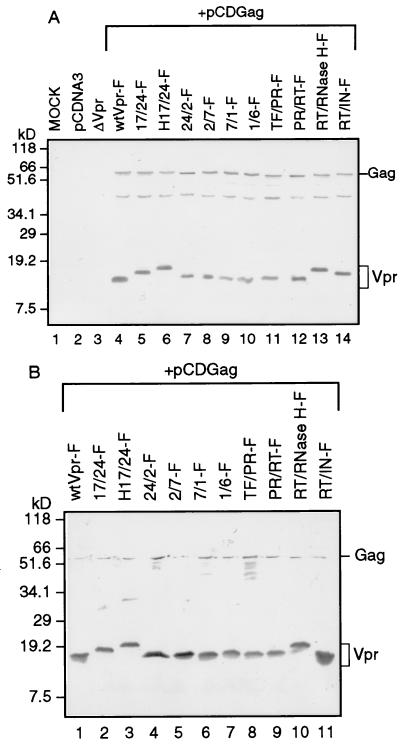
To verify that the Vpr-C proteins retain the ability to incorporate into virus-like particles directed by HIV-1 Gag and to monitor the expression of Vpr-C in cells, we employed a vaccinia virus T7 RNA polymerase expression system (vTF7-3). vTF7-3-infected HeLa cells were transfected with wild-type Vpr or Vpr-C expression plasmids containing the Flag epitope (F) in combination with the Gag expression vector pCDGag by the Lipofectin method. Immunoblot analysis of Vpr-F, Vpr-C-F, and Gag was performed in both cell lysates and culture media with anti-Flag and anti-Gag antiserum 24 hr posttransfection (Fig. 4). Results of the cell lysate showed the presence of both Gag and Vpr (Fig. 4A). The culture medium immunoblot indicated that the Vpr-C proteins are incorporated into virus-like particles (Fig. 4B). As expected, the mock, control plasmid pCDNA-3 and pCDVprΔ cells failed to show a corresponding protein.
Figure 4.
Immunoblot analysis of HeLa cells transfected with Vpr-C constructs. (A) Analysis of cell lysates showed that Vpr-C and Gag proteins are produced and migrate to the expected 18-kDa and 55-kDa positions, respectively. (B) Analysis of cell supernatants revealed that the Vpr-C retains its ability to be incorporated into virus-like particles.
Effect of Vpr-C in HIV-1 Infection of CEM Cells.
Upon characterization of Vpr-C constructs, the Vpr-C protein coding sequences were introduced into HIV-1-NL43 proviral DNA (NLVpr-C) (Fig. 2B). To avoid disruption of the overlap of vpr with tat at the 3′ end in the NLVpr-C construct, the unique EcoRI and SalI restriction endonuclease cleavage sites were used to introduce the 3′ end of chimeric Vpr from the recombinant plasmids.
The proviral DNA NL4-3 and NL-Vpr-C were transfected into rhabdomayosarcoma cells to generate viruses for evaluating the effect of Vpr-C at the level of viral replication. Virus released into the medium was collected 5 days posttransfection and quantitated by an RT assay (40). Equivalent RT activity-containing virus particles were added to CEM cultures, and infected cells were monitored for virus replication for nearly 30 days. Cultures infected with wild-type pNL4-3 showed peak virus production at 15–21 days postinfection, as is generally observed with a spreading infection. Similar results were noted using virus derived from NLVpr-F (data not shown). The replication of the virus particles derived from proviral DNA containing NL-Vpr-C was altered in comparison to the control (Table 1). The chimeric viruses exhibited a delayed kinetics of virus spread and very little replication was evident for up to 14 days, suggesting the presence of a mixture of infectious and noninfectious virus particles. Strikingly, the NLVpr-24/2 virus showed no viral replication for up to 28 days. In general, the infectivity assays carried out in CEM cells represent a spreading infection and the results generated may reflect a cumulative effect over multiple rounds of infection.
Table 1.
Effect of Vpr-C on virus replication
| Virus derived from designated proviral DNA | RT activity in culture supernatant, cpm/μl
|
||
|---|---|---|---|
| 14 days after infection | 21 days after infection | 28 days after infection | |
| pNL43* | 1082 | 1678 | 1380 |
| NLVpr 1/6 | 49 | 219 | 828 |
| NLVpr 24/2 | 0 | 67 | 0 |
| NLVpr 2/7 | 287 | 1883 | 1755 |
| NLVpr PR/RT | 29 | 2845 | 2450 |
| NLVpr 17/24 | 28 | 2133 | 1366 |
| NLVpr RT/RNase | 193 | 2116 | 1248 |
| NLVpr 7/1 | 299 | 1965 | 2132 |
| NLVpr TF/PR | 681 | 652 | 1016 |
The replication pattern of virus derived from NLVPR-F was similar to pNL4-3.
Utilization of Single-Round Replication Assay to Evaluate the Effect of Vpr-C.
To precisely evaluate the effect of Vpr-C in a single cycle of virus replication, a single-round replication assay was established as described (41–44). For this purpose, NL4-3 and NLVpr-C proviral DNA were cleaved at the NheI restriction enzyme cleavage site where a hygromycin gene cassette (SV-Hygr) under the control of the SV40 early promoter was inserted (NLVpr-C HYGRO) (Fig. 2B) Because the SV-Hygr cassette was inserted in the env gene leading to the disruption of Env expression, virus particles containing amphotropic envelope were generated by trans-complementation using the MLV envelope expression plasmid by cotransfection.
The virus particles present in the supernatant were used to infect HeLa T4 cells and the infected cells were selected in media containing hygromycin B as described (41–44). The number of Hygr-resistant colonies in the presence of the antibiotic reflects the ability of a pseudotyped particle to infect and integrate into the cellular genome. The results are presented in Table 2 and correlate well with the multiple rounds of replication assay.
Table 2.
Effect of Vpr-C in a single round of replication
| Proviral Vpr clones | Titers CFU/ml* | % (+) inhibition† | % (+) up-regulation† |
|---|---|---|---|
| ED84‡ | 1536 | ||
| NL 1/6 | 1088 | 30 | |
| NL 2/7 | 928 | 40 | |
| NL H 17/24 | 1088 | 30 | |
| NL PR/RT | 2608 | 169 | |
| NL TF/PR | 1072 | 31 | |
| NL 17/24 | 1248 | 29 | |
| NL 7/1 | 1936 | 126 | |
| NL 24/2 | 0 | 100 |
No Hygr colonies were observed for any of the proviral clones when the trans-complementation was performed without pSV-A-MLV-env, pSV-A-MLV-env by itself, or mock transfected.
Extent of inhibition and upregulation was calculated in comparison to pED84 control proviral DNA.
The virus derived from NLVPR-F proviral DNA showed replication results similar to virus from pED84.
In an attempt to increase the efficiency of Vpr-C as pseudosubstrates for protease, we have introduced amino acids constituting a flexible hinge region (Gly-Gly-Ser-Ser-Gly) immediately 5′ to the cleavage signal of the 17/24 construct (Fig. 2A). This construct was chosen to receive a hinge based on its weak performance and from previous reports that the Tyr-Pro scissile bond it contains acts as a late site for cleavage making it one of the less efficient sites for protease cleavage (50). Results generated from the H17/24 chimera indicate an enhancement of the inhibitory affect in the single round infection assay; however, it still lacks the total inhibition seen with Vpr-24/2.
DISCUSSION
Currently, there are several drugs that have been approved by the Federal Drug Administration to treat HIV-1-infected individuals, all of which either target the viral RT or protease enzymatic activities of the virus. The continued treatment of virus-infected individuals with these drugs has led to the identification of viruses that exhibit partial to full resistance to treatment as a result of specific changes in the target enzymes (51). In the absence of a successful vaccine to prevent HIV-1 infection, various alternative approaches have been proposed and are being actively investigated (8–11). These include the capsid fusion approach where a toxic gene product can be fused to the capsid protein for inactivating the virion components, chimeric receptor molecules targeted to Env, and the use of trans-dominant mutants targeting Gag, Rev, Tat, and Env for inactivating the virus at different stages of the life cycle (52–57).
The virion association of nonstructural proteins encoded by HIV-1 provides a unique opportunity to attack the virus particle in trans, and are advantageous over structural protein based antiviral approaches (52–54, 57). Along these lines, Kappes and coworkers (58, 59) have generated chimeric proteins based on HIV-1 Vpr and HIV-2 Vpx utilizing staphylococcal nuclease and wild-type and mutated HIV-1 protease fused in-frame to these proteins. In our studies, we have generated a chimeric protein based on Vpr utilizing the conserved protease cleavage site sequences from the Gag and Gag–Pol precursor polyproteins as a fusion partner. These sequences are efficiently cleaved by HIV-1 protease when presented as peptide substrates (46–49, 60). The interesting features of the chimeric proteins generated here are the minimal addition of residues (10 residues), no toxicity due to added sequences, and the likelihood of Vpr-C to behave like the wild-type Vpr protein due to the minimal increase in size. The strategy outlined here is novel in that it brings the chimeric protein closer to the target protein in the virus particle. It is likely that the ability of Vpr-C to serve as a pseudosubstrate for HIV-1 protease can lead to the exhaustion of protease activity. This would, in essence, reduce the effectiveness of protease to participate in the maturation of the virus particle by not acting on the bona fide viral precursor proteins.
The results generated with Vpr-C showed that the chimeric protein retains the ability to get incorporated into the virus particle. In the context of the proviral DNA, the Vpr-C suppressed HIV-1 replication both in multiple- and single-round replication assays. As the single-round replication assay involves the establishment of cells resistant to hygromycin, one concern was whether the wild-type Vpr or Vpr-C would complicate the results due to cell cycle arrest. This does not seem to be the case due to the positioning of the selectable marker (hygromycin gene) which is under the control of the SV40 promoter. The concordance between the results from the single- and multiple-round replication assays also argues against such a possibility. In addition, further support for such an interpretation is seen with the variable replication results observed with the different Vpr-C-containing viruses.
Considering the stability of chimeric vpr sequences in the viral genome over several rounds of replication (data not shown),the kinetics of the replication pattern suggests that the viral population may contain a mixture of replication competent and defective viruses. Surprisingly, we noted that some Vpr-C proteins did not have any influence on viral replication and some even showed an up-regulation of viral replication (Tables 1 and 2). Complete inhibition was observed only with the Vpr chimera 24/2 and moderate inhibition was observed with Vpr chimeras 1/6, 2/7, 17/24, and trans-frame protease (TF/PR). The cleavage of the 24/2 site is important in that it serves as a regulator for the sequential processing of the Gag precursor, and is unique in that it is the only site of the nine recognized by the protease to have a glutamic acid at the P2′ position (61).
The inhibition of viral replication observed with viruses containing Vpr-C may be due to the ability of Vpr-C to overwhelm the protease activity. Biochemical analysis of virus particles with respect to the status of the precursor proteins is likely to provide information regarding the mechanism of inhibition. Earlier biochemical studies showed that 2,500 Gag and ≈5–10% of Gag–Pol molecules in relation to Gag are present in each virion (1, 31). Though the exact number of Vpr molecules present in the virus particle has not been determined for HIV-1, studies on HIV-2 indicate that Vpx, a protein related to Vpr, is present in equimolar concentration to that of p28 in virus particles (62). Such a scenario in HIV-1 is likely to lead to the presence of an enormous number of Vpr-C pseudosubstrates for protease to act on within the virus particle and may interfere with the processing of the authentic viral precursor proteins. It has been shown that partial inhibition of Gag and Gag–Pol processing results in aberrantly assembled viruses (2). The variable inhibition we observed regarding viral replication suggests that the ability of Vpr-C to serve as a pseudosubstrate for protease may vary as observed with oligopeptide substrates corresponding to cleavage sites (46, 47, 50, 60, 63). Furthermore, previous work on protease cleavage in the context of corresponding peptides and protein precursors has revealed the importance of either residues or conformational determinants within the Gag and Gag–Pol precursors that affect the order of cleavage and the actual cleavage event of the target substrates (50, 61, 64, 65). Because the cleavage signals in Vpr-C are presented out of the context of the Gag precursors, the absence of both up and downstream determinants may have prevented Vpr-C from being a substrate and competitor for the protease cleavage site. This would ultimately lead to the lack of influence on viral maturation and infectivity.
The threshold level of protease required for maturation of the virus particle is not known. Studies involving protease inhibitors have shown that the enzyme needs only a 50-fold reduction in activity whereas a 25-fold reduction still allows for processing and subsequently the production of infectious virus (66). It is plausible that the constructs showing no effect did not offer enough competition to prevent processing from occurring. Likewise the partial inhibitory constructs may not have reached the 50-fold reduction threshold, but went farther than those chimeras with no apparent effect. The high level of replication observed with virus derived from certain Vpr-C containing proviral DNA is intriguing. Because equal amounts of virus were used as innoculum to infect CEM cells, it is likely that some Vpr-C pseudosubstrates enhanced the level of virus production by activating protease. Alternatively, the increased protease activity may act on an as yet unidentified step in viral replication.
We have shown that a novel class of HIV-1 agents can be generated with the desirable end result of eliminating virus infection. This class of agents, which we have termed “anti-HIV agents from within”, is conceptually unique, and the studies carried out with the virus derived from proviral DNA containing Vpr-C provided evidence in support of this strategy. The disadvantage in the use of Vpr as a fusion partner for generating a chimeric protein is its ability to arrest cells at the G2 stage of the cell cycle. In this regard, work from our laboratory and others has demonstrated that the cell cycle arrest can be abolished by introducing changes at the C terminus of Vpr, and yet still retain virion incorporation (ref. 22 and unpublished data). This modified Vpr may enable the generation of chimeric proteins for use with gene therapy approaches. In addition, the Vpr-C proteins will also prove to be useful for dissecting the steps involved in virus maturation.
Acknowledgments
We would like to express our thanks to Dr. Antiono Panganiban (University of Wisconsin, Madison) for the gift of pED84. pSV-A-MLV-env was obtained from the AIDS Research and Reference Reagent Program of the National Institutes of Health. This work was supported by funds from the National Institutes of Health (AI29306) (A.S.), a grant from the Commonwealth of Pennsylvania to the Biotechnology Foundation, Inc., and a grant from the Biomedical Research Support Committee and institutional funds (T.A.R.).
ABBREVIATIONS
- Vpr-C
chimeric Vpr
- RT
reverse transcriptase
- RIPA
radioimmunoprecipitation assay
- SV40
simian virus 40
- SV-Hygr
SV40 hygromycin gene cassette
- MLV
murine leukemia virus
References
- 1.Levy J A. Microbiol Rev. 1993;57:183–289. doi: 10.1128/mr.57.1.183-289.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Debouck C. AIDS Res Hum Retroviruses. 1992;8:153–164. doi: 10.1089/aid.1992.8.153. [DOI] [PubMed] [Google Scholar]
- 3.Ridky T, Leis J. J Biol Chem. 1995;270:29621–29623. doi: 10.1074/jbc.270.50.29621. [DOI] [PubMed] [Google Scholar]
- 4.Miller R H, Turk S R, Black R J, Bridges S, Sarver N. AIDS Res Hum Retroviruses. 1996;12:859–864. doi: 10.1089/aid.1996.12.859. [DOI] [PubMed] [Google Scholar]
- 5.Mellors J M. Nat Med. 1996;2:274–295. doi: 10.1038/nm0396-274. [DOI] [PubMed] [Google Scholar]
- 6.Vella, S. (1995) J. Acquired Immune Defic. Syndr. Hum. Retrovirol. 10, Suppl. 1, S58–S61. [PubMed]
- 7.Crowe S, Cooper D A, Chambers D E. MJA. 1996;164:290–295. [PubMed] [Google Scholar]
- 8.Collier A C, Coombs R W, Schoenfield D A, Bassett R L, Timpoone J, Baruch A, Jones M, Facey K, Whitacre C, McAuliffe V J, Friedman H M, Merigan T C, Reichman R C, Hooper C, Corey L. New Engl J Med. 1996;334:1011–1017. doi: 10.1056/NEJM199604183341602. [DOI] [PubMed] [Google Scholar]
- 9.Larder B A, Kohli A, Bloor S, Kemp S D, Harrigan P R, Schooley R T, Lange J M A, Pennington K N, St. Clair M H the Protocol 34,225–02 Collaborative Group. J Virol. 1996;70:5922–5929. doi: 10.1128/jvi.70.9.5922-5929.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jablonowski, H. (1995) J. Acquired Immune Defic. Syndr. Hum. Retrovirol. 10, Suppl. 1, S52–S56. [PubMed]
- 11.Larder, B. A. (1995) J. Acquired Immune Defic. Syndr. Hum. Retrovirol. 10, Suppl. 1, S28–S33. [PubMed]
- 12.Cohen E A, Dehni G, Sodroski J G, Haseltine W A. J Virol. 1990;64:3097–3099. doi: 10.1128/jvi.64.6.3097-3099.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu H, Wu X, Newman M, Shaw G M, Hahn B, Kappes J C. J Virol. 1995;699:7630–7638. doi: 10.1128/jvi.69.12.7630-7638.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Camaur D, Trono D. J Virol. 1996;70:6106–6111. doi: 10.1128/jvi.70.9.6106-6111.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Welker R, Kottler H, Kalbitzer H R, Kräusslich H-G. Virology. 1996;219:228–236. doi: 10.1006/viro.1996.0240. [DOI] [PubMed] [Google Scholar]
- 16.Pandori M W, Fitch N J S, Craig H M, Richman D D, Spina C A, Guatelli J C. J Virol. 1996;70:4283–4290. doi: 10.1128/jvi.70.7.4283-4290.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Myers G, Korber B, Wain-Hobson S, Smith R F. Human Retroviruses and AIDS. Los Alamos, NM: Los Alamos National Lab.; 1993. [Google Scholar]
- 18.Lu Y L, Spearman P, Ratner L. J Virol. 1993;67:6542–6550. doi: 10.1128/jvi.67.11.6542-6550.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Re F, Braten D, Frank E K, Luban J. J Virol. 1995;69:6859–6864. doi: 10.1128/jvi.69.11.6859-6864.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jowett J B, Planelles V, Poon B, Shah N P, Chen M, Chen I S. J Virol. 1995;69:6304–6313. doi: 10.1128/jvi.69.10.6304-6313.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rogel M E, Wu L I, Emmerman M. J Virol. 1995;69:882–888. doi: 10.1128/jvi.69.2.882-888.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He J, Choe S, Walker R, DiMarzio P, Morgan D O, Landau N. J Virol. 1995;69:6705–6711. doi: 10.1128/jvi.69.11.6705-6711.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao L J, Wang L, Mukherjee S, Narayan O. J Biol Chem. 1994;269:32131–32137. [PubMed] [Google Scholar]
- 24.Lu Y L, Spearman P, Ratner L. J Virol. 1993;67:6542–6550. doi: 10.1128/jvi.67.11.6542-6550.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heinzinger N K, Bukrinsky M I, Haggerty S A, Ragland A M, Kewalramani V, Lee M A, Gendelman H E, Ratner L, Stevenson M, Emmerman M. Proc Natl Acad Sci USA. 1994;91:7311–7315. doi: 10.1073/pnas.91.15.7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Piller S C, Ewart G D, Premkumar A, Cox G B, Gage P W. Proc Natl Acad Sci USA. 1996;93:111–115. doi: 10.1073/pnas.93.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bouhamdan M, Benichou S, Rey F, Navarro J M, Agostini I, Spore B, Camonis J, Slupphaug G, Vigne R, Benarous R, Sire J. J Virol. 1996;70:697–704. doi: 10.1128/jvi.70.2.697-704.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Refaeli Y, Levy D N, Weiner D B. Proc Natl Acad Sci USA. 1995;92:3621–3625. doi: 10.1073/pnas.92.8.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Balliet J W, Kolson D L, Eiger G, Kim F M, McGann K A, Srinivasan A, Collman R. Virology. 1994;200:623–631. doi: 10.1006/viro.1994.1225. [DOI] [PubMed] [Google Scholar]
- 30.Westervelt P, Henkel T, Trowbridge D B, Orenstein J, Heuser J, Gendelman H E, Ratner L. J Virol. 1992;68:6161–6169. doi: 10.1128/jvi.66.6.3925-3931.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mahalingam S, Khan S A, Murali R, Jabbar M A, Monken C E, Collman R G, Srinivasan A. Proc Natl Acad Sci USA. 1995;92:3794–3798. doi: 10.1073/pnas.92.9.3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mahalingam S, Khan S A, Jabbar M A, Monken C E, Collman R G, Srinivasan A. Virology. 1995;207:297–302. doi: 10.1006/viro.1995.1081. [DOI] [PubMed] [Google Scholar]
- 33.Mahalingam, Collman R G, S, Patel M, Monken C E, Srinivasan A. Virology. 1995;210:495–500. doi: 10.1006/viro.1995.1368. [DOI] [PubMed] [Google Scholar]
- 34.Mahalingam, Collman R G, S, Patel M, Monken C E, Srinivasan A. Virology. 1995;212:331–339. doi: 10.1006/viro.1995.1490. [DOI] [PubMed] [Google Scholar]
- 35.Mahalingam, Patel M, Collman R G, Srinivasan A. Virology. 1995;214:647–652. doi: 10.1006/viro.1995.0079. [DOI] [PubMed] [Google Scholar]
- 36.Yao X J, Subbramanian R A, Rougeau N, Boisvert F, Bergeron D, Cohen E A. J Virol. 1995;69:7032–7044. doi: 10.1128/jvi.69.11.7032-7044.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paxton W, Connor R I, Landau N. J Virol. 1993;67:7229–7237. doi: 10.1128/jvi.67.12.7229-7237.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Di Marzio P, Choe S, Ebright M, Knoblauch R, Landau N. J Virol. 1995;69:7909–7916. doi: 10.1128/jvi.69.12.7909-7916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hunter E. Semin Virol. 1994;5:71–83. [Google Scholar]
- 40.Nagashunmugam T, Velpandi A, Goldsmith C S, Zaki S R, Kalyanaraman V S, Srinivasan A. Proc Natl Acad Sci USA. 1992;89:4114–4118. doi: 10.1073/pnas.89.9.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rizvi T A, Panganiban A. AIDS Res Hum Retrovirus. 1992;8:89–95. doi: 10.1089/aid.1992.8.89. [DOI] [PubMed] [Google Scholar]
- 42.Rizvi T A, Schmidt R D, Lew K, Keeling M E. Virology. 1996;222:457–463. doi: 10.1006/viro.1996.0444. [DOI] [PubMed] [Google Scholar]
- 43.Rizvi T A, Lew K A, Murphy E C, Schmidt R D. Virology. 1996;224:517–532. doi: 10.1006/viro.1996.0558. [DOI] [PubMed] [Google Scholar]
- 44.Delwart E L, Buchschacher G L, Jr, Freed E O, Panganiban A T. AIDS Res Hum Retrovirus. 1992;8:1669–1677. doi: 10.1089/aid.1992.8.1669. [DOI] [PubMed] [Google Scholar]
- 45.Page K A, Landau N R, Littman D R. J Virol. 1990;64:5270–5276. doi: 10.1128/jvi.64.11.5270-5276.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tözsér J, Bláha I, Copeland T D, Wondrak E M, Oroszlan S. FEBS Lett. 1991;281:77–80. doi: 10.1016/0014-5793(91)80362-7. [DOI] [PubMed] [Google Scholar]
- 47.Darke P L, Nutt R F, Brady S F, Garsky V M, Ciccarone T M, Leu C, Lumma P K, Freidinger R M, Veber D F, Sigal I S. Biochem Biophys Res Commun. 1988;156:297–303. doi: 10.1016/s0006-291x(88)80839-8. [DOI] [PubMed] [Google Scholar]
- 48.Billich S, Knoop M T, Hansen J, Strop P, Sedlacek J, Mertz R, Moelling K. J Biol Chem. 1988;263:17905–17908. [PubMed] [Google Scholar]
- 49.Moore M L, Btyan W M, Fakhoury S A, Magaard V W, Huffman W F, Dayton B D, Meek T D, Hyland L, Dreyer G B, Metcalf B W, Strickler J E, Gorniak J G, Debouck C. Biochem Biophys Res Commun. 1989;159:420–425. doi: 10.1016/0006-291x(89)90008-9. [DOI] [PubMed] [Google Scholar]
- 50.Tritch R J M, Cheng Y S, Yin F H, Erickson-Viitanen S. J Virol. 1991;65:922–930. doi: 10.1128/jvi.65.2.922-930.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Katz R A, Skalka A M. Annu Rev Biochem. 1994;63:133–173. doi: 10.1146/annurev.bi.63.070194.001025. [DOI] [PubMed] [Google Scholar]
- 52.Natsoulis G, Boeke J D. Nature (London) 1991;352:632–635. doi: 10.1038/352632a0. [DOI] [PubMed] [Google Scholar]
- 53.Pastan I, Chaudary V, Fitzgerald D J. Annu Rev Biochem. 1992;61:331–354. doi: 10.1146/annurev.bi.61.070192.001555. [DOI] [PubMed] [Google Scholar]
- 54.Trono D, Feinberg M B, Baltimore D. Cell. 1989;59:113–120. doi: 10.1016/0092-8674(89)90874-x. [DOI] [PubMed] [Google Scholar]
- 55.Malim M H, Freimuth W W, Liu J, Boyle T J, Lyerly H K, Cullen B R, Nabel G J. J Exp Med. 1992;76:1197–1201. doi: 10.1084/jem.176.4.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pearson L, Garcia J, Wu F, Modesti N, Nelson J, Gaynor R. Proc Natl Acad Sci USA. 1990;85:5079–5083. doi: 10.1073/pnas.87.13.5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Freed E O, Delwart E L, Buchschacher G L, Jr, Panganiban A T. Proc Natl Acad Sci USA. 1992;89:70–74. doi: 10.1073/pnas.89.1.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu X, Liu H, Xiao H, Kim J, Seshaiah S, Natsoulis G, Boeke J D, Hahn B H, Kappes J C. J Virol. 1995;69:3389–3398. doi: 10.1128/jvi.69.6.3389-3398.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wu X, Liu H, Xiao, Conway J A, Kappes J C. J Virol. 1996;70:3378–3384. doi: 10.1128/jvi.70.6.3378-3384.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cameron C E, Grinde B, Jentoft J, Leis J, Weber I T, Copeland T D, Wlodawer A. J Biol Chem. 1992;267:23735–23741. [PubMed] [Google Scholar]
- 61.Pettit S C, Moody M D, Wehbie R S, Kaplan A H, Nantermet P V, Klein C A, Swanstrom R. J Virol. 1994;68:8017–8027. doi: 10.1128/jvi.68.12.8017-8027.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Henderson L E, Sowder T D, Copeland R E, Beneveniste R E, Oroszlan S. Science. 1988;241:199–201. doi: 10.1126/science.3388031. [DOI] [PubMed] [Google Scholar]
- 63.LeGrice S F J, Ette R, Mills J, Mous J. J Biol Chem. 1989;264:14902–149008. [PubMed] [Google Scholar]
- 64.Partin K, Zybarth G, Ehrlich L, DeCrombrugghe M, Wimmer E, Carter C. Proc Natl Acad Sci USA. 1991;88:4776–4780. doi: 10.1073/pnas.88.11.4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sheng N, Erickson-Viitanen S. J Virol. 1994;68:6207–6214. doi: 10.1128/jvi.68.10.6207-6214.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rosé J R, Babé, Lilia M, Craik C S. J Virol. 1995;69:2751–2758. doi: 10.1128/jvi.69.5.2751-2758.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]