Abstract
A guanylyl cyclase (GC-D) was recently shown to be expressed in a subclass of neurons within the neuroepithelim of the rat, but given that only a single cyclase was discovered, whether it represents an odorant/pheromone receptor as has been suggested for the large family of seven-transmembrane receptors remains unclear. Through cloning and expression of cDNA we now demonstrate that at least 29 genomic or cDNA sequences found in Caenorhabditis elegans represent guanylyl cyclases. Many of the membrane forms retain cysteine residues conserved within the extracellular, ligand-binding domain of known cyclase receptors. Of eight orphan cyclase receptor::GFP (green fluroescence protein) fusion constructs for which signals were obtained, all were expressed in specific sensory neurons. Furthermore, a cyclase/GFP fusion protein (GCY-10/GFP) was principally expressed in the sensory cilium, suggesting these cyclases function as primary chemosensory receptors. For the first time, we also found that chemosensory neurons (ASE), known to be bilaterally symmetric, demonstrate absolute right or left sidedness with respect to the expression of three different cyclases. Thus, the guanylyl cyclases represent an unexpectedly large and new family of sensory neuron receptors that may complement the 7-transmembrane family of odorant/pheromone receptors.
Until the diacetyl receptor was discovered using a genetic approach in Caenorhabditis elegans (1), the major evidence that a family of seven-transmembrane proteins were chemosensory receptors was a large number of different gene products being expressed in distinct olfactory neurons (2–4). With respect to pheromone/odorant receptor candidates, other than the seven-transmembrane type, none have been found, although of the six plasma membrane forms of guanylyl cyclase found in mammals (5, 6), one (GC-D) is located within the neuroepithelium of olfactory tissue and is expressed in a punctate pattern in a broad medial zone of the rat nose, a pattern of expression similar to the seven-transmembrane, chemosensory/odorant receptors (5). Recently, the sensory neurons expressing GC-D have been shown to be unique and to project to atypical glomeruli within the olfactory bulb, suggesting that GC-D is a pheromone receptor (7).
Initial searches of the genome database of C. elegans (8) suggested a large family of guanylyl cyclases (now as many as 29). Given the daunting task of determining whether or not the mammalian olfactory guanylyl cyclase is an odorant/pheromone receptor, and whether the cyclase family might contain other members within specific neurons, we designed experiments to determine whether the genomic sequences of C. elegans encode proteins that possess guanylyl cyclase activity, whether specific expression of these cyclases occurs in distinct sensory neurons, akin to the seven-transmembrane receptors, and whether the site of expression within a sensory neuron is consistent with that of a chemosensory/pheromone receptor. We demonstrate that cDNA representing apparent genes for guanylyl cyclase in C. elegans, in fact, encodes guanylyl cyclase activity and that mRNA for the various cyclases is expressed in specific and different sensory neurons (the differential expression of three guanylyl cyclases demonstrated that neurons which detect chemoattractant signals are not symmetric), and that a cyclase protein is localized to the tip of the nose. The expression of a moderately large family of guanylyl cyclase receptors within distinct sensory neurons replicates the criteria first used to define a family of seven-transmembrane proteins as pheromone/odorant receptors.
MATERIALS AND METHODS
Genome Database Search and Sequence Analysis.
We initially used the conserved catalytic domain of mammalian guanylyl cyclases to search GenBank and identified six C. elegans guanylyl cyclases; they were gcy-1 to gcy-5 and gcy-12. We subsequently used the catalytic domains of the worm guanylyl cyclases to identify the remainder of the guanylyl cyclases by tblastn program in the National Center for Biotechnology Information. Nucleotide sequences of the cosmids encoding putative guanylyl cyclases were obtained from Sanger Center Network. All putative clones (named gcy-xn in Table 1) contain the KVET/S and PRYCLF conserved motif within the catalytic domain.
Table 1.
Summary of putative guanylyl cyclase genes in C. elegans
| Genes | Predicted coding regions | Chromosomal locations with flanking markers |
|---|---|---|
| gcy-1 | AH6.1 | II, kin-15, spe-2 |
| gcy-2 | R134.2 | II, kin-15, spe-2 |
| gcy-3 | R134.1 | II, kin-15, spe-2 |
| gcy-4 | ZK970.5 | II, gpd-4, gpd-1 |
| gcy-5 | ZK970.6 | II, gpd-4, gpd-1 |
| gcy-6 | B0024.6 | V, msp-72ps, col-12 |
| gcy-7 | F52E1.4 | V, gpa-2, his-23 |
| gcy-8 | C49H3.2 | IV, col-4, vit-6 |
| gcy-9 | ZK455.2 | V, egl-15, hum-4 |
| gcy-10 | R01E6.1 | X, unc-9, col-9 |
| gcy-11 | C30G4.3 | X, mec-4, mlc-1 |
| gcy-12 | F08B1.2 | II, msp-45, vhp-1 |
| gcy-13 | F23H12.6 | V, col-37, sel-1 |
| gcy-14 | ZC412.2 | V, cyp-1, sdc-3 |
| gcy-31* | T07D1.1 | X, lin-32, fox-1 |
| gcy-32* | C06B3.8 | V, ceh-24, him-5 |
| gcy-33* | F57F5.2 | V, osm-6, col-37 |
| gcy-x1†‡†‡ | cDNA | I, unc-73, unc-38 |
| gcy-x2† | C17F4 | II, vet-8, lin-31 |
| gcy-x3† | ZC239 | II, vet-8, lin-31 |
| gcy-x4† | F22E5 | II, cya-2, vet-7 |
| gcy-x5† | F21H7 | V, cyb-2, rrs-1 |
| gcy-x6† | W03F11 | I, gsa-1, unc-73 |
| gcy-x7† | T26C12 | IV, lin-22, osm-9 |
| gcy-x8† | C06A12 | IV, hsp-1 |
| gcy-x9† | T04D3 | I, vet-6, TCbn2 |
| gcy-x10† | M04G12 | V, eat-6, gut-2 |
| gcy-x11† | C04H5 | II, rsn-3, unc-52 |
| gcy-x12† | ZK896 | IV, unc-31, unc-30 |
The gene nomenclatures of different guanylyl cyclases are assigned roughly in their order of discovery in the genome sequencing project.
Putative soluble guanylyl cyclases.
gcy-x’s have not received a gene nomenclature.
A cDNA clone that has not been identified in the genome sequencing project (E.B., Arora, V., S.Y., Wedel, B., and D.L.G., unpublished data).
cDNA Cloning and Expression of gcy-12.
A mixed staged nematode Uni-ZAP XR cDNA library was purchased from Stratagene, and a plasmid library of approximately 1 million clones was generated. A 350-bp cDNA fragment close to the N terminus of gcy-12 was obtained by PCR, and it was used as a probe to screen the cDNA library by plaque hybridization. gcy-12 cDNA was subcloned into pCMV5 expression vector and transfected into COS-M6 cells by DEAE-dextran method (9). Guanylyl cyclase assays were carried out in a volume of 100 μl, which contained 30–40 μg of membrane proteins, 1% Triton X-100, 1 mM NaN3, 100 mM NaCl, 0.2 mM 3-isobutyl-1-methylxanthine, 3.5 mM MnCl2, 100 μM GTP, and about 1 μCi [α-32P]GTP (NEN; 3000 Ci/mmol, 10 mCi/ml; 1 Ci = 37 GBq). The reactions were incubated at different temperatures for 15 min and stopped by the addition of 0.5 ml ZnAc (110 mM) and NaCO3 (110 mM). cGMP from each reaction was purified on alumina columns, and labeled cyclic nucleotides were quantified by scintillation counting as described (10).
Construction of Promoter::GFP (Green Fluroescence Protein) Transgenes.
For the transcription fusions, promoters of different guanylyl cyclases were amplified by PCR from C. elegans genomic DNA or from cosmids obtained from the Sanger Center. The 3′ PCR primers were designed such that the first two amino acids of the predicted coding region were included. A restriction site (generally PstI or BamHI) was introduced in-frame with the coding region. The 5′ PCR primers were designed to anneal to a sequence ≈4 kb upstream of the predicted coding region for gcy-12; 3 kb for gcy-5; ≈2 kb for gcy-6, gcy-8, and gcy-10; 1.2 kb for gcy-7; 1 kb for gcy-33; and 700 bp for gcy-32. Shorter promoters were used for gcy-6, -7, -8, -10, -32, and -33 and because of predicted genes upstream. Germ-line transformation was by microinjection of a lin-15 rescue plasmid with each of the above promoter::GFP constructs into lin-15(n765ts) nematodes (11–14). Specific GFP signals were not observed in transgenic animals for gcy-1::GFP, gcy-4::GFP, gcy-31::GFP, gcy-14::GFP, gcy-9::GFP, or gcy-13::GFP. The absence of GFP signals could be a result of wrong promoter region selection or of very low levels of expression.
To make the translation fusion of gcy-10, a 5.6-kb ClaI/BglII genomic fragment of gcy-10 was subcloned from cosmid R01E6 into plasmid pPD 95.75 containing the promoter of gcy-10. The resulting construct removed the last two amino acids from the carboxyl terminus of gcy-10.
Identification of Neurons Expressing GFP.
Pictures of the sites of GFP expressions were taken under fluorescence images. Identification of cells expressing GFP were made by a combination of the morphology of the neurons, and relative positions of the GFP cells compared with the 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine (DiI)-filled cells (ASK, ADL, ASI, AWB, ASH, and ASJ in the head; PHA and PHB in the tail). In particular, gcy-10::GFP animals had GFP signals in DiI-filled AWB. gcy-12::GFP animals had GFP signals in DiI-filled PHA.
RESULTS AND DISCUSSION
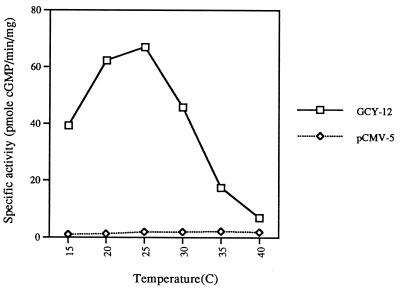
Although the predicted amino acid sequence from segments of each of the putative guanylyl cyclase genes of C. elegans suggests they are guanylyl cyclases, evidence that apparent guanylyl cyclases from invertebrates are actually cyclases has most often met with failure (15, 16). Of those sequences that appear to encode a guanylyl cyclase, a cDNA clone isolated based on the sequence of gcy-12 was subsequently expressed in COS-M6 cells. The C. elegans clone expressed a protein with guanylyl cyclase activity (Fig. 1). Activity was clearly dependent on temperature, with higher temperatures leading to inactivation of the cyclase. A guanylyl cyclase cDNA not yet represented in the genome database (GCY-X1 in Table 1) was also isolated and expressed, and cyclase activity was again evident, but in this case principally only as a chimera: GCY-X1 was fused to the extracellular and protein kinase region of GC-B, a mammalian receptor cyclase for C-type natriuretic peptide (CNP) (E.B., V. Arora, S.Y., B. Wedel, and D.L.G., unpublished data). Basal activity was evident in COS cell membranes and CNP-stimulated enzyme activity. Thus, not only did the C. elegans cyclase express activity as a chimera, but it possessed the ability to be activated by a mammalian hormone when fused to the receptor region of the mammalian cyclase. These results also suggest that the C. elegans cyclases are orphan receptors. Given the high degree of relatedness of each of the genes within the putative cyclase catalytic domain, it can be concluded that there are at least 29 guanylyl cyclases in C. elegans; The cosmids containing these genes and the flanking markers are summarized in Table 1.
Figure 1.
Guanylyl cyclase activity of GCY-12 expressed in COS-M6 cells as a function of assay temperature. Cyclase activity was estimated as described.
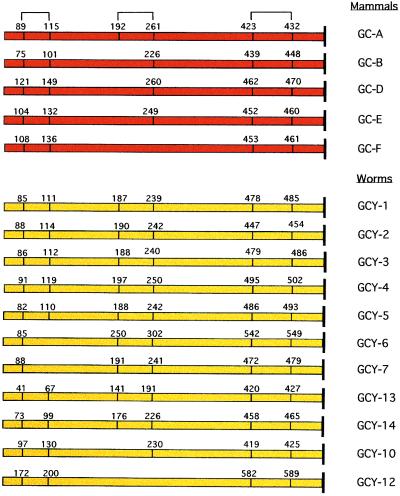
That the cyclases likely represent orphan receptors is not only suggested from their overall similarity in domain structure and alignment to the known mammalian receptors (each membrane form contains an apparent extracellular domain, a single transmembrane segment, and intracellular protein kinase-like and cyclase catalytic domains), and the ability of GCY-X1 to be activated when fused with a mammalian receptor cyclase, but also by their retention of conserved cysteine residues within the extracellular domain (Fig. 2). These cysteine residues are known to form disulfide bridges that appear essential for atrial natriuretic peptide (ANP) binding to the C-type ANP receptor (17, 18). It is presumed that the pressure to retain these conserved cysteine residues is for the purpose of conferring a conformation required for ligand binding.
Figure 2.
The conservation of cysteine residues within the C. elegans guanylyl cyclase extracellular domain. The predicted disulfide bonds in GC-A are based on analysis of the atrial natriuretic peptide (ANP) clearance receptor (17, 18).
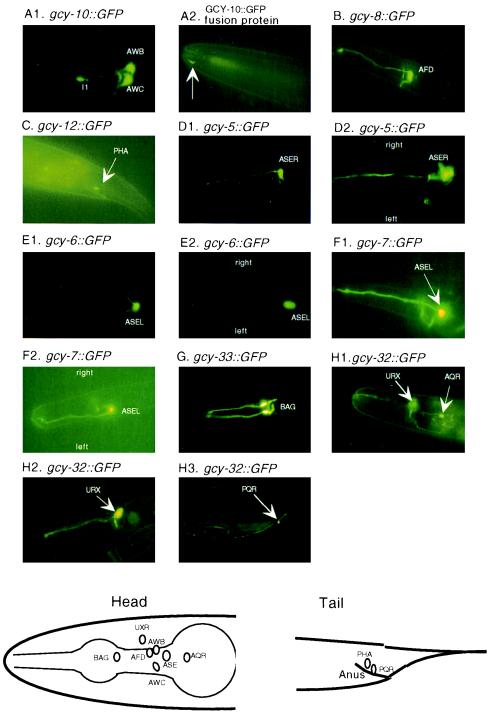
To understand the general functions of such a large family of cyclases, we constructed promoter::GFP transgenic worms and analyzed the expression of the reporter gene. Of the eight transgenes that resulted in detectable GFP signals, all were specific to sensory neurons or interneurons (Fig. 3). The AWC neurons appear to detect odorants and to respond to attractants such as 2-butanone and benzaldehyde (19), and gcy-10::GFP expression was evident in olfactory neurons AWB, AWC, and interneuron I1, with the most intense expression in AWC (Fig. 3A). AFD neurons are at least in part responsible for thermosensation (20), and gcy-8::GFP expression was confined to these neurons (Fig. 3B). gcy-12 appeared to express in PHA neurons (Fig. 3C), which are also thought to be sensory. One of the most intriguing results was the expression of gcy-5, gcy-6, and gcy-7. The ASE neurons, which are thought to detect chemoattractants (21), have been considered as bilaterally symmetric; however, the GFP expression data clearly demonstrate a distinct sidedness (Fig. 3 D–F). gcy-5 expression was always in ASER, while gcy-6 and gcy-7 are ASEL-specific. Two apparent soluble cyclases, gcy-32 and gcy-33, were also found to be expressed in neurons (Fig. 3 G and H). gcy-33::GFP was expressed in sensory neuron BAG, while gcy-32 was expressed in URX and in AQR/PQR. AQR and PQR are a pair of equivalent neurons that migrate anteriorly (AQR) and posteriorly (PQR) during development. A schematic diagram that summarizes the expression information is shown at the bottom of Fig. 3.
Figure 3.
Expression of guanylyl cyclase::GFP in C. elegans. (A1) Expression pattern of gcy-10::GFP (transcriptional fusion). (A2) Subcellular localization of GCY-10/GFP fusion protein at the tip of the nose (arrow). (B) Expression pattern of gcy-8::GFP. (C) Expression pattern of gcy-12::GFP. (D1) Expression pattern of gcy-5::GFP (lateral view). (D2 Expression pattern of gcy-5::GFP (dorsal view). (E1) Expression pattern of gcy-6::GFP (lateral view). (E2) Expression pattern of gcy-6::GFP (dorsal view). (F1) Expression pattern of gcy-7::GFP (lateral view). (F2) Expression pattern of gcy-7::GFP (dorsal view). (G) Expression pattern of gcy-33::GFP. (H 1–3) Expression pattern of gcy-32::GFP. Schematic diagrams of the expression information of the above promoter::GFP transgenes are shown at the bottom for head and tail.
A guanylyl cyclase was previously discovered in the rat olfactory neuroepithelium, where it is specifically expressed on the apical membrane of a small population of neurons (5). These neurons are now known to be unique compared with most or all other sensory neurons in the rat olfactory neuroepithelium (7). Although it was speculated that the cyclase could be an odorant receptor, the finding of only one member of the family suggested either another function, or that the cyclase receptor family, unlike the seven-transmembrane family, was involved in the detection of only a small subset of odorants/pheromones. That the nematode contains a large family of guanylyl cyclase receptors and that multiple guanylyl cyclase receptors are expressed in different and specific sensory neurons now suggests that these enzymes function as chemosensory/odorant receptors. Indeed, a fusion protein between GFP and gcy-10 was found predominantly at the tip of the nose (Fig. 3A2), much like the expression of a known odorant receptor, odr-10 (1). Recently, the genes encoding chemotaxis mutants tax-2 and tax-4 have been cloned (22, 23); they are subunits of a cyclic nucleotide-gated channel, and the tax-4 gene product has been shown to be more sensitive to cGMP activation. In fact, many of the neurons reported to express tax-4 also express guanylyl cyclases. These neurons include AWB, AWC, AFD, ASE, BAG, and URX. Therefore, the cyclic nucleotide-gated channel may represent a principal target for cGMP within these neurons.
The finding of bilateral asymmetry for the ASE neurons with respect to the expression of gcy-5, -6, and -7 is of particular interest. Because the ASE neurons contain sensory cilia within a short distance of each other in the nose, and because they appear to detect chemoattractants, the sidedness to the expression could provide an efficient single input with respect to chemical gradient detection, while affording the nematode increased expression of a diversity of receptors. The presence of two different cyclases, gcy-6 and -7, in ASEL also suggests that they are chemosensory/odorant receptors for different environmental ligands as opposed to having key roles in axonal guidance.
The large number of orphan receptors in the nematode also raises the question of whether a large number of guanylyl cyclase receptors are yet to be discovered in the mammal.
ABBREVIATIONS
- GFP
green fluorescence protein
- GC-D
guanylyl cyclase-D
References
- 1.Sengupta P, Chou J H, Bargmann C I. Cell. 1996;84:899–909. doi: 10.1016/s0092-8674(00)81068-5. [DOI] [PubMed] [Google Scholar]
- 2.Dulac C, Axel R. Cell. 1995;83:195–206. doi: 10.1016/0092-8674(95)90161-2. [DOI] [PubMed] [Google Scholar]
- 3.Buck L, Axel R. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- 4.Vassar R, Ngai J, Axel R. Cell. 1993;74:309–318. doi: 10.1016/0092-8674(93)90422-m. [DOI] [PubMed] [Google Scholar]
- 5.Fulle H-J, Vassar R, Foster D C, Yang R-B, Axel R, Garbers D L. Proc Natl Acad Sci USA. 1995;92:3571–3575. doi: 10.1073/pnas.92.8.3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garbers D L. Cell. 1992;71:1–4. doi: 10.1016/0092-8674(92)90258-e. [DOI] [PubMed] [Google Scholar]
- 7.Julifs D M, Fülle H-J, Zhao A, Houslay M D, Garbers D L, Beavo J A. Proc Natl Acad Sci USA. 1997;94:3388–3395. doi: 10.1073/pnas.94.7.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sulston J, Du Z, Thomas K, Wilson R, Hillier L, Staden R, Halloran N, Green P, Thierry-Mieg J, Qiu L, Dear S, Coulson A, Craxton M, Durbin R, Berks M, Metzstein M, Hawkins T, Ainscough R, Waterston R. Nature (London) 1992;356:37–41. doi: 10.1038/356037a0. [DOI] [PubMed] [Google Scholar]
- 9.Cullen B R. Methods Enzymol. 1987;152:684–704. doi: 10.1016/0076-6879(87)52074-2. [DOI] [PubMed] [Google Scholar]
- 10.Domino S E, Tubb D J, Garbers D L. Methods Enzymol. 1991;195:345–355. doi: 10.1016/0076-6879(91)95179-n. [DOI] [PubMed] [Google Scholar]
- 11.Fire A, Harrison S W, Dixon D. Gene. 1990;93:189–198. doi: 10.1016/0378-1119(90)90224-f. [DOI] [PubMed] [Google Scholar]
- 12.Huang L S, Tzou P, Sternberg P W. Mol Biol Cell. 1994;5:395–412. doi: 10.1091/mbc.5.4.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mello C C, Kramer J M, Stinchcomb D, Ambros V. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chalfie M, Tu Y, Euskirchen G, Ward W W, Prasher D C. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 15.Singh S, Lowe D G, Thrope D S, Rodriguez H, Kuang W-J, Dangott L J, Chinkers M, Goeddel D V, Garbers D L. Nature (London) 1988;334:708–712. doi: 10.1038/334708a0. [DOI] [PubMed] [Google Scholar]
- 16.McNeil L, Chinkers M, Forte M. J Biol Chem. 1995;270:7189–1796. doi: 10.1074/jbc.270.13.7189. [DOI] [PubMed] [Google Scholar]
- 17.Stults J T, O’ Connell K L, Garcia C, Wong S, Engel A M, Garbers D L, Lowe D G. Biochemistry. 1994;33:11372–11381. doi: 10.1021/bi00203a036. [DOI] [PubMed] [Google Scholar]
- 18.Mizuno T, Iwashina M, Itakura M, Hagiwara H, Hirose S. J Biol Chem. 1993;268:5162–5167. [PubMed] [Google Scholar]
- 19.Bargmann C I, Hartwieg E, Horvitz H R. Cell. 1993;74:515–527. doi: 10.1016/0092-8674(93)80053-h. [DOI] [PubMed] [Google Scholar]
- 20.Mori I, Ohshima Y. Nature (London) 1995;376:344–348. doi: 10.1038/376344a0. [DOI] [PubMed] [Google Scholar]
- 21.Bargmann C I, Horvitz H R. Neuron. 1991;7:729–742. doi: 10.1016/0896-6273(91)90276-6. [DOI] [PubMed] [Google Scholar]
- 22.Coburn C M, Bargmann C I. Neuron. 1996;17:695–706. doi: 10.1016/s0896-6273(00)80201-9. [DOI] [PubMed] [Google Scholar]
- 23.Hidetoshi K, Mori I, Rhee J-S, Akaike N, Ohshima Y. Neuron. 1996;17:707–718. doi: 10.1016/s0896-6273(00)80202-0. [DOI] [PubMed] [Google Scholar]