Abstract
Changes in lactose concentration and feed rate altered bacterial growth and population levels in a whey-processing chemostat. The bacterial population and methane production levels increased in relation to increased lactose concentrations comparable to those in raw whey (6%) and converted over 96% of the substrate to methane, carbon dioxide, and cells. Sequential increases in the chemostat dilution rate demonstrated excellent biomethanation performance at retention times as low as 25 h. Retention times shorter than 25 h caused prevalent bacterial populations and methane production to decrease, and intermediary carbon metabolites accumulated in the following order: acetate, butyrate, propionate, lactate, ethanol, and lactose. Bacterial species dominated in the chemostat as a function of their enhanced substrate uptake and growth kinetic properties. The substrate uptake kinetic properties displayed by the mixed chemostat population were equivalent to those of individual species measured in pure culture, whereas the growth kinetic properties of species in mixed culture were better than those measured in pure culture. A designed starter culture consisting of Leuconostoc mesenteroides, Desulfovibrio vulgaris, Methanosarcina barkeri, and Methanobacterium formicicum displayed biomethanation performance, which was similar to that of a diverse adapted mixed-culture inoculum, in a continuous contact digestor system to which 10 g of dry whey per liter was added. Preserved starter cultures were developed and used as inocula for the start-up of a continuous anaerobic digestion process that was effective for biomethanation of raw whey at a retention time of 100 h.
Full text
PDF

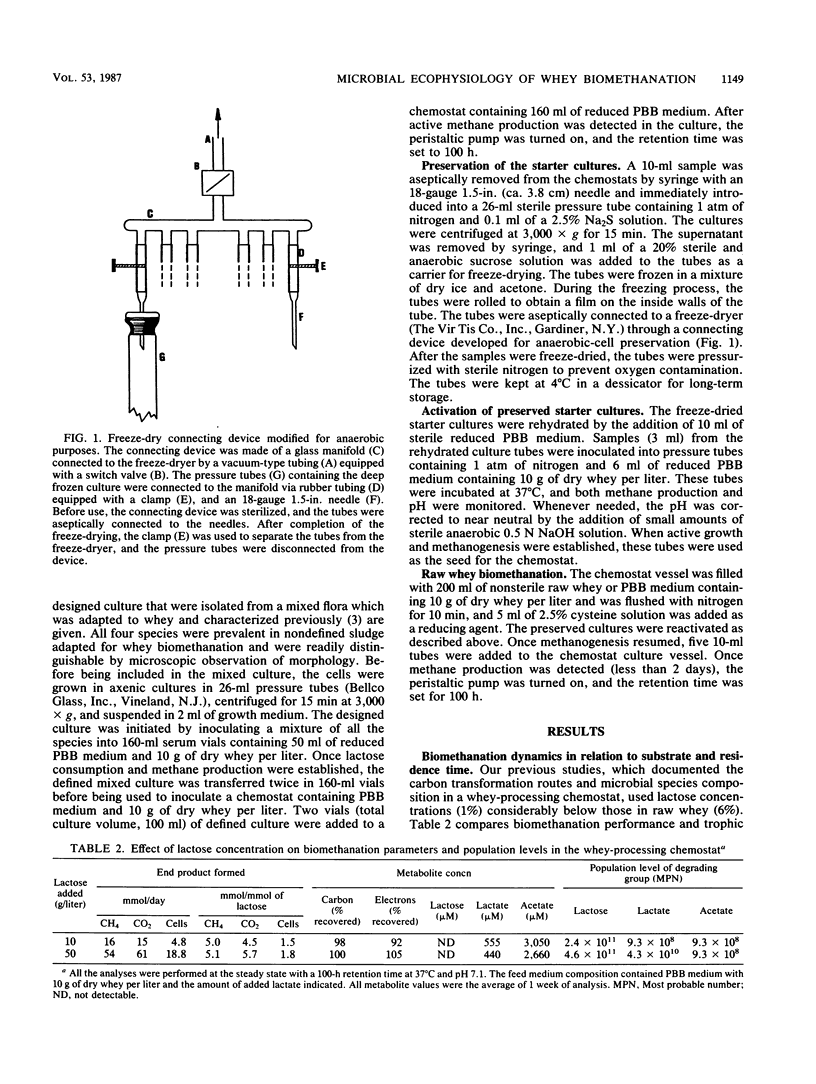

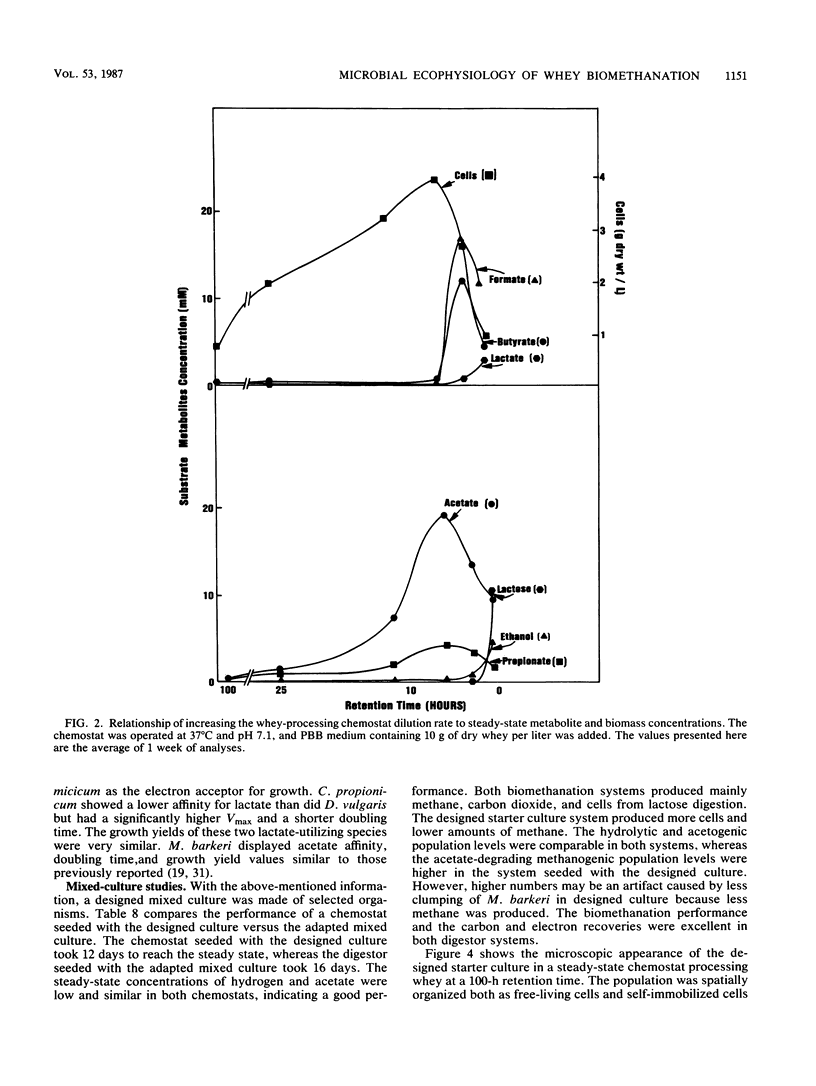
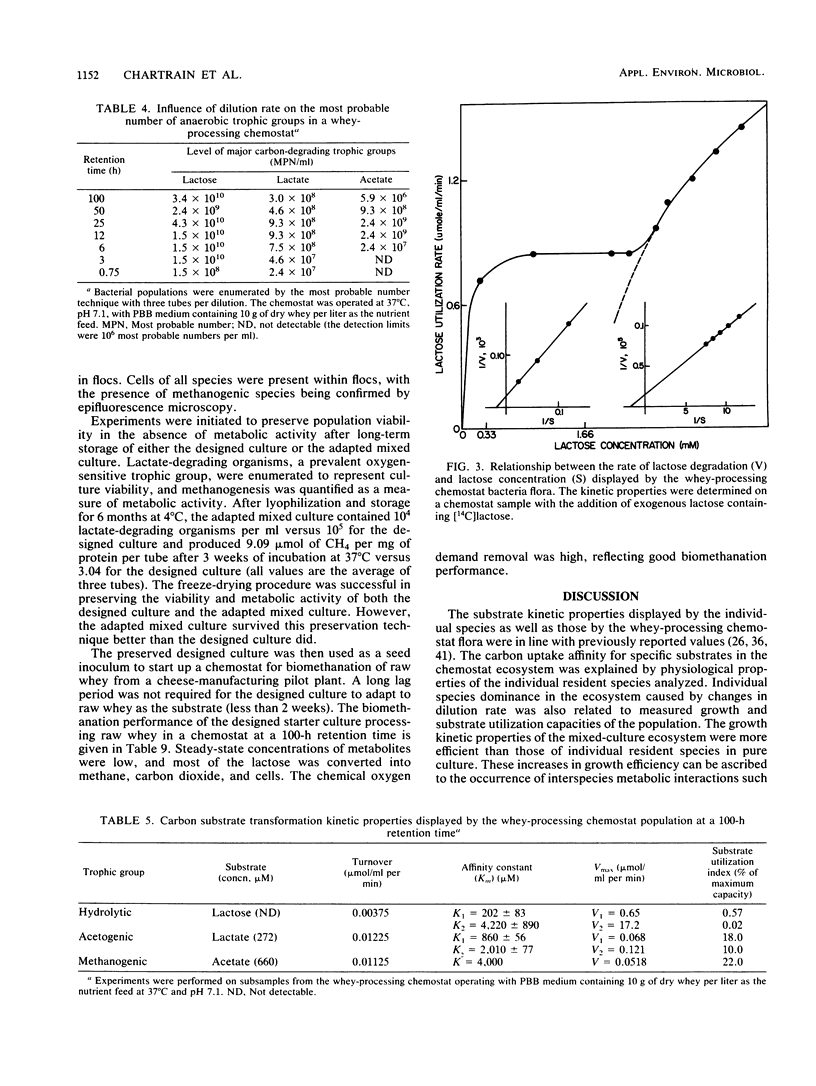


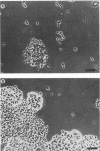
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chartrain M., Zeikus J. G. Microbial ecophysiology of whey biomethanation: characterization of bacterial trophic populations and prevalent species in continuous culture. Appl Environ Microbiol. 1986 Jan;51(1):188–196. doi: 10.1128/aem.51.1.188-196.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartrain M., Zeikus J. G. Microbial ecophysiology of whey biomethanation: intermediary metabolism of lactose degradation in continuous culture. Appl Environ Microbiol. 1986 Jan;51(1):180–187. doi: 10.1128/aem.51.1.180-187.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Wolin M. J. Influence of CH4 production by Methanobacterium ruminantium on the fermentation of glucose and lactate by Selenomonas ruminantium. Appl Environ Microbiol. 1977 Dec;34(6):756–759. doi: 10.1128/aem.34.6.756-759.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiebig K., Gottschalk G. Methanogenesis from Choline by a Coculture of Desulfovibrio sp. and Methanosarcina barkeri. Appl Environ Microbiol. 1983 Jan;45(1):161–168. doi: 10.1128/aem.45.1.161-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones W. J., Guyot J. P., Wolfe R. S. Methanogenesis from sucrose by defined immobilized consortia. Appl Environ Microbiol. 1984 Jan;47(1):1–6. doi: 10.1128/aem.47.1.1-6.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaspar H. F., Wuhrmann K. Kinetic parameters and relative turnovers of some important catabolic reactions in digesting sludge. Appl Environ Microbiol. 1978 Jul;36(1):1–7. doi: 10.1128/aem.36.1.1-7.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klass D. L. Methane from anaerobic fermentation. Science. 1984 Mar 9;223(4640):1021–1028. doi: 10.1126/science.223.4640.1021. [DOI] [PubMed] [Google Scholar]
- Latham M. J., Wolin M. J. Fermentation of cellulose by Ruminococcus flavefaciens in the presence and absence of Methanobacterium ruminantium. Appl Environ Microbiol. 1977 Sep;34(3):297–301. doi: 10.1128/aem.34.3.297-301.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laube V. M., Martin S. M. Conversion of Cellulose to Methane and Carbon Dioxide by Triculture of Acetivibrio cellulolyticus, Desulfovibrio sp., and Methanosarcina barkeri. Appl Environ Microbiol. 1981 Sep;42(3):413–420. doi: 10.1128/aem.42.3.413-420.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mountfort D. O., Asher R. A., Bauchop T. Fermentation of Cellulose to Methane and Carbon Dioxide by a Rumen Anaerobic Fungus in a Triculture with Methanobrevibacter sp. Strain RA1 and Methanosarcina barkeri. Appl Environ Microbiol. 1982 Jul;44(1):128–134. doi: 10.1128/aem.44.1.128-134.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. R., Mah R. A. Growth and methanogenesis by Methanosarcina strain 227 on acetate and methanol. Appl Environ Microbiol. 1978 Dec;36(6):870–879. doi: 10.1128/aem.36.6.870-879.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ten Brummeler E., Pol L. W., Dolfing J., Lettinga G., Zehnder A. J. Methanogenesis in an Upflow Anaerobic Sludge Blanket Reactor at pH 6 on an Acetate-Propionate Mixture. Appl Environ Microbiol. 1985 Jun;49(6):1472–1477. doi: 10.1128/aem.49.6.1472-1477.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varel V. H., Hashimoto A. G., Chen Y. R. Effect of temperature and retention time on methane production from beef cattle waste. Appl Environ Microbiol. 1980 Aug;40(2):217–222. doi: 10.1128/aem.40.2.217-222.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weimer P. J., Zeikus J. G. Fermentation of cellulose and cellobiose by Clostridium thermocellum in the absence of Methanobacterium thermoautotrophicum. Appl Environ Microbiol. 1977 Feb;33(2):289–297. doi: 10.1128/aem.33.2.289-297.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter J. U., Wolfe R. S. Methane formation from fructose by syntrophic associations of Acetobacterium woodii and different strains of methanogens. Arch Microbiol. 1980 Jan;124(1):73–79. doi: 10.1007/BF00407031. [DOI] [PubMed] [Google Scholar]
- Zehnder A. J., Huser B. A., Brock T. D., Wuhrmann K. Characterization of an acetate-decarboxylating, non-hydrogen-oxidizing methane bacterium. Arch Microbiol. 1980 Jan;124(1):1–11. doi: 10.1007/BF00407022. [DOI] [PubMed] [Google Scholar]
- Zinder S. H., Anguish T., Cardwell S. C. Effects of Temperature on Methanogenesis in a Thermophilic (58 degrees C) Anaerobic Digestor. Appl Environ Microbiol. 1984 Apr;47(4):808–813. doi: 10.1128/aem.47.4.808-813.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinder S. H., Cardwell S. C., Anguish T., Lee M., Koch M. Methanogenesis in a Thermophilic (58 degrees C) Anaerobic Digestor: Methanothrix sp. as an Important Aceticlastic Methanogen. Appl Environ Microbiol. 1984 Apr;47(4):796–807. doi: 10.1128/aem.47.4.796-807.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinder S. H., Mah R. A. Isolation and Characterization of a Thermophilic Strain of Methanosarcina Unable to Use H(2)-CO(2) for Methanogenesis. Appl Environ Microbiol. 1979 Nov;38(5):996–1008. doi: 10.1128/aem.38.5.996-1008.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]