Abstract
The gene unc-76 (unc, uncoordinated) is necessary for normal axonal bundling and elongation within axon bundles in the nematode Caenorhabditis elegans. The UNC-76 protein and two human homologs identified as expressed sequence tags are not similar to previously characterized proteins and thus represent a new protein family. At least one of these human homologs can function in C. elegans, suggesting that it, like UNC-76, acts in axonal outgrowth. We propose that the UNC-76 protein, which is found in cell bodies and processes of all neurons throughout development, either has a structural role in the formation and maintenance of axonal bundles or transduces signals to the intracellular machinery that regulates axonal extension and adhesion.
Axons in developing nervous systems navigate through a varied set of extracellular environments to reach their targets. Most axons grow along other axons for much of their lengths, and the association of axons in specific bundles, or fascicles, is likely to play a major role in nervous system assembly. Although many cell-surface proteins involved in fascicle formation have been identified (1), relatively little is known about the intracellular mechanisms by which surface interactions lead to the elongation of axons specifically along other axonal surfaces.
Genetic screens for fasciculation-defective mutants can, in principle, identify molecules necessary for fasciculation without presuppositions as to their biochemical nature or subcellular localization. Analysis of strains of the nematode Caenorhabditis elegans with mutations causing locomotory defects (uncoordinated or unc mutants) has revealed a group of three genes that, when mutant, affect the growth of axons in fascicles, but not along nonneuronal substrates (cells of the lateral hypodermis and the overlying basement membrane; refs. 2–4). Mutations in this fascicle-specific group of genes, unc-34, unc-71, and unc-76, cause two types of defects: many axons fail to extend fully within the axon bundles of the dorsal and ventral nerve cords, and many fail to remain in their normal fascicles (2–4). The best-characterized example of these defects is provided by the axons of the left and right hermaphrodite-specific neuron (HSN) motor neurons. In fascicle-specific mutants, these axons end prematurely within the left and right fascicles of the ventral nerve cord, respectively, and they often fail to remain on opposite sides of the cord (refs. 3, 4; L.B. and H.R.H., unpublished observations). If a second mutation causes the HSN axons to be rerouted along a lateral process tract instead of the ventral nerve cord, the axons in these mutants grow to their normal lengths, indicating that unc-34 and unc-76 affect the interaction of these axons with the ventral cord environment rather than the ability of the HSN axons to grow beyond a certain length (4).
Among the mutants with fascicle-specific defects, unc-76 mutant animals have the most severe abnormalities in locomotion and HSN outgrowth (3, 4). To understand the basis of these defects, we have undertaken a genetic and molecular analysis of the unc-76 gene.
MATERIALS AND METHODS
Genetic Methods and Strains.
Strains were constructed and maintained according to standard methods (5). Most mutant strains used in this paper were described in refs. 5–7; J. Culotti (Mt. Sinai Hospital, Toronto) and S. Siddiqui (Toyohashi University) provided unc-76(ev424), and E. Hedgecock (Johns Hopkins University) provided unc-76(rh116). Mutations that failed to complement unc-76(e911) were obtained by treating wild-type (N2) or egl-1(n986dm) males with ethyl methanesulfonate (5), mating them with + sdc-3(y52) unc-76(e911)/unc-61(e228) ++; dpy-3(e27) hermaphrodites, and isolating severely Unc non-Dpy F1 hermaphrodites. After three generations of backcrossing to N2, mutants were stained with antiserotonin antisera (3), and HSN axon length was estimated to the nearest tenth of the distance between the vulva and the posterior end of the pharynx. Because a large-scale screen for suppressors of the Unc phenotype of unc-76(e911) produced only smg suppressors (L.B. and H.R.H., unpublished observations; ref. 8), the effects of smg-1 on other unc-76 alleles were tested in strains of genotype smg-1(e1228) him-2(e1065); unc-76.
Molecular Analysis of unc-76.
Molecular biological and immunological procedures were performed according to standard methods (9–11). Cosmids obtained as part of the C. elegans genome project (12, 13) and plasmids containing fragments of the rescuing cosmid C56C4 were injected at 50 μg/ml into the gonads of unc-76(e911) mutant hermaphrodites (14), and the Unc phenotype was scored in the F1 and F2 generations. unc-76 cDNA clones were obtained by screening 220,000 plaques from a mixed-stage C. elegans cDNA library (15) with the 32P-labeled insert from p76–8. DNA from exons and splice junctions of each mutant unc-76 strain was amplified by PCR (16) for sequence determination. Database searches were performed at the National Center for Biotechnology Information with the blast program (17).
Clones for FEZ1, FEZ1-T, and FEZ2 (accession numbers R61145R61145, R25079R25079, and R21583R21583, respectively) were obtained by the Washington University–Merck EST Project (unpublished results) and provided to us by the I.M.A.G.E. consortium (18). p76HsA-5 contained a 1.5-kb HindIII-BsrBI FEZ1 cDNA fragment driven by a 1.05-kb unc-76 promoter fragment (Y. Jin, personal communication) in pPD49.26 (19). p76HsA-5 was injected into unc-76(e911) animals, or, together with a dpy-20-rescuing plasmid (20), into dpy-20(e1282ts); unc-76(e911) animals, and rescued lines were stained with anti-GABA (γ-aminobutyric acid) antisera (3). p86/76–1 contained a 5-kb SpeI-StyI fragment from the unc-86::lacZ fusion SA2 (provided by G. Ruvkun; Massachusetts General Hospital) fused to a BglII-BclI unc-76 fragment, which was fused in turn to lacZ. In p86-L1, the same unc-86 fragment was fused directly to lacZ. N2-derived lines carrying either fusion plasmid with the rol-6 plasmid pRF4 (21) were stained with a monoclonal anti-β-galactosidase antibody (Promega; ref. 22).
Anti-UNC-76 Antibodies.
Three rabbits were immunized with the following series of UNC-76 fusion proteins produced in Escherichia coli: a maltose binding protein fusion to amino acids 13–385, a glutathione S-transferase fusion to amino acids 48–385, and a fusion of amino acids 12–385 to His6. Sera were affinity-purified on either UNC-76-His6 or UNC-76::TrpE fusion proteins bound to nitrocellulose. Worms were fixed 16 h with 4% paraformaldehyde in PBS at 4°C. Half of the worms were frozen and thawed 2–3 times in liquid nitrogen, and then all worms were partially broken in a Dounce homogenizer, washed in PBS, stained as described for anti-serotonin staining (3), and washed in 1 μg/ml diamidinophenolindole (Sigma). Serum from one rabbit (no. 275) was used for all experiments shown.
RESULTS
Genetic Analysis of unc-76.
To determine the effects of a complete loss of unc-76 function, we obtained five new unc-76 alleles in a screen for mutations that failed to complement unc-76(e911). The new mutations, n2367, n2397, n2398, n2399, and n2457, arose at a frequency (1/3,300 mutagenized genomes) close to the average frequency (1/2,000) of inducing a loss-of-function mutation using the same ethyl methanesulfonate mutagenesis procedure (5, 23–24). The defects caused by all unc-76 alleles appeared to be restricted to fascicles; HSN cell body migration and ventral axonal outgrowth along the lateral hypodermis were nearly normal (data not shown). All unc-76 alleles, except n2398, caused similar HSN axonal outgrowth defects (Table 1), with HSN axons extending an average of 64–79% of the distance from the vulva to the head (wild type = 100%). Animals carrying the allele n2398 had a mean HSN length of 89% and were slightly less uncoordinated than the rest. The mean HSN lengths in animals carrying e911, n2367, n2397, n2457, or ev424 in trans to the deficiency yDf8 were all similar to one another (73–77%) and to those observed in animals homozygous for each mutant allele. The mutation smg-1(e1228), which is believed to elevate the levels of unstable RNA species (25), suppressed the effects of two of the eight alleles, n2398 and e911. These observations are consistent with a model in which n2398 is a weak allele, ev424, rh116, n2397, n2367, n2399, and n2457 are severe loss-of-function or null alleles, and e911 is similar in its consequences to the strong alleles but not fully null, because smg suppression can restore partial function. Analyses of DNA and protein from unc-76 mutants support this model (see below).
Table 1.
HSN axon length in unc-76 mutants
| Genotype | HSN length, % wild type | n |
|---|---|---|
| N2 | 100 | >1000 |
| e911 | 66 ± 3 | 55 |
| e911/yDf8 | 73 ± 3 | 46 |
| ev424 | 64 ± 4 | 47 |
| ev424/yDf8 | 73 ± 4 | 64 |
| rh116 | 65 ± 5 | 56 |
| n2367 | 79 ± 4 | 57 |
| n2367/yDf8 | 77 ± 7 | 31 |
| n2397 | 77 ± 3 | 90 |
| n2397/yDf8 | 76 ± 6 | 42 |
| n2398 | 89 ± 2 | 75 |
| n2399 | 73 ± 4 | 68 |
| n2457 | 72 ± 5 | 40 |
| n2457/yDf8 | 77 ± 5 | 44 |
HSN axon length (±95% confidence limits) in unc-76 mutants. n, number of axons scored. The small number of HSNs with cell bodies in an abnormal posterior position or with axons in a lateral position were not included because these displacements affect outgrowth (4). The genotypes of the deficiency heterozygotes are dpy-11(e224)unc-76(e911)/unc-42(e270)yDf8 and for ev424, n2367, n2397 and n2398, sma-1(e30)unc-76/yDf8.
Isolation of unc-76 Genomic and cDNA Clones.
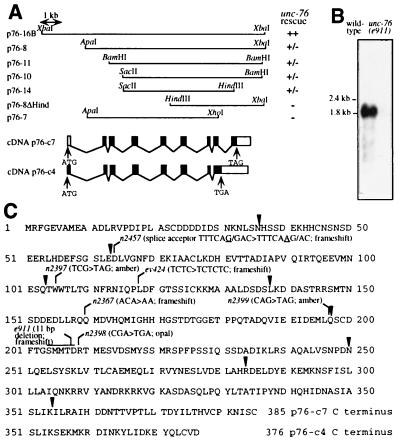
We cloned the unc-76 gene by identifying cosmid clones able to restore wild-type locomotion to uncoordinated unc-76(e911) animals after germ-line transformation. unc-76 is located less than 0.1 map units to the right of sdc-3 (26). Any of five overlapping cosmids, C56C4, T25A9, C08C1, C01G10, and C13G10, located to the right of sdc-3 on the C. elegans physical map (12–13) rescued the Unc phenotype of unc-76(e911) animals, while cosmids flanking this group (C25D7 to the left and T06H10, T01G5, and C28G7 to the right) did not. A 10.7-kb XbaI fragment from C56C4, represented in p76–16B, completely restored normal locomotion of unc-76(e911) animals, while transformed lines carrying any of several subclones as small as 5.5 kb (p76–14) showed wild-type movement among young larvae but increasing uncoordination of older animals (Fig. 1A).
Figure 1.
The unc-76 gene. (A) Structures of unc-76 genomic and cDNA clones. ++, restoration of locomotion of unc-76(e911) animals to that of the wild type; −, no effect on locomotion; ±, rescue in young larvae but poorer locomotion in older animals. Exons (black bars) and introns (thin lines) of the cDNA clones p76-c4 and p76-c7 are aligned with the genomic map. (B) unc-76 RNA. The 32P-labeled insert from p76-c4 was hybridized to 10 μg of poly(A)+ RNA from wild-type and unc-76(e911) mutant embryos. (C) UNC-76 protein sequences deduced from p76-c4 and p76-c7. The sequences are identical through amino acid 354, and the alternative C termini are shown beginning at amino acid 351. Exon boundaries (arrowheads) and positions affected by unc-76 mutations are indicated. The sequence deleted from e911 is ATCCGTCATCA.
Inserts from two clones isolated from a mixed-stage C. elegans cDNA library, p76-c4 (2.6 kb) and p76-c7 (1.7 kb), hybridized to sequences throughout the rescuing genomic DNA (data not shown) and to a major transcript of 1.8 kb in RNA from wild-type embryos (Fig. 1B). This transcript was reduced in abundance in unc-76(e911) embryos. A low-abundance 2.4-kb transcript that hybridized to the p76-c4 and the p76-c7 inserts was observed in long exposures of blots of wild-type, but not mutant, RNA (data not shown).
Analysis of unc-76 Sequence and RNA Structure.
Comparison of genomic and cDNA sequences showed that the two cDNA clones, each of which contained a single large open reading frame (Fig. 1C), differed primarily in their 5′ ends (p76-c4 extended 119 nucleotides further 5′ than p76-c7) and in the presence or absence of an intron near the 3′ end. RNase protection experiments showed that p76-c7 corresponds to the major splice form (intron 8 removed) in mixed-stage RNA (data not shown). Primer extension and reverse transcriptase-PCR experiments with mixed-stage RNA indicated that the two major unc-76 transcript types contain the leader sequence SL1 (27) trans-spliced 5 or 23 nucleotides upstream of the 5′ end of p76-c7 (data not shown), consistent with protein products starting at amino acids 1 or 8 in the sequences shown in Fig. 1C. These experiments also indicated that at least one other class of transcripts begins approximately 500 bp further 5′ and extends at least to the beginning of exon 2. Such transcripts would not alter the predicted UNC-76 protein products.
We identified mutations in the coding regions or splice junctions in seven of the eight unc-76 alleles (Fig. 1C). All are predicted to generate truncated protein products; three mutations introduced premature stop codons (n2397, n2398, and n2399), and four caused frameshifts (e911, ev424, n2367, and n2457). The mutant allele encoding the longest truncated protein, n2398, caused the least severe HSN and locomotory defects.
UNC-76 and Its Human Homologs Define a New Protein Family.
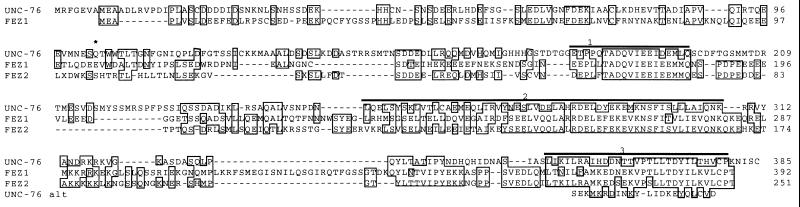
The 376 and 385 amino acid forms of the UNC-76 protein (encoded by p76-c4 and p76-c7, respectively) showed no strong similarity to any previously characterized proteins, but three groups of similar human sequences were identified in the expressed sequence tag database, dbEST (28). We determined the sequences of the longest available cDNA clones from each group and found that they are probably derived from two genes. Two other laboratories recently have identified rat unc-76 homologs in independent yeast two-hybrid screens (see Discussion). To reflect the names given by these groups, this family of UNC-76-like proteins has been called FEZ, for fasciculation and elongation protein; zygin/zeta-1, and we have named the two human genes we identified FEZ1 and FEZ2.
Alignment of the 251 amino acid FEZ2 fragment inferred from partial cDNA clones with the complete 392 amino acid FEZ1 protein and the C. elegans UNC-76 proteins (Fig. 2) showed that UNC-76 is 35% identical (46% similar) to FEZ1 and 34% identical (45% similar) to FEZ2 in the region of overlap. The two human proteins are 49% identical (56% similar) to each other in the region represented in both proteins. Three regions in the C-terminal halves are most similar, with amino acids 179–197, 251–307, and 354–381 (numbered according to the C. elegans protein), showing 68%, 46%, and 54% identity, respectively, among all three proteins. The N-terminal halves are more divergent but still show substantial similarity.
Figure 2.
Alignment of C. elegans UNC-76 protein with two human homologs. Identical amino acids are boxed. Amino acids 1–98 of FEZ2 were inferred from the overlapping clone DY1C1TG01 (accession number F15259F15259), which probably lacks sequences encoding the N terminus of FEZ2. Longer FEZ2 clones were not present in dbEST. The alternatively spliced C terminus from p76-c4 is labeled UNC-76 alt. X, amino acid was ambiguous in the reported DY1C1TG01 sequence. ∗, C terminus of FEZ1-T. Bars labeled 1–3, regions of highest conservation.
A third class of human cDNA clones contained sequence identical to that of FEZ1 through codon 104, followed by a TAA stop codon and 1.3 kb of unrelated sequence. The truncated protein encoded by this clone, which we have named FEZ1-T, is probably a product of alternative splicing, because the cDNA sequences diverge after an AG dinucleotide. The nematode unc-76 gene is spliced at the equivalent position.
None of the FEZ family proteins appeared by Kyte-Doolittle hydrophobicity analysis (29) to have a hydrophobic region sufficiently long to serve as a signal sequence or transmembrane domain (data not shown), suggesting that the proteins are intracellular. Secondary structure predictions suggested that nine conserved regions (UNC-76 amino acids 48–61, 66–83, 105–121, 182–196, 237–248, 257–282, 292–307, 351–360, and 369–378) could form amphipathic helices. No other structural or functional motifs were identified.
UNC-76 Protein Is Present in All Axons Throughout Development.
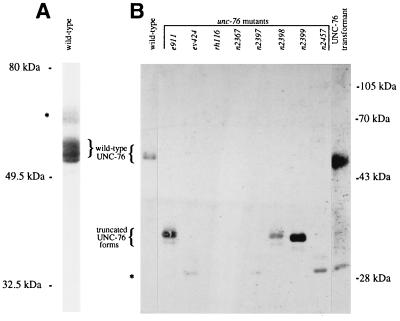
On immunoblots, affinity-purified UNC-76 antisera from each of three rabbits (see Materials and Methods) recognized a set of at least three 51- to 54-kDa proteins (Fig. 3) that were not recognized by preimmune sera (data not shown). These proteins were absent from all eight unc-76 mutants and were restored by transformation of unc-76(e911) mutant animals with the rescuing clone p76–16B. Three mutants (e911, n2398, and n2399) showed novel truncated forms 19–20 kDa smaller than the wild-type proteins, as predicted from the mutant DNA sequences (Fig. 3B).
Figure 3.
Immunoblots of UNC-76 protein. Proteins from mixed-stage populations of worms were separated on (A) 7.5% and (B) 10% polyacrylamide gels, blotted with affinity-purified anti-UNC-76 serum and reacted with a peroxidase-conjugated anti-rabbit antiserum and the ECL chemiluminescent reagent (Amersham). The UNC-76 transformant carried the rescuing plasmid p76–16B in an unc-76(e911) background and contained the e911 mutant form of UNC-76 protein, not visible in this exposure, in addition to the wild-type form. Bands indicated by asterisks probably do not represent UNC-76 protein, because they appeared in only a subset of samples from wild-type and unc-76 mutant animals in other experiments (data not shown) and were unaffected by unc-76 mutations.
Indirect immunofluorescence microscopy of wild-type worms stained with affinity-purified sera from each rabbit showed intense staining in all major nerve bundles (the nerve ring and dorsal and ventral nerve cords) and all minor longitudinal and circumferential process tracts and bundles of sensory processes (Fig. 4). Staining was visible in neuronal cell bodies but not in nuclei (Fig. 4 B and E–G). No neuronal fluorescence was observed in worms stained with any of the three preimmune sera (data not shown), and staining was reduced or nearly eliminated in all eight mutant unc-76 strains (Fig. 4C). The residual staining in unc-76(e911) animals was eliminated by preincubation of antisera with maltose binding protein::UNC-76 or UNC-76::His6 fusion proteins (data not shown), indicating that this staining represented mutant UNC-76 protein.
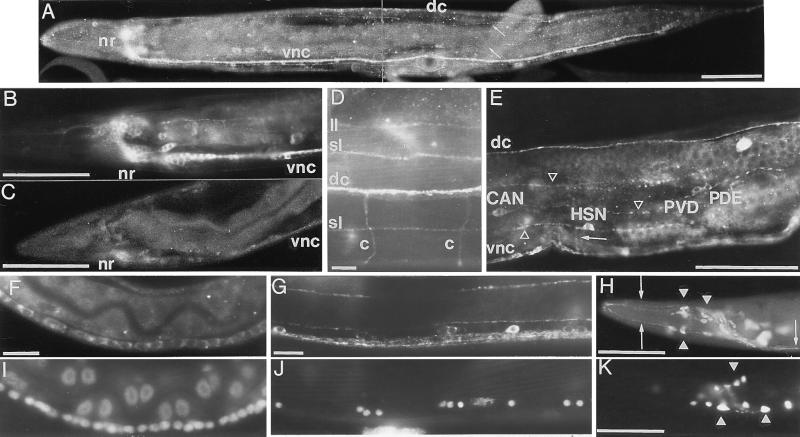
Figure 4.
Localization of UNC-76 and UNC-76::β-galactosidase fusion proteins. Anterior is to the left, and dorsal is at the top. (A) UNC-76 protein in a wild-type adult is visible in the nerve ring (nr), ventral cord (vnc), dorsal cord (dc), and dorsal and ventral sublateral process tracts (arrows). UNC-76 protein in a wild-type adult (B) is more abundant than in an unc-76(ev424) mutant adult (C). (D) Dorsal region of a wild-type adult just anterior to the vulva. Staining is visible in the dorsal nerve cord (dc), dorsal sublateral tracts (sl), a left lateral tract (ll), and two motor commissures (c). (E) Midbody region of a wild-type adult. Staining is visible in cell bodies of PVDR, PDER, HSNR, and CANR as well as the dorsal (dc) and ventral (vnc) nerve cords, several longitudinal process tracts (arrowheads), and the circumferentially directed HSN axon (arrow). UNC-76 staining near the midbody region of L2 (F) and adult (G) ventral nerve cords and the corresponding nuclei visualized by diamidinophenolindole (I and J). (H) An UNC-86::UNC-76::β-galactosidase fusion protein encoded by p86/76–1 is localized to axons (arrow) as well as cell bodies (arrowheads) in a small set of neurons in the head of an adult stained with anti-β-galactosidase antibodies. (K) The equivalent UNC-86::β-galactosidase fusion protein lacking UNC-76 sequences (encoded by p86-L1) is confined primarily to cell bodies (arrowheads). Cell identities in H and K were not determined. [Bars = 50 μm (A–C, E, H, and K) and 10 μm (D, F, G, I, and J).]
Outside the pharynx, all neurons, but no other cells, stained with anti-UNC-76 antisera. Specifically, no staining was observed in cells that flank the ventral or dorsal nerve cords, such as body wall muscles or hypodermal cells. Pharyngeal UNC-76 expression was not visible, but pharyngeal cells were probably not made accessible to antibodies during fixation because they lacked the background stain seen in the rest of the animal.
Axonal segments that are not fasciculated appear normal in unc-76 mutant animals, in contrast to the segments of the same axons that grow in fascicles (2–4). Nonetheless, we observed UNC-76 protein throughout the axons of wild-type animals, including some nonfasciculated axon segments known to be normal in unc-76 mutant animals (all motor neuron commissures and the ventral-directed segments of the HSN, PDE, and PVD axons; Fig. 4 D–E). UNC-76 protein-containing commissural and longitudinal axons on the lateral body wall usually crossed without apparent changes of direction (Fig. 4D), indicating that the simple presence of UNC-76 protein does not cause bundling.
UNC-76 staining was visible throughout the nervous system of animals at all developmental stages from newly hatched larvae through adults. Embryos were not sufficiently permeabilized to allow anti-UNC-76 antibody staining, but unc-76::lacZ fusions that appeared to be expressed normally in larvae and adults were expressed in a few cells, the identities of which were not determined, in embryos of about 200 cells, before the outgrowth of the first axons (data not shown). UNC-76 staining in axons was strong throughout development, whereas cell body staining was strong in young larvae, but weaker in adults except for cell bodies in the head and tail ganglia (Figs. 4 B and F–G). Of neurons with laterally positioned cell bodies, only the CAN and HSN cell bodies consistently contained UNC-76 protein in adults (Fig. 4E).
The presence of C terminally truncated UNC-76 proteins in axons of mutant worms (Fig. 4C) suggested that axon-targeting activity of UNC-76 resides in the N-terminal third of the protein. Consistent with these observations, we found that amino acids 13–186 of UNC-76 can confer axonal localization to an otherwise nuclear-localized fusion of amino acids 1–83 of the C. elegans UNC-86 protein (22) to β-galactosidase (Fig. 4 H and K). Amino acids 1–197 of UNC-76 have been used with several promoters to direct the green fluorescent protein (30) or β-galactosidase to axons in C. elegans (L.B., Y. Jin and H.R.H., unpublished observations; refs. 31–34) and in zebrafish (J. Dynes and J. Ngai, personal communication).
Rescue of unc-76(e911) by FEZ1.
To determine whether the human FEZ1 protein can function in C. elegans, we transformed unc-76(e911) and dpy-20(e1282ts); unc-76(e911) worms with the FEZ1 cDNA under the control of a 1-kb fragment of the C. elegans unc-76 promoter. FEZ1 partially rescued the unc-76(e911) locomotory defect (Fig. 5 A–D): FEZ1-carrying animals moved with an irregular sinusoidal waveform and were somewhat sluggish compared with wild-type animals, but were considerably more active and coordinated than unc-76 mutant animals. Fasciculation of GABA-containing motor axons in the ventral nerve cord was also subtly improved in FEZ1-rescued dpy-20(e1282ts); unc-76(e911) animals (Fig. 5 E–G). Although axons aberrantly exited and re-entered bundles at about the same frequency in mutant and rescued animals (data not shown), axons in the rescued worms seemed to remain in closer proximity than did axons in the mutants, suggesting improved overall fasciculation in the ventral nerve cord. The severely defective outgrowth of the axons of the HSNs and of the sensory neurons PHA and PHB observed in unc-76(e911) animals was not improved by FEZ1 (data not shown).
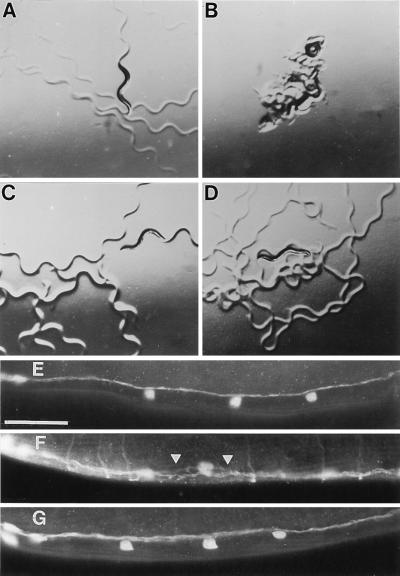
Figure 5.
Restoration of locomotion and ventral cord fasciculation in unc-76(e911) worms by FEZ1. (A–D) Animals were photographed after 1 h on slightly dried agar plates without bacteria. (A) Wild type; (B) unc-76(e911); (C) unc-76(e911) carrying the C. elegans unc-76 gene in p76–16B; (D) unc-76(e911) carrying the human FEZ1 gene in p76-HsA-5. (E-G) Ventral nerve cords just posterior to the heads of adults stained with anti-GABA antisera. Anterior is at the left. (E) Wild type; (F) unc-76(e911); (G) dpy-20(e1292ts); unc-76(e911) carrying the human FEZ1 gene in p76-HsA-5 and the C. elegans dpy-20 gene. Arrowheads, regions of defasciculation. Axons perpendicular to ventral nerve cord in F are motor commissures, out of the plane of focus in E and G. (Bar = 5 μm.)
DISCUSSION
unc-76 Function Is Necessary for Normal Fascicle Structure.
C. elegans unc-76 mutants have two types of axonal defects: axons in fascicles often do not reach their full lengths, and many axons that grow in fascicles fail to bundle tightly together (2–4). In unc-76 mutants, many of the axons with abnormal growth in fascicles nonetheless extend normally around the body wall, unaccompanied by other axons, suggesting that unc-76 is required specifically for axon-axon interactions. We have shown that these defects are caused by unc-76 alleles that encode severely truncated proteins and are not substantially different in unc-76/unc-76 homozygotes and unc-76/deficiency heterozygotes, indicating that this phenotype is likely to result from an absence of unc-76 function.
unc-76 Encodes a New Class of Axonal Protein.
We cloned the unc-76 gene by rescue of the uncoordinated phenotype of unc-76(e911) animals after germ-line transformation. Evidence that the 10.7-kb rescuing clone contains the unc-76 gene includes the observations that mutant unc-76 strains contain alterations in DNA sequence, mRNA levels, and size and abundance of the protein predicted from analysis of cDNA clones that correspond to this genomic DNA fragment.
The major predicted UNC-76 protein is highly similar to two human proteins, identified as expressed sequence tags, which we have called FEZ1 and FEZ2, for fasciculation and elongation proteins; zygin/zeta-1 (see below). The FEZ1 gene was able to restore partial locomotion and axonal fasciculation to C. elegans unc-76 mutants in germ-line transformation experiments, indicating that both the function and the structure of the FEZ proteins have been conserved in evolution. Amino acids 1–197 of the C. elegans UNC-76 protein can target reporter proteins to axons in both C. elegans and zebrafish, suggesting that proteins that interact with this region in FEZ family members likewise have been conserved.
The proteins of the FEZ family have little similarity to previously characterized proteins and thus define a new class of proteins involved in axonal outgrowth. The N-terminal halves of the FEZ proteins are highly acidic, and large regions of the proteins are predicted to form amphipathic helices. However, the proteins lack motifs indicating specific functions. The absence of a potential signal sequence or transmembrane domain suggests that the FEZ proteins are intracellular. We observed C. elegans UNC-76 protein in the cell bodies and axons of nearly all neurons but not in nuclei or in nonneuronal cells. Membrane and cytoplasmic staining could not be distinguished in these experiments.
While most FEZ1-related clones in dbEST were derived from brain libraries (15/17 FEZ1 and 4/5 FEZ1-T clones), all but two of the 15 FEZ2 clones were derived from nonneural tissues (placenta, melanocytes, spleen, lung, heart, testis, and muscle). This distribution indicates that the FEZ family functions in nonneuronal cell types as well as in neurons.
Possible Functions of the FEZ Family Proteins.
The severe defects in newly hatched unc-76 mutant larvae suggest a function for UNC-76 early during nervous system development, whereas the persistence of UNC-76 in axons through adulthood suggests a continuing role. We propose two possible UNC-76 functions consistent with its likely intracellular localization and the fascicle-specific defects of unc-76 mutants. First, the UNC-76 protein could play a structural role in the formation and/or maintenance of fascicles, e.g., by its intracellular association with a cell-surface adhesion molecule, the axonal membrane, or the cytoskeleton. Second, UNC-76 could transduce signals from cell-surface molecules to the intracellular machinery that regulates axonal extension and adhesion. In either case, the requirement for UNC-76 protein only in regions of axon-axon contact despite its presence throughout the axon could reflect the restricted distribution of cell-surface adhesion molecules or their ligands.
Consistent with both roles, two interactions of FEZ family members recently have been suggested from the results of two-hybrid screens in yeast. A rat homolog of FEZ1, called zeta 1, was found to bind to the C1 regulatory domains of protein kinase C types ζ and ɛ (S. Kuroda and U. Kikkawa, personal communication), and rat FEZ1 and FEZ2 homologs, named zygins I and II, were identified as synaptotagmin-binding proteins (T. Sudhof, personal communication). The protein kinase C family has been implicated in a wide range of cellular functions, including modulation of membrane structure (35). Synaptotagmins are membrane proteins required for synaptic vesicle exocytosis and are believed to function in other membrane-fusion events in nonneuronal cells (36). If the interactions of these proteins with FEZ family members are confirmed for endogenous proteins in vivo, the UNC-76 protein and other FEZ family members could provide points of convergence between signaling pathways and assembly of the cell membrane, perhaps as regulators of membrane insertion in growing axons or in motile organelles of nonneuronal cells.
An understanding of the biochemical functions of FEZ family members awaits further characterization of their interactions with other proteins, both those discussed above and others that might interact with the axon-targeting N-terminal region of the UNC-76 protein and the corresponding portion of the putative nonneuronal FEZ2. Our finding that the function of FEZ family members has been conserved between C. elegans and humans suggests that this novel protein family and the proteins with which its members interact are important components of the network of molecules that regulate cellular morphology.
Acknowledgments
We thank Y. Jin, U. Kikkawa, B. Klein, S. Kuroda, B. Meyer, J. Ngai, M. Nonet, and T. Sudhof for sharing unpublished data, S. Zhang for secondary structure predictions, L. Delissio and B. James for DNA sequencing, M. Metzstein for initiating the dbEST search, S. Cameron for GABA staining, M. DiPersio, M. Koelle, B. Sawin, and K. Stark for critical reading of this manuscript, and members of the Horvitz laboratory for helpful discussions. Oligonucleotides were prepared by the Massachusetts Institute of Technology Biopolymers Facility. Some strains used in this work were provided by the Caenorhabditis Genetics Center (University of Minnesota, St. Paul), which is funded by the National Institutes of Health Center for Research Resources. This work was supported by United States Public Health Service research grant GM24663. L.B. was supported by predoctoral fellowships from the National Science Foundation and the Markey Foundation. H.R.H. is an Investigator of the Howard Hughes Medical Institute.
ABBREVIATIONS
- GABA
γ-aminobutyric acid
- HSN
hermaphrodite-specific neuron
Footnotes
References
- 1.Tessier-Lavigne M, Goodman C S. Science. 1996;274:1123–1133. doi: 10.1126/science.274.5290.1123. [DOI] [PubMed] [Google Scholar]
- 2.Hedgecock E M, Culotti J G, Thomson J N, Perkins L A. Dev Biol. 1985;111:158–170. doi: 10.1016/0012-1606(85)90443-9. [DOI] [PubMed] [Google Scholar]
- 3.Desai C, Garriga G, McIntire S L, Horvitz H R. Nature (London) 1988;336:638–646. doi: 10.1038/336638a0. [DOI] [PubMed] [Google Scholar]
- 4.McIntire S L, Garriga G, White J, Jacobson D, Horvitz H R. Neuron. 1992;8:307–322. doi: 10.1016/0896-6273(92)90297-q. [DOI] [PubMed] [Google Scholar]
- 5.Brenner S. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Babu P, Brenner S. Mutat Res. 1981;82:269–273. doi: 10.1016/0027-5107(81)90156-1. [DOI] [PubMed] [Google Scholar]
- 7.Hodgkin J, Edgley M, Riddle D L, Albertson D G the Community of C. elegans Researchers, editors. In: The Nematode Caenorhabditis elegans. Wood W B, editor. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. pp. 491–586. [Google Scholar]
- 8.Hodgkin J, Papp A, Pulak R, Ambros V, Anderson P. Genetics. 1989;123:301–313. doi: 10.1093/genetics/123.2.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 10.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 11.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current Protocols in Molecular Biology. New York: Wiley; 1991. [Google Scholar]
- 12.Coulson A, Sulston J, Brenner S, Karn J. Proc Natl Acad Sci USA. 1986;83:7821–7825. doi: 10.1073/pnas.83.20.7821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Coulson A, Waterston R, Kiff J, Sulston J, Kohara Y. Nature (London) 1988;335:184–186. doi: 10.1038/335184a0. [DOI] [PubMed] [Google Scholar]
- 14.Fire A, Kondo K, Waterston R. Nucleic Acids Res. 1990;18:4269–4270. doi: 10.1093/nar/18.14.4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim S K, Horvitz H R. Genes Dev. 1990;4:357–371. doi: 10.1101/gad.4.3.357. [DOI] [PubMed] [Google Scholar]
- 16.Williams B D, Schrank B, Huynh C, Showkeen R, Waterston R H. Genetics. 1992;131:609–624. doi: 10.1093/genetics/131.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 18.Lennon G G, Auffray D, Polymeropoulos M, Soares M B. Genomics. 1996;33:151–152. doi: 10.1006/geno.1996.0177. [DOI] [PubMed] [Google Scholar]
- 19.Fire A, Harrison S W, Dixon D. Gene. 1990;93:189–198. doi: 10.1016/0378-1119(90)90224-f. [DOI] [PubMed] [Google Scholar]
- 20.Han M, Sternberg P W. Genes Dev. 1991;5:2188–2198. doi: 10.1101/gad.5.12a.2188. [DOI] [PubMed] [Google Scholar]
- 21.Kramer J M, French R P, Park E C, Johnson J J. Mol Cell Biol. 1990;10:2081–2089. doi: 10.1128/mcb.10.5.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Finney M, Ruvkun G. Cell. 1990;63:895–905. doi: 10.1016/0092-8674(90)90493-x. [DOI] [PubMed] [Google Scholar]
- 23.Meneely P M, Herman R K. Genetics. 1979;92:99–115. doi: 10.1093/genetics/92.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greenwald I S, Horvitz H R. Genetics. 1980;96:147–164. doi: 10.1093/genetics/96.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pulak R, Anderson P. Genes Dev. 1993;7:1885–1897. doi: 10.1101/gad.7.10.1885. [DOI] [PubMed] [Google Scholar]
- 26.Klein R D, Meyer B J. Cell. 1993;72:349–364. doi: 10.1016/0092-8674(93)90113-5. [DOI] [PubMed] [Google Scholar]
- 27.Krause M, Hirsh D. Cell. 1987;49:753–761. doi: 10.1016/0092-8674(87)90613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boguski M S, Lowe T M, Tolstoshev C M. Nat Genet. 1993;4:332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- 29.Kyte J, Doolittle R F. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 30.Chalfie M, Tu Y, Euskirchen G, Ward W W, Prasher D C. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 31.Miller D M, III, Niemeyer C J, Chitkara P. Genetics. 1993;135:741–753. doi: 10.1093/genetics/135.3.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller D M, III, Niemeyer C J. Development. 1995;121:2877–2886. doi: 10.1242/dev.121.9.2877. [DOI] [PubMed] [Google Scholar]
- 33.Maricq A V, Peckol E, Driscoll M, Bargmann C I. Nature (London) 1995;378:78–81. doi: 10.1038/378078a0. [DOI] [PubMed] [Google Scholar]
- 34.Wightman B, Clark S G, Taskar A M, Forrester W C, Maricq A V, Bargmann C I, Garriga G. Development. 1996;122:671–682. doi: 10.1242/dev.122.2.671. [DOI] [PubMed] [Google Scholar]
- 35.Newton A. J Biol Chem. 1995;270:28495–28498. doi: 10.1074/jbc.270.48.28495. [DOI] [PubMed] [Google Scholar]
- 36.Li C, Ullrich B, Zhang J, Anderson R G W, Brose N, Sudhof T C. Nature (London) 1995;375:594–599. doi: 10.1038/375594a0. [DOI] [PubMed] [Google Scholar]