Abstract
Human mesenchymal stem cells (hMSCs) have been shown to trans-differentiate into neuronal-like cells by culture in neuronal induction media, although the mechanism is not well understood. Topography can also influence cellular responses including enhanced differentiation of progenitor cells. As extracellular matrix (ECM) in vivo comprises topography in the nanoscale, we hypothesize that nanotopography could influence stem cell differentiation into specific non-default pathways, such as transdifferentiation of hMSC. Differentiation and proliferation of hMSCs were studied on nano-gratings of 350nm width. Cytoskeleton and nuclei of hMSCs were aligned and elongated along the nano-gratings. Gene profiling and immunostaining showed significant up-regulation of neuronal markers such as microtubule-associated protein 2 (MAP2) compared to unpatterned and micro-patterned controls. The combination of nanotopography and biochemical cues such as retinoic acid further enhanced the upregulation of neuronal marker expressions, but nanotopography showed a stronger effect compared to retinoic acid alone on unpatterned surface. This study demonstrated the significance of nanotopography in directing differentiation of adult stem cells.
Keywords: Nanotopography, human mesenchymal stem cells, neuronal differentiation, nano-imprinting, cytoskeleton rearrangement
Introduction
Topography of extracellular microenvironment can influence cellular responses from attachment and migration to differentiation and production of new tissue [1-5]. Cells in their natural environment interact with extracellular matrix (ECM) components in the nanometer scale [6]. Recent findings underscore the phenomenon that mammalian cells do respond to nanoscale features on a synthetic surface[7-12]. Nanoscaled topography of synthetic materials has been receiving increasing attention because of its resemblance to in vivo surroundings. We have demonstrated that the morphology, proliferation and cell migration was significantly influenced when bovine pulmonary smooth muscle cells were cultured on a nano-imprinted gratings of 350 nm linewidth, in sub-cellular dimensions [13]. As surface nano-topography could induce pronounced changes to cell shape, and consequently gene expression, topographical cues could potentially mediate differentiation of stem stems into various cell types such as neuron and muscle.
Human MSCs were initially believed to be restricted to mesenchymal lineages. After Woodbury et al. demonstrated the differentiation of rat and human bone marrow stromal cells into neurons[14], neuronal differentiation of hMSCs has been studied by other groups. The transdifferentiation can be induced by neuronal induction medium [14-17] and/ or cell contact with neurons [18]. The mechanism responsible for the transdifferentiation of mesenchymal stem cells to neurons, which are non-mesenchymal derivatives, is not well understood.
In the present study, we have used human MSC as a model system to study nano-topography-induced cell differentiation. When cultured on nanopatterns with gratings of 350 nm linewidth in MSC proliferation medium, the cell bodies and nuclei of the hMSC were significantly elongated. Gene expression and microarray study showed that the neuronal and muscular gene markers were significantly upregulated. Further investigation in the neuronal marker expression showed that mature neuronal markers such as microtubule associated protein 2 (MAP2) and β-Tubulin III (Tuj1) were also detected. The study showed that nanotopography, with or without the presence of biochemical signals, played an important role in regulating stem cell differentiation.
Materials and Methods
Production of nanograting with soft lithography
The nanopattern was first produced by nano-imprinting as previously described [13]. The nanopattern was reproduced on poly(dimethylsiloxan) (PDMS) using soft lithography on the nanoimprinted poly(methyl methacrylate) (PMMA) -coated Si master mold. The gratings on the nanoimprinted PMMA master molds were 350nm in depth, either with 350nm width and 700nm pitch, 1μm width and 2μm pitch, or 10μm in width and 20μm pitch. Patterned PDMS samples were coated with bovine collagen I (BD Biosciences) at 20μg/cm2 to increase cell adhesion on the hydrophobic PDMS surface.
Human mesenchymal stem cell culture
Human mesenchymal stem cells (Poietics™ hMSC from human bone marrow, CD105+, CD166+, CD29+, CD44+, CD14−, CD34−, CD45−, Cambrex, NJ) were cultured and expanded in MSCGM medium (Cambrex). hMSCs used in the experiments were at passage 6−9. The hMSCs were seeded on the nanopatterned or unpatterned PDMS at 6×103 cell/cm2. Glass cover slip (grade 1, Fisher Scientific) and unpatterned PDMS were used as controls.
For neuronal induction culture with retinoic acid, a stock solution of retinoic acid (RA, Sigma) was prepared in dimethylsulfoxide to 1.8mg/ml. After hMSCs were seeded onto the samples, RA was added to the culture medium at a final concentration of 300ng/ml (1μM) or 9μg/ml (30μM).
For F-actin cytoskeleton disruption, 2μM of cytochalasin D (Sigma) was added into the MSCGM medium one day after cell-seeding. The concentration used (2μM) was decided after determining the LD50 of cytochalasin D for hMSCs in 6-day exposure of MSCGM using the WST-1 cytotoxicity assay (Roche).
Scanning electronic microscopy preparation
Samples with hMSCs cultured were fixed in 4% paraformaldehyde, washed in 0.1 M sodium cacodylate, and post-fixed in 2% OsO4 in 0.1 M Na cacodylate, pH 7.2. After post-fixation, the samples were dehydrated in a graded ethanol series. Critical point-dried samples were sputter-coated with a 5 nm coating of chromium and viewed with a LEO FESEM (LEO 1550) (LEO Electronic Microscopy Inc.) at 1 kV.
Fluorescent staining of F-actin
F-actin was fluorescent-stained in samples fixed in 4% paraformaldehyde with Oregon Green 488 phalloidin (Molecular Probes, Eugene, OR) as previously described[13], and the nucleus was stained with 4',6-Diamidino-2-phenylindole (DAPI, Molecular Probes) stain. Samples were inspected by confocal microscopy.
Elongation and alignment characterization
Five samples of each nanopatterned PDMS, unpatterned PDMS and glass cover slip seeded with hMSCs were fixed at day 7. Eight separate regions of each sample were photographed (20× or 40×). The images were analyzed with ImageJ NIH image processing software (Bethesda, MD).
The elongation (E) parameter describes the extent to which the equimomental eclipse is lengthened or stretched out[8]. It was calculated as the ratio of the long axis over the short axis minus one. Alignment is defined as to how well the long axis of an elongated cell, with E >4, is oriented with respect to the grating. Cells were considered aligned if the angle between the long axis and the grating was <15 degrees. The percentage of cell alignment and the E-factor were measured. For each type of sample an average of 300 cells was counted.
Immunofluorescent staining
Fixed samples were stained according to a standard immunofluorescent staining procedure. The primary antibodies used were mouse anti-Tuj1 antibody diluted 1:1000 (Covance) in tris-buffered saline (TBS) and rabbit anti-GFAP antibody diluted 1:2000 (Dako), mouse anti-nestin (Dako) diluted 1:1000, or mouse anti-synaptophysin (Dako) 1:1000 and chicken anti-MAP2 (Abcam) diluted 1:10000. The secondary antibodies used were Alexa-Fluor546 goat anti-mouse antibody at 1:750, and Alexa-Fluor488 goat anti-rabbit antibody at 1:750 (Molecular Probes), Alexa-Fluor488 anti-chicken antibody at 1:750 and Alexa-Fluor546 goat anti-mouse antibody at 1:750. DAPI was used as a counter-stain for the nucleus. Samples were inspected by confocal microscopy.
For 5-bromo-2-deoxyuridine (BrdU) staining, BrdU (Sigma) was added to the medium 4 h before fixation. Samples were fixed in 4% paraformaldehyde and permeabilized with a 15-min incubation in 1% Triton X100 and a 10-min incubation in 4N HCl. The samples were stained with mouse anti-BrdU antibody (Developmental Studies Hybridoma Bank) with Alexa Fluor 546-conjugated goat anti-mouse antibody as the secondary antibody. Four samples of each patterned and unpatterned PDMS were examined. Thirty separate regions of each sample were photographed. The images were analyzed with ImageJ (NIH software).
Apoptosis detection with TUNEL Assay
Fixed samples were permeablized with 0.1% Triton X and 0.1% sodium citrate in PBS. Apoptosis was detected with DeadEnd™ Fluorometric TUNEL system (Promega, Madison, WI). Samples were inspected by fluorescent microscopy.
mRNA expression profiles
Total RNA was isolated from hMSCs at 7 and 14 days. The RNA isolated from hMSCs cultured on tissue culture polystyrene (TCPS) served as controls. RNA was isolated with a Qiagen RNeasy Mini Kit. Synthesis and amplification of DNA were performed with the Qiagen one-step RT-PCR kit. Twenty-four pairs of primers were tested; their sequences are listed in supplement table 1 [19]. Human β-actin was used as the endogenous control. Thirty-five cycles of PCR were carried out with an annealing temperature of 54°C and incubation for 30s. PCR reactions were resolved on 1.2% agarose gel in TAE buffer.
Real-time PCR
The levels of MAP2 and β-actin in hMSCs cultured on patterned-PDMS and unpatterned PDMS were measured by real-time PCR. hMSCs cultured on TCPS served as a control. Reverse transcription was performed with a Sensiscript RT kit (Qiagen) with a poly-A 16 oligomer. PCR was performed using TaqMan® Universal PCR Master Mix (Applied Biosystems) with human MAP2 Taqman® Gene Expression Assays primer and probes (Applied Biosystems) and human β-actin (Applied Biosystems) as endogenous control. The PCR cycling consisted of 40 cycles of amplification of the template DNA with a primer annealing at 60°C (15 s). The real-time PCR was performed on an Applied Bioscience Thermocycler (7900). The RNA levels were normalized to the β-actin mRNA level. The data are presented as the normalized RNA level divided by the normalized RNA level expressed in the hMSCs control on TCPS at day 7.
Microarray analysis
The RNA (3 μg from each sample set) was converted to labeled cDNA probe with AmpoLabeling-LPR kit (Superarray) following the manufacturer protocol for chemiluminescent detection (Superarray).
The labeled cDNA probes of hMSCs cultured on nanopatterned-PDMS, or plain-PDMS and TCPS were hybridized to human neurotrophin and receptors cDNA microarrary, human extracellular matrix and adhesion molecules cDNA microarray and human stem cell cDNA microarray. Hybridization was performed in a regular hybridization chamber, as recommended by the manufacturer. The recommended washes were followed. Signal of the various cDNA microarray was detected by chemiluminescent kit, and the image was acquired by exposure onto X-ray film for 1−2 minutes. Data from the microarray were normalized to the averaged signals of housekeeping genes that were spotted as internal controls on each slide. For comparison of expression level, the relative expression of each gene was either considered as up-regulated (nanopattern:unpattern signal ratio >2.0), down-regulated (nanopattern:unpattern signal ratio<0.5), unchanged, or not expressed. Selected up- or down-regulated genes were verified by real-time PCR.
Micro-scale versus Nano-scale topography
The effect of microtopography and nanotopography was examined on nano- / micro-gratings with various widths of 350nm, 1μm and 10μm. The proliferation and differentiation of hMSCs were monitored with BrdU incorporation and MAP2 gene expression level.
Data Analysis
All data are presented as mean ± SD. Student’s t-test and ANOVA were used to evaluate the statistical significance where indicated. Significance level was set at p < 0.01.
Results
hMSC morphology on the nanopattern
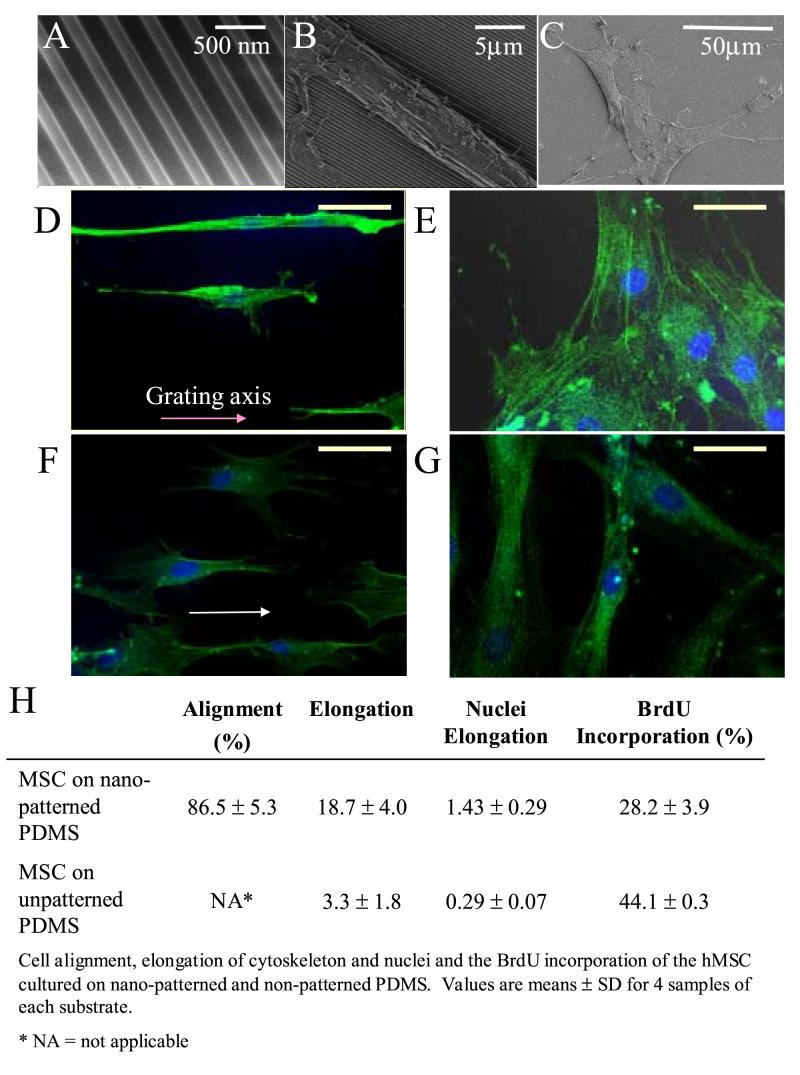
When we cultured hMSCs on poly(dimethylsiloxan) (PDMS) with gratings of 350nm linewidth (Figure 1A) in proliferation medium, the cell bodies and nuclei of the hMSCs were elongated and aligned along the grating axis (Figure 1B, D). In contrast, hMSCs cultured on unpatterned-PDMS showed neither elongation nor orientation at either low or high cell densities (Figure 1C, E). The F-actin fibers were mostly stretched along the long axis of the cells. Cell alignment could be observed within 2 hours after cell seeding, as soon as cell attachment begun, while cell elongation (when factor E was significantly higher than the control) could be observed after 3−4 hours. (Data not shown) In the presence of 1μM retinoic acid (RA), the hMSCs on the nanopattern produced more protrusions while the cell body and the protrusions remained aligned along the gratings (Figure 1F). However, no morphological change was observed on the hMSCs cultured on unpatterned-PDMS in 1μM RA.
Figure 1.
Changes in morphology and proliferation of human mesenchymal stem cells (hMSCs) cultured on nano-gratings. Scanning electron micrographs of (A) PDMS nano-patterned by replica molding; hMSCs cultured on (B) nano-patterned PDMS and (C) unpatterned PDMS. Confocal micrographs of F-actin-stained hMSCs on (D) nano-patterned PDMS and (E) unpatterned PDPS in hMSC proliferation medium; (F) nano-patterned PDMS and (G) unpatterned PDMS cultured in presence of 1μM of retinoic acid (RA). Bar = 500nm for A, 5 μm for B, 50 μm for C-G. (H) Cell alignment, elongation of cytoskeleton and nuclei and the BrdU incorporation of the hMSCs cultured on nano-patterned and unpatterned PDMS. Values are means ± SD for 4 samples of each substrate. * NA = not applicable
Morphological characterization and cell proliferation
The percentage of cell alignment and elongation of the cytoskeleton and nuclei on different surfaces is shown in Figure 1H. The alignment of cells in nanopatterned-PDMS was 86.5 ± 5.3%. Both the cytoskeleton and the nuclei of the hMSCs were elongated on the nanopattern. The hMSCs cultured on nanopatterned-PDMS were significantly elongated compared to unpatterned-control (E=18.7±4.0 vs. 3.3±1.8, p<0.05). Meanwhile, the E-factor of the nuclei of hMSCs cultured on nanopatterned PDMS was 1.43 ± 0.29 versus 0.29 ± 0.07 for hMSCs on unpatterned PDMS. The proliferation of hMSCs on the nanopattern was significantly lower, 26.9 ± 3.1% of BrdU incorporation during 4 hours of incubation, compared to 35.7 ± 7.6% on unpatterned surfaces (p < 0.05).
Immunostaining of neuronal markers
In addition to morphological changes, mature neuronal markers such as MAP2 and β-Tubulin III (Tuj1) were also detected in hMSCs cultured on nanopatterned-samples, with or without RA-induction. hMSCs cultured on unpatterned-surface showed these expressions only with RA in the culture medium (Figure 2A-B). GFAP expression was only weakly detected on the samples. Nestin expression was detected on all samples, although the expression level was weak. Synaptophysin expression was also detected in hMSCs cultured on nanopattern, with or without RA, but not on unpatterned-control, suggesting synapse formation in the cells cultured on nanopattern (Figure 2C).
Figure 2.
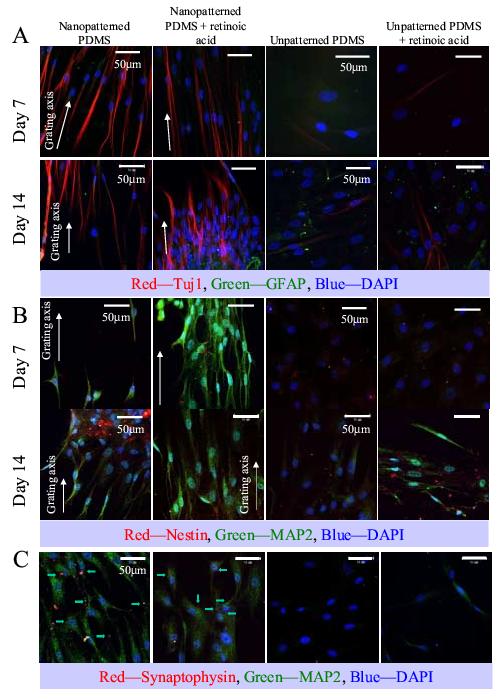
Immunofluorescent staining of (A) Tuj1 and GFAP, (B) MAP2 and nestin and (C) synaptophysin and MAP2 of hMSCs cultured on nano-patterned PDMS, nano-patterned PDMS in the presence of retinoic acid (RA), unpatterned PDMS and unpatterned PDMS in the presence of RA. (A) Tuj1 is shown in red, GFAP in green; (B) nestin is shown in red, MAP2 in green and (C) synaptophysin is shown in red and marked by arrows, while MAP2 is shown in green. In all panels the DAPI nuclei counter-stain is shown in blue, bar = 50 μm. The direction of the gratings on the nano-patterned PDMS is indicated with a white arrow. Images were taken at representative areas of the samples.
When hMSCs cultured on the samples approached high confluence, cell elongation was observed in random areas on unpatterned samples. This could contribute to the observation of isolated cells with weak Tuj1 expression on unpatterned samples without RA at day 14.
Gene expression analysis
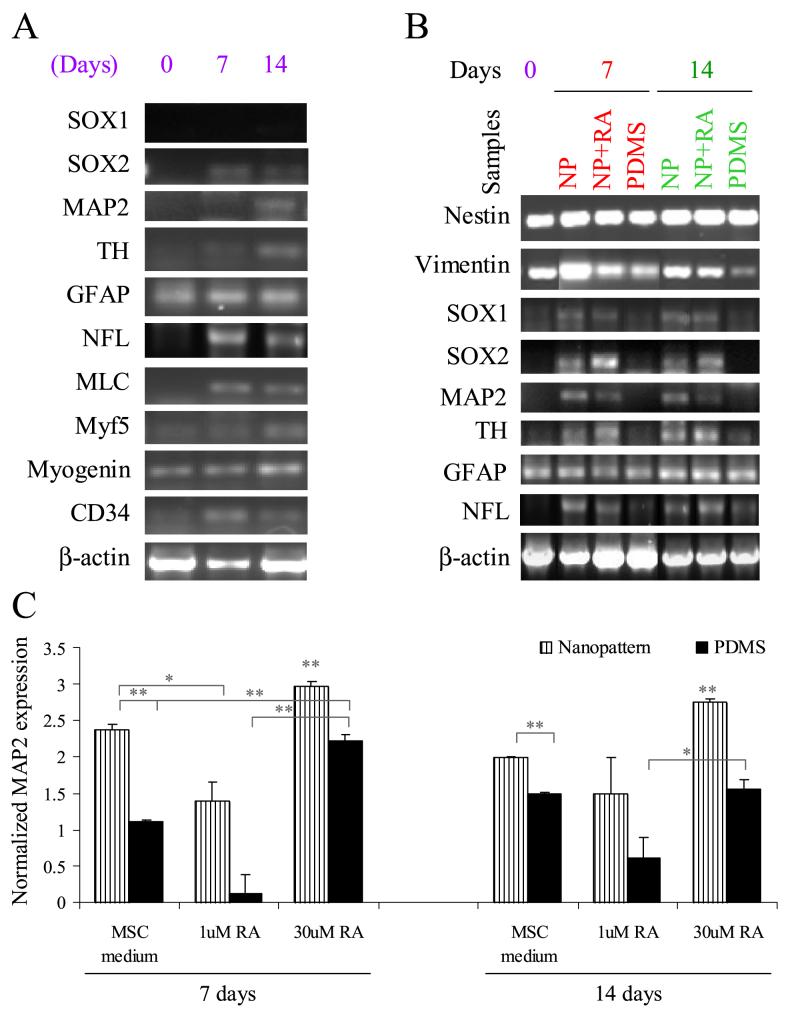
RT-PCR showed alteration of gene expression by the hMSCs cultured on the nanopatterns in comparison to unpatterned tissue-culture polystyrene (TCPS) and PDMS controls at day 7 and 14 (Figure 3A). The neuronal markers SOX2, MAP2, neurofilament light peptide (NFL) and tyrosine hydroxylase (TH) were up-regulated on the nanopatterned-PDMS in hMSCs proliferation medium. However, expression of SOX1 was not detected in either nanopatterned or control samples. Muscle markers such as myosin light chain and myf5 were up-regulated, as was the vascular marker CD34. GFAP, myogenin and the β-actin control were unaffected by the nanopattern.
Figure 3.
(A) Multi-lineage gene expression analysis of hMSCs cultured on nano-patterned PDMS and the unpatterned control in proliferation medium at day 7 and day 14. (B) Neuronal gene markers analysis of hMSCs cultured on nano-patterned (NP) PDMS in proliferation medium or in the presence of retinoic acid (RA), and unpatterned PDMS (PDMS). hMSCs cultured on TCPS were used as the day 0 control. (C) Quantitative analysis of MAP2 expression in hMSCs cultured on nano-patterned PDMS in proliferation medium or in the presence of RA and on unpatterned PDMS and tissue culture polystyrene (TCPS) surface control on day 7 and day 14. * p < 0.01 and ** p < 0.001, two-tailed t-test. The expression of MAP2 was first normalized with the expression of endogenous control β-actin, then normalized with the MAP2 expression of unpatterned hMSC control at day 7.
Next, we examined the expression of neuronal gene markers in the neuronal induction culture with RA (Figure 3B). The expression of the neuronal markers, such as SOX1 and SOX2, was future enhanced, but the expression of MAP2, NFL and TH was not affected by the RA-induction. GFAP, nestin, vimentin and control β-actin were detected under all culture conditions. MAP2 expression was further analyzed by quantitative real-time PCR analysis (Figure 3C). The expression of MAP2 was significantly enhanced in the hMSCs cultured on the nanopatterned-PDMS with or without RA in comparison with unpatterned-controls at day 7 (p<0.001, two-tailed t-test). Similar trend was observed at day 14. When hMSCs were cultured in the presence of a high concentration of RA (30μM), the expression of MAP2 on nanopatterne-samples with RA was higher than the samples with 1μM RA on day 7 and 14 days. Similarly, the MAP2 expression of the hMSCs cultured on unpatterned-PDMS in the presence of 30μM of RA was enhanced. Interestingly, MAP2 expression was observed on the unpatterned controls either with or without RA at 14 days with immunostaining in Figure 2, the quantitative transcriptional expression was increased compared to 7 days but not significantly up-regulated. The low level of expression might be due to non-specific differentiation of hMSCs or caused by the cell alignment upon confluence.
A broad gene profiling of 474 gene were analyzed with focus-microarray (Table 1) and the expression of selected genes were verified by real-time PCR (Supplement table 2). Genes related to neuronal differentiation and neuronal function, such as brain-derived neurotrophic factor, synaptotagmin I, tubulin β3, were regulated by nanotopography, in accord with the immuno-staining and morphological observation. Interestingly, among the changes in neurotrophin and receptor genes, genes related to glial differentiation, such as interleukin 6, Stat1 and Stat3, were downregulated, implying nanotopography-induced differentiation might have a preference in the neuronal pathway over the glial differentiation pathway.
Table 1.
Up-/down-regulation of gene expression in microarray analysis
| Up-regulation NP/Control > 2.0 | Downregulation: NP/Control < 0.5 |
|---|---|
|
Extracellular Matrix | |
| Caspase 8, apoptosis-related cysteine protease | Caspase 9, apoptosis-related cysteine protease |
| Fibrinogen, B beta polypeptide | Caveolin 1, caveolae protein, 22kDa |
| Fibronectin 1 | Collagen, type I, alpha 1 |
| Integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) | Collagen, type IV, alpha 2 |
| Cathepsin B | |
| Integrin, alpha 5 (fibronectin receptor, alpha polypeptide) | Cathepsin D (lysosomal aspartyl protease) |
| Integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61) | Intercellular adhesion molecule 1 (CD54), human rhinovirus receptor |
| Integrin, beta 5 | Integrin, alpha 1, alpha 4, alpha 8, beta 8 |
| Laminin, beta 1 | Laminin, gamma 1 (formerly LAMB2) |
| Matrix metalloproteinase 13 (collagenase 3) | Plasminogen activator, urokinase |
| Matrix metalloproteinase 14 (membrane-inserted) | Secreted protein, acidic, cysteine-rich (osteonectin) |
| Neuronal cell adhesion molecule | Secreted phosphoprotein 1 (osteopontin, bone sialoprotein I, early T-lymphocyte activation 1) |
| Thrombospondin 1 | |
| Tissue inhibitor of metalloproteinase 2 | |
| Vitronectin (serum spreading factor, somatomedin B, complement S-protein) | |
|
Stem cells | |
| BMP 2, BMP4, BMP15, BMP receptor, type IA | ATP-binding cassette, sub- family G (WHITE), member 2 |
| Catenin (cadherin-associated protein) a2 | Actin, alpha2, smooth muscle, aortab |
| Cadherin 2, type 1, N-cadherin (neuronal) | Bone morphogenetic protein receptor, type IA, type I |
| Cyclin-dependent kinase inhibitor 1B (p27, Kip1), 2D (p19, inhibits CDK4) | TAP binding protein (tapasin) |
| Fibroblast growth factor 1, 7 (keratinocyte growth factor), 17.19 | |
| Cystatin C (amyloid angiopathy and cerebral hemorrhage) | Interleukin 6 (interferon, beta 2) |
| EGF (beta-urogastrone) | Integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor) |
| FGF 15, 2 (basic), 20, 3, 5 | Integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor), alpha 4, alpha 8 |
| Frizzled homolog 3, 9 | |
| Growth differentiation factor 5, 8 | Mdm2, transformed 3T3 cell double minute 2, p53 binding |
| Gap junction protein, a7 | NGF |
| Insulin-like growth factor 2 (somatomedin A) | Platelet-derived growth factor receptor, alpha polypeptide |
| Integrin, b 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12), b 3, b 5 | Platelet/endothelial cell adhesion molecule (CD31 antigen) |
| Retinoblastoma-like 1 (p107) | |
| Keratin 8 | Tenascin C (hexabrachion) |
| Leukemia inhibitory factor receptor | Transforming growth factor, beta 1 |
| Microtubule-associated protein 1B | |
| Nodal homolog | |
| Neural cell adhesion molecule1 (NCAM1) | |
| Neurofilament, light polypeptide | |
| Neurotrophic tyrosine kinase, receptor, type 2 | |
| Synaptotagmin I | |
| Transforming growth factor, beta I, beta receptor II (70/80kDa) | |
| Sox3, 13, 18 | |
| Tubulin, b 3 | |
| Wnt-5A, 6, 8A, 7B | |
|
Neurotrophin | |
| Apoptotic protease activating factor | Fibroblast growth factor receptor 1 (fms-related tyrosine kinase 2, Pfeiffer syndrome) |
| Brain-derived neurotrophic factor | |
| MADS box transcription enhancer factor 2, polypeptide C (myocyte enhancer factor 2C) | GDNF family receptor alpha 1 |
| Glia maturation factor, beta | |
| Neuregulin 1 | Interleukin 6 (interferon, beta 2) |
| Neuregulin 2 | Interleukin 6 signal transducer (gp130, oncostatin M receptor) |
| Proenkephalin | Nerve growth factor, beta polypeptide |
| Prepronociceptin | Neurotrophin 3 |
| Pleiotrophin (heparin binding growth factor 8, neurite growth-promoting factor 1) | Pancreatic polypeptide receptor 1 |
| Persephin | |
| Fibroblast growth factor receptor substrate 3 | Signal transducer and activator of transcription 1, 91kDa |
| Transforming growth factor, beta 1 (Camurati-Engelmann disease) | Signal transducer and activator of transcription 3 (acute-phase response factor)- Stat3 |
| VGF nerve growth factor inducible | Transforming growth factor, alpha |
| CD40 antigen (TNF receptor superfamily member 5) Stat5 | |
The gene expression related to the ECM, adhesion molecule signaling and cell-cycle were also regulated, providing evidence that the induction of neuronal differentiation was associated with alteration of ECM signaling or cytoskeleton arrangement.
Nano-versus Micro-scale topography
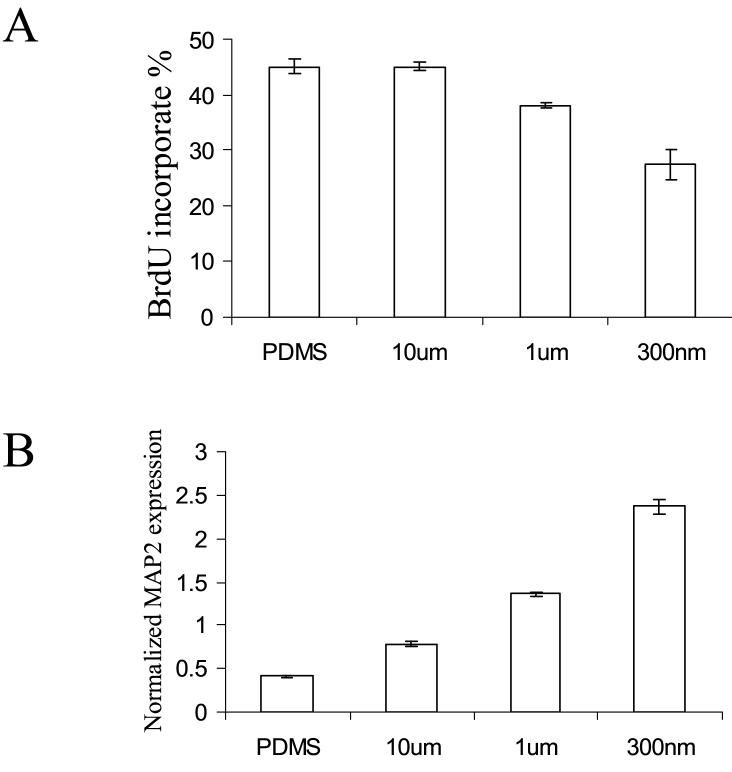
Although stem cells interact with ECM components in nano-scale, the question of whether nanometer scale is necessary to produce significant effect on stem cell behavior has been scarcely studied. The size effect was examined on nano-/micro-gratings with various widths of 350nm, 1μm and 10μm. A significant width dependency of the proliferation and differentiation was observed on nanopatterns (Figure 4). The BrdU incorporation percentage was significantly downregulated, while the MAP2 expression was also significantly upregulated in hMSCs cultured on the nano-gratings. The results demonstrated nanopattern could have a more significant effect in stem-cell behavior compared to micro-pattern in terms of differentiation and proliferation.
Figure 4.
Proliferation and differentiation of the hMSCs on gratings with various widths. (A) Brdu incorporation percentage after a 4-hour pre-fix incubation and (B) quantitative analysis of MAP2 expression in hMSCs cultured on gratings with widths of 350nm, 1μm and 10μm.
Discussion
In this study, we demonstrated that nanotopography alone could induce a significant up-regulation of neuronal marker, hence suggesting the induction into neuronal lineage, of human mesenchymal stem cells. When the hMSCs were cultured on nano-gratings, which size was at least one order of magnitude smaller than the cell body, the hMSCs significantly aligned and elongated on the nano-gratings. The cytoskeleton as well as the nuclei of the cells were significantly aligned and elongated along the direction of the grating axis. In addition to morphological differences, upregulation of neuronal markers such as Tuj1, MAP2 and GFAP were observed. Collectively these findings established the importance of nanotopographical cues alone on the neuronal differentiation in hMSCs.
MSCs were believed to differentiate into mesenchyme-restricted lineages until Woodbury et al. demonstrated the differentiation of rat and human bone marrow stromal cells into neurons [14] in 2000. Thereafter, neuronal differentiation of the hMSCs has been studied by other groups. The mechanism responsible for the transdifferentiation of mesenchymal stem cells to neurons, which are non-mesenchymal derivatives, is not well understood. Because of the ready availability of hMSCs, their differentiation to functional neurons would be of interest to field of cell therapy. In this study, we demonstrated that nanotopography could up-regulate markers of neuronal, muscular and vascular lineages (Figure 3), suggesting the possible effect of nanotopographical cues in guiding the multipotent MSC. Since the up-regulation in neuronal markers was the most significant among the lineages examined, we further investigated the neuronal marker expression and demonstrated that neuronal differentiation could be induced by topographical cues alone even in culture medium containing no biochemically inducing factors.
Cytoskeleton rearrangement and nuclei elongation would play an important role in creating the signal transduction for the transdifferentiation. The elongation of cytoskeleton and nucleus have been correlated with changes in gene expression profile and cell differentiation in other studies[20-23]. Forces are transferred to the nucleus through actin-intermediate filament system during changes in cell shape [24]. The mechanical tension can rearrange the centromere through deformation of the nucleus[20] and mechano-transduction can affect the cellular structure and phenotypes in MSCs[25]. Cell elongation can also induce protein expression in embryonic mesenchymal smooth muscle precursor cells [23]. Similarly, various studies have shown that cell elongation and distortion of nucleus would result in changes of gene and protein expression [20, 22].
The microarray analysis revealed an alteration of gene expression involved in neuronal differentiation and neuronal function and signaling, for example, neuronal cell adhesion molecule (NCAM), brain-derived neurotrophic factor, synaptotagmin I, tubulin β3 and neuregulin during the culture on nanopatterns. Gene expression in ECM signaling was also affected, likewise the gene expression involved in cell cycle. Taken together, these data provided evidence that alteration of ECM signaling or cytoskeleton arrangement could lead to a change in gene regulation directing cell differentiation. Changes in gene regulation by nanotopography in fibroblasts have also been observed on random pattern of nano-scaled islands produced with polymer-demixing techniques[26]. The difference in cytoskeleton arrangement, cell adhesion and protein expression, such as Rac and other signaling proteins, on fibroblasts cultured on nano-islands versus flat surface are observed in cDNA microarray gene expression analysis. Further investigation would be needed to identify which components in the cytoskeleton and which signaling pathways govern the topography-directed differentiation.
The results of the present study suggest that the topographical cues could be combined with chemical cues to create a synergistic condition for neuronal differentiation. The combination of RA-induction and nanotopographical induction showed the highest MAP2 expression in the quantitative PCR study; however, the effect of nanopattern alone was greater than the effect produced by 30μM of RA on unpatterned-control, suggesting that the effect of nanotopographical induction could be stronger than biochemical induction by RA. Interestingly, we observed a dose-dependent phenomenon. In conjunction with nanotopography, high concentration of RA promoted neuronal differentiation while the low dose appeared to have the opposite effect. The suppression might be due to the interference of nanotopography-induced signal with the RA signaling. Although the concentration of 1μM of RA has been shown to be sufficient in inducing neuronal differentiation in embryonic stem cells[27] and neuronal stem cells[28], a concentration of 30μM RA is necessary to achieve the neuronal differentiation of hMSCs [29]. A higher concentration of the RA might be needed to overcome the lineage restriction of MSCs to become neuronal-like cells.
Cell morphology and gene regulation can be influenced by cell stress and / or disruption of cytoskeleton[30]. To examine this contribution in our findings, we examined apoptosis of the hMSCs with TUNEL assay. No evidence of apoptosis was observed on the nanopatterned- or unpatterned-PDMS samples (Supplement Figure 1). To examine if the up-regulation of neuronal marker expression was a result caused merely by reduced proliferation, hMSCs were cultured in reduced serum (from 10% to 1%) condition. The hMSCs cultured on TCPS in reduced serum medium did not show a significant expression of Tuj1 and MAP2 in immunostaining after a 7 day culture period (Supplement Figure 2). We also disrupted the F-actin cytoskeleton by cytochalasin-D to study the effects of cytoskeleton on neuronal gene expression (Supplement Figure 3). Neither the cytochalasin-D treatment nor the reduced serum condition enhanced neuronal markers, including TH and MAP2, in nanopatterned or unpatterned samples. To ascertain the archetypical characteristics of these hMSCs, their ability to undergo in vitro osteogenesis and chondrogenesis, demonstrating differentiation to mesenchymal cell fates, has also been confirmed (Supplement figure 4). Collectively, the data indicate that the neuronal differentiation observed in this study is nanotopography-specific.
The neuronal differentiation of hMSCs had been evaluated only by the morphological changes and the gene and protein expression of neuronal markers in this study. The functional assessment including synaptic transmission and electrophysiological properties of these differentiated hMSCs would be of interest for future studies.
Nanotopography has been demonstrated to enhance differentiation of progenitor cells into their programmed pathway[1, 2]. Nevertheless, the novelty of this work lies in the application of nanotopography to direct adult stem cells to differentiate into a non-default pathway. Because of the availability MSCs, their differentiation to functional neurons would be of interest to many for cell-therapy. Also of significance is the finding that the topographical cue has to be at the nanoscale in order to exert a significant influence on the directed differentiation. The results demonstrated in a systematic manner that nanopattern, in comparison to micropattern, exerted a significantly stronger effect on stem cell behavior in terms of morphology, proliferation, and differentiation.
Acknowledgement
The authors would like to acknowledge NIH for funding support (EB003447) and Dr. RM Reano for his help in providing the nanoimprinted molds.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary Material
Supplement Table 1. RT-PCR primers for gene profile analysis of hMSCs cultured on nano-gratings. Primer sequences for neurofilament light (NFL) were obtained from NCBI NM_006158; β-actin sequences were provided by Clontech; and the other sequences were obtained from [19].
Supplement Table 2. Quantitative real-time PCR analysis of the selected up- / down-regulation of genes observed in microarray analysis.
Supplement Figure 1. TUNEL assay of hMSCs cultured on (A) nanopatterned and (B) unpattern PDMS, and (C) the positive control with DNA fragmentation induced by DNase I and (D) negative control on glass cover slip. Bar = 50μm
Supplement Figure 2. F-actin staining (A) and immunostaining of (B) Tuj1 and GFAP and (C) MAP2 and nestin of hMSCs cultured on unpatterned PDMS in the reduced-serum (1%) condition at day 7. Bar = 100μm.
Supplement Figure 3. Neuronal gene markers analysis of hMSCs cultured on nano-patterned or unpatterned PDMS with 6 days (6D) or 1 day (1D) exposure to 2μM of cytochalasin D, in MSC proliferation medium, or in reduced-serum condition (1% serum).
Supplement Figure 4. (A) chondrogenic gene markers and (B) osteogenic gene markers analysis of the hMSCs cultured in chondrogenic medium (CM) or osteogenic induction medium (OM), respectively, and the control sample cultured in MSC proliferation medium (PM) on TCPS for 4 weeks.
References
- 1.Dalby MJ, McCloy D, Robertson M, Agheli H, Sutherland D, Affrossman S, Oreffo RO. Osteoprogenitor response to semi-ordered and random nanotopographies. Biomaterials. 2006;27:2980–2987. doi: 10.1016/j.biomaterials.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 2.Silva GA, Czeisler C, Niece KL, Beniash E, Harrington DA, Kessler JA, Stupp SI. Selective differentiation of neural progenitor cells by high-epitope density nanofibers. Science. 2004;303:1352–1355. doi: 10.1126/science.1093783. [DOI] [PubMed] [Google Scholar]
- 3.Curtis A, Wilkinson C. Nantotechniques and approaches in biotechnology. Trends Biotechnol. 2001;19:97–101. doi: 10.1016/s0167-7799(00)01536-5. [DOI] [PubMed] [Google Scholar]
- 4.Curtis AS, Wilkinson CD. Reactions of cells to topography. J Biomater Sci Polym Ed. 1998;9:1313–1329. doi: 10.1163/156856298x00415. [DOI] [PubMed] [Google Scholar]
- 5.Wilkinson CDW. Nanostructures in Biology. Microelectron. Eng. 1995;27:61–65. [Google Scholar]
- 6.Abrams GA, Goodman SL, Nealey PF, Franco M, Murphy CJ. Nanoscale topography of the basement membrane underlying the corneal epithelium of the rhesus macaque. Cell Tissue Res. 2000;299:39–46. doi: 10.1007/s004419900074. [DOI] [PubMed] [Google Scholar]
- 7.Dalby MJ, Riehle MO, Johnstone H, Affrossman S, Curtis AS. In vitro reaction of endothelial cells to polymer demixed nanotopography. Biomaterials. 2002;23:2945–2954. doi: 10.1016/s0142-9612(01)00424-0. [DOI] [PubMed] [Google Scholar]
- 8.Andersson AS, Backhed F, von Euler A, Richter-Dahlfors A, Sutherland D, Kasemo B. Nanoscale features influence epithelial cell morphology and cytokine production. Biomaterials. 2003;24:3427–3436. doi: 10.1016/s0142-9612(03)00208-4. [DOI] [PubMed] [Google Scholar]
- 9.Thapa A, Webster TJ, Haberstroh KM. Polymers with nano-dimensional surface features enhance bladder smooth muscle cell adhesion. J Biomed Mater Res. 2003;67A:1374–1383. doi: 10.1002/jbm.a.20037. [DOI] [PubMed] [Google Scholar]
- 10.Dalby MJ, Gadegaard N, Riehle MO, Wilkinson CD, Curtis AS. Investigating filopodia sensing using arrays of defined nano-pits down to 35 nm diameter in size. Int J Biochem Cell Biol. 2004;36:2015–2025. doi: 10.1016/j.biocel.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 11.Miller DC, Thapa A, Haberstroh KM, Webster TJ. Endothelial and vascular smooth muscle cell function on poly(lactic-co-glycolic acid) with nano-structured surface features. Biomaterials. 2004;25:53–61. doi: 10.1016/s0142-9612(03)00471-x. [DOI] [PubMed] [Google Scholar]
- 12.Dalby MJ, Riehle MO, Johnstone HJ, Affrossman S, Curtis AS. Polymer-Demixed Nanotopography: Control of Fibroblast Spreading and Proliferation. Tissue Eng. 2002;8:1099–1108. doi: 10.1089/107632702320934191. [DOI] [PubMed] [Google Scholar]
- 13.Yim EK, Reano RM, Pang SW, Yee AF, Chen CS, Leong KW. Nanopattern-induced changes in morphology and motility of smooth muscle cells. Biomaterials. 2005;26:5405–5413. doi: 10.1016/j.biomaterials.2005.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woodbury D, Schwarz EJ, Prockop DJ, Black IB. Adult rat and human bone marrow stromal cells differentiate into neurons. J Neurosci Res. 2000;61:364–370. doi: 10.1002/1097-4547(20000815)61:4<364::AID-JNR2>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 15.Sanchez-Ramos J, Song S, Cardozo-Pelaez F, Hazzi C, Stedeford T, Willing A, Freeman TB, Saporta S, Janssen W, Patel N, Cooper DR, Sanberg PR. Adult bone marrow stromal cells differentiate into neural cells in vitro. Exp Neurol. 2000;164:247–256. doi: 10.1006/exnr.2000.7389. [DOI] [PubMed] [Google Scholar]
- 16.Deng W, Obrocka M, Fischer I, Prockop DJ. In vitro differentiation of human marrow stromal cells into early progenitors of neural cells by conditions that increase intracellular cyclic AMP. Biochem Biophys Res Commun. 2001;282:148–152. doi: 10.1006/bbrc.2001.4570. [DOI] [PubMed] [Google Scholar]
- 17.Qian L, Saltzman WM. Improving the expansion and neuronal differentiation of mesenchymal stem cells through culture surface modification. Biomaterials. 2004;25:1331–1337. doi: 10.1016/j.biomaterials.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 18.Wislet-Gendebien S, Hans G, Leprince P, Rigo JM, Moonen G, Rogister B. Plasticity of cultured mesenchymal stem cells: switch from nestin-positive to excitable neuron-like phenotype. Stem Cells. 2005;23:392–402. doi: 10.1634/stemcells.2004-0149. [DOI] [PubMed] [Google Scholar]
- 19.Shamblott MJ, Axelman J, Littlefield JW, Blumenthal PD, Huggins GR, Cui Y, Cheng L, Gearhart JD. Human embryonic germ cell derivatives express a broad range of developmentally distinct markers and proliferate extensively in vitro. Proc Natl Acad Sci U S A. 2001;98:113–118. doi: 10.1073/pnas.021537998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dalby MJ, Riehle MO, Yarwood SJ, Wilkinson CD, Curtis AS. Nucleus alignment and cell signaling in fibroblasts: response to a micro-grooved topography. Exp Cell Res. 2003;284:274–282. doi: 10.1016/s0014-4827(02)00053-8. [DOI] [PubMed] [Google Scholar]
- 21.Itano N, Okamoto S, Zhang D, Lipton SA, Ruoslahti E. Cell spreading controls endoplasmic and nuclear calcium: a physical gene regulation pathway from the cell surface to the nucleus. Proc Natl Acad Sci U S A. 2003;100:5181–5186. doi: 10.1073/pnas.0531397100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas CH, Collier JH, Sfeir CS, Healy KE. Engineering gene expression and protein synthesis by modulation of nuclear shape. Proc Natl Acad Sci U S A. 2002;99:1972–1977. doi: 10.1073/pnas.032668799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Relan NK, Yang Y, Beqaj S, Miner JH, Schuger L. Cell elongation induces laminin alpha2 chain expression in mouse embryonic mesenchymal cells: role in visceral myogenesis. J Cell Biol. 1999;147:1341–1350. doi: 10.1083/jcb.147.6.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maniotis AJ, Chen CS, Ingber DE. Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc Natl Acad Sci U S A. 1997;94:849–854. doi: 10.1073/pnas.94.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Putnam AJ, Schultz K, Mooney DJ. Control of microtubule assembly by extracellular matrix and externally applied strain. Am J Physiol Cell Physiol. 2001;280:C556–564. doi: 10.1152/ajpcell.2001.280.3.C556. [DOI] [PubMed] [Google Scholar]
- 26.Dalby MJ, Yarwood SJ, Riehle MO, Johnstone HJ, Affrossman S, Curtis AS. Increasing fibroblast response to materials using nanotopography: morphological and genetic measurements of cell response to 13-nm-high polymer demixed islands. Exp Cell Res. 2002;276:1–9. doi: 10.1006/excr.2002.5498. [DOI] [PubMed] [Google Scholar]
- 27.Levenberg S, Huang NF, Lavik E, Rogers AB, Itskovitz-Eldor J, Langer R. Differentiation of human embryonic stem cells on three-dimensional polymer scaffolds. Proc Natl Acad Sci U S A. 2003;100:12741–12746. doi: 10.1073/pnas.1735463100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palmer TD, Markakis EA, Willhoite AR, Safar F, Gage FH. Fibroblast Growth Factor-2 Activates a Latent Neurogenic Program in Neural Stem Cells from Diverse Regions of the Adult CNS. J. Neurosci. 1999;19:8487–8497. doi: 10.1523/JNEUROSCI.19-19-08487.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cho KJ, Trzaska KA, Greco SJ, McArdle J, Wang FS, Ye JH, Rameshwar P. Neurons Derived From Human Mesenchymal Stem Cells Show Synaptic Transmission and Can Be Induced to Produce the Neurotransmitter Substance P by Interleukin-1{alpha} Stem Cells. 2005;23:383–391. doi: 10.1634/stemcells.2004-0251. [DOI] [PubMed] [Google Scholar]
- 30.Neuhuber B, Gallo G, Howard L, Kostura L, Mackay A, Fischer I. Reevaluation of in vitro differentiation protocols for bone marrow stromal cells: disruption of actin cytoskeleton induces rapid morphological changes and mimics neuronal phenotype. J Neurosci Res. 2004;77:192–204. doi: 10.1002/jnr.20147. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplement Table 1. RT-PCR primers for gene profile analysis of hMSCs cultured on nano-gratings. Primer sequences for neurofilament light (NFL) were obtained from NCBI NM_006158; β-actin sequences were provided by Clontech; and the other sequences were obtained from [19].
Supplement Table 2. Quantitative real-time PCR analysis of the selected up- / down-regulation of genes observed in microarray analysis.
Supplement Figure 1. TUNEL assay of hMSCs cultured on (A) nanopatterned and (B) unpattern PDMS, and (C) the positive control with DNA fragmentation induced by DNase I and (D) negative control on glass cover slip. Bar = 50μm
Supplement Figure 2. F-actin staining (A) and immunostaining of (B) Tuj1 and GFAP and (C) MAP2 and nestin of hMSCs cultured on unpatterned PDMS in the reduced-serum (1%) condition at day 7. Bar = 100μm.
Supplement Figure 3. Neuronal gene markers analysis of hMSCs cultured on nano-patterned or unpatterned PDMS with 6 days (6D) or 1 day (1D) exposure to 2μM of cytochalasin D, in MSC proliferation medium, or in reduced-serum condition (1% serum).
Supplement Figure 4. (A) chondrogenic gene markers and (B) osteogenic gene markers analysis of the hMSCs cultured in chondrogenic medium (CM) or osteogenic induction medium (OM), respectively, and the control sample cultured in MSC proliferation medium (PM) on TCPS for 4 weeks.