Abstract
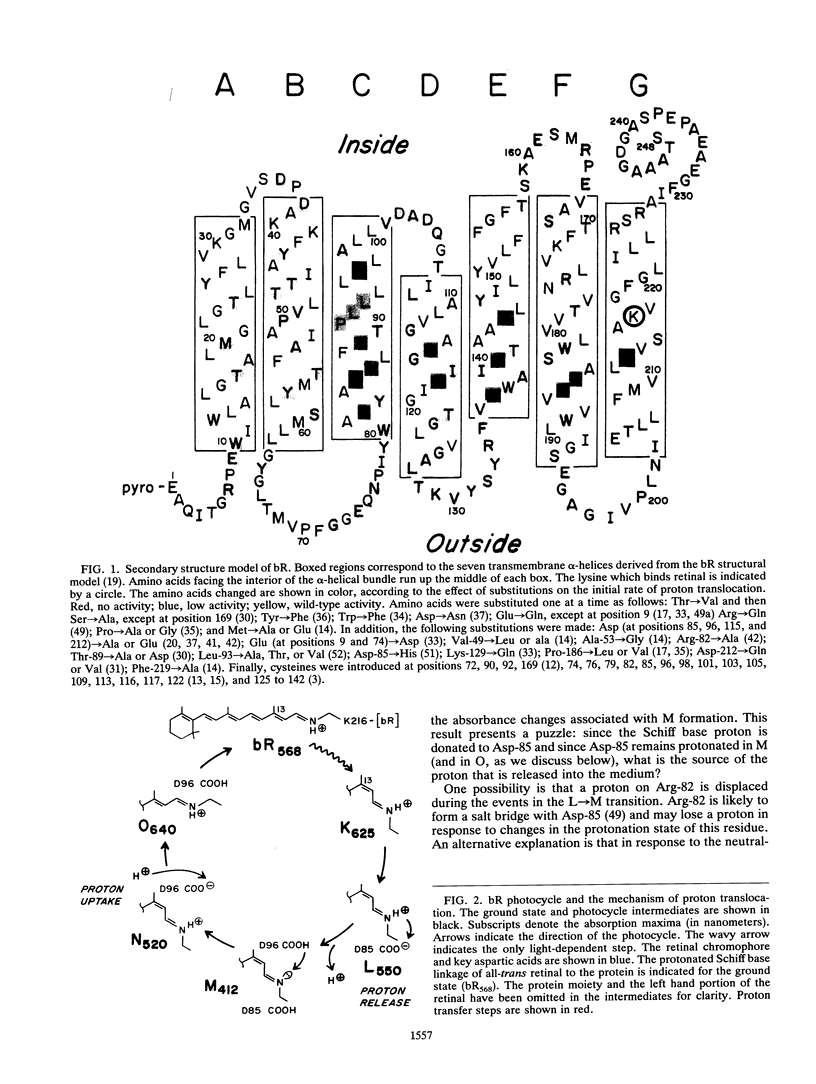
Site-specific mutagenesis has identified amino acids involved in bR proton transport. Biophysical studies of the mutants have elucidated the roles of two membrane-embedded residues: Asp-85 serves as the acceptor for the proton from the isomerized retinylidene Schiff base, and Asp-96 participates in reprotonation of this group. The functions of Arg-82, Leu-93, Asp-212, Tyr-185, and other residues that affect bR properties when substituted are not as well understood. Structural characterization of the mutant proteins will clarify the effects of substitutions at these positions. Current efforts in the field remain directed at understanding how retinal isomerization is coupled to proton transport. In particular, there has been more emphasis on determining the structures of bR and its photointermediates. Since well-ordered crystals of bR have not been obtained, continued electron diffraction studies of purple membrane offer the best opportunity for structure refinement. Other informative techniques include solid-state nuclear magnetic resonance of isotopically labeled bR (56) and electron paramagnetic resonance of bR tagged with nitroxide spin labels (2, 3, 13, 15). Site-directed mutagenesis will be essential in these studies to introduce specific sites for derivatization with structural probes and to slow the decay of intermediates. Thus, combining molecular biology and biophysics will continue to provide solutions to fundamental problems in bR.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abercrombie D. M., Khorana H. G. Regeneration of native bacteriorhodopsin structure following acetylation of epsilon-amino groups of Lys-30, -40, and -41. J Biol Chem. 1986 Apr 15;261(11):4875–4880. [PubMed] [Google Scholar]
- Altenbach C., Flitsch S. L., Khorana H. G., Hubbell W. L. Structural studies on transmembrane proteins. 2. Spin labeling of bacteriorhodopsin mutants at unique cysteines. Biochemistry. 1989 Sep 19;28(19):7806–7812. doi: 10.1021/bi00445a042. [DOI] [PubMed] [Google Scholar]
- Altenbach C., Marti T., Khorana H. G., Hubbell W. L. Transmembrane protein structure: spin labeling of bacteriorhodopsin mutants. Science. 1990 Jun 1;248(4959):1088–1092. doi: 10.1126/science.2160734. [DOI] [PubMed] [Google Scholar]
- Ames J. B., Mathies R. A. The role of back-reactions and proton uptake during the N----O transition in bacteriorhodopsin's photocycle: a kinetic resonance Raman study. Biochemistry. 1990 Aug 7;29(31):7181–7190. doi: 10.1021/bi00483a005. [DOI] [PubMed] [Google Scholar]
- Bousché O., Sonar S., Krebs M. P., Khorana H. G., Rothschild K. J. Time-resolved Fourier transform infrared spectroscopy of the bacteriorhodopsin mutant Tyr-185-->Phe: Asp-96 reprotonates during O formation; Asp-85 and Asp-212 deprotonate during O decay. Photochem Photobiol. 1992 Dec;56(6):1085–1095. doi: 10.1111/j.1751-1097.1992.tb09732.x. [DOI] [PubMed] [Google Scholar]
- Braiman M. S., Mogi T., Marti T., Stern L. J., Khorana H. G., Rothschild K. J. Vibrational spectroscopy of bacteriorhodopsin mutants: light-driven proton transport involves protonation changes of aspartic acid residues 85, 96, and 212. Biochemistry. 1988 Nov 15;27(23):8516–8520. doi: 10.1021/bi00423a002. [DOI] [PubMed] [Google Scholar]
- Braiman M. S., Stern L. J., Chao B. H., Khorana H. G. Structure-function studies on bacteriorhodopsin. IV. Purification and renaturation of bacterio-opsin polypeptide expressed in Escherichia coli. J Biol Chem. 1987 Jul 5;262(19):9271–9276. [PubMed] [Google Scholar]
- Cao Y., Váró G., Chang M., Ni B. F., Needleman R., Lanyi J. K. Water is required for proton transfer from aspartate-96 to the bacteriorhodopsin Schiff base. Biochemistry. 1991 Nov 12;30(45):10972–10979. doi: 10.1021/bi00109a023. [DOI] [PubMed] [Google Scholar]
- Cline S. W., Doolittle W. F. Efficient transfection of the archaebacterium Halobacterium halobium. J Bacteriol. 1987 Mar;169(3):1341–1344. doi: 10.1128/jb.169.3.1341-1344.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn R., McCoy J., Simsek M., Majumdar A., Chang S. H., Rajbhandary U. L., Khorana H. G. The bacteriorhodopsin gene. Proc Natl Acad Sci U S A. 1981 Nov;78(11):6744–6748. doi: 10.1073/pnas.78.11.6744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flitsch S. L., Khorana H. G. Structural studies on transmembrane proteins. 1. Model study using bacteriorhodopsin mutants containing single cysteine residues. Biochemistry. 1989 Sep 19;28(19):7800–7805. doi: 10.1021/bi00445a041. [DOI] [PubMed] [Google Scholar]
- Greenhalgh D. A., Altenbach C., Hubbell W. L., Khorana H. G. Locations of Arg-82, Asp-85, and Asp-96 in helix C of bacteriorhodopsin relative to the aqueous boundaries. Proc Natl Acad Sci U S A. 1991 Oct 1;88(19):8626–8630. doi: 10.1073/pnas.88.19.8626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett N. R., Stern L. J., Chao B. H., Kronis K. A., Khorana H. G. Structure-function studies on bacteriorhodopsin. V. Effects of amino acid substitutions in the putative helix F. J Biol Chem. 1987 Jul 5;262(19):9277–9284. [PubMed] [Google Scholar]
- Heberle J., Dencher N. A. Surface-bound optical probes monitor protein translocation and surface potential changes during the bacteriorhodopsin photocycle. Proc Natl Acad Sci U S A. 1992 Jul 1;89(13):5996–6000. doi: 10.1073/pnas.89.13.5996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson R., Baldwin J. M., Ceska T. A., Zemlin F., Beckmann E., Downing K. H. Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy. J Mol Biol. 1990 Jun 20;213(4):899–929. doi: 10.1016/S0022-2836(05)80271-2. [DOI] [PubMed] [Google Scholar]
- Holz M., Drachev L. A., Mogi T., Otto H., Kaulen A. D., Heyn M. P., Skulachev V. P., Khorana H. G. Replacement of aspartic acid-96 by asparagine in bacteriorhodopsin slows both the decay of the M intermediate and the associated proton movement. Proc Natl Acad Sci U S A. 1989 Apr;86(7):2167–2171. doi: 10.1073/pnas.86.7.2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K. S., Bayley H., Liao M. J., London E., Khorana H. G. Refolding of an integral membrane protein. Denaturation, renaturation, and reconstitution of intact bacteriorhodopsin and two proteolytic fragments. J Biol Chem. 1981 Apr 25;256(8):3802–3809. [PubMed] [Google Scholar]
- Karnik S. S., Nassal M., Doi T., Jay E., Sgaramella V., Khorana H. G. Structure-function studies on bacteriorhodopsin. II. Improved expression of the bacterio-opsin gene in Escherichia coli. J Biol Chem. 1987 Jul 5;262(19):9255–9263. [PubMed] [Google Scholar]
- Khorana H. G. Bacteriorhodopsin, a membrane protein that uses light to translocate protons. J Biol Chem. 1988 Jun 5;263(16):7439–7442. [PubMed] [Google Scholar]
- Khorana H. G., Gerber G. E., Herlihy W. C., Gray C. P., Anderegg R. J., Nihei K., Biemann K. Amino acid sequence of bacteriorhodopsin. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5046–5050. doi: 10.1073/pnas.76.10.5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krebs M. P., Hauss T., Heyn M. P., RajBhandary U. L., Khorana H. G. Expression of the bacterioopsin gene in Halobacterium halobium using a multicopy plasmid. Proc Natl Acad Sci U S A. 1991 Feb 1;88(3):859–863. doi: 10.1073/pnas.88.3.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanyi J. K. Proton transfer and energy coupling in the bacteriorhodopsin photocycle. J Bioenerg Biomembr. 1992 Apr;24(2):169–179. doi: 10.1007/BF00762675. [DOI] [PubMed] [Google Scholar]
- Lozier R. H., Xie A., Hofrichter J., Clore G. M. Reversible steps in the bacteriorhodopsin photocycle. Proc Natl Acad Sci U S A. 1992 Apr 15;89(8):3610–3614. doi: 10.1073/pnas.89.8.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinetti T., Subramaniam S., Mogi T., Marti T., Khorana H. G. Replacement of aspartic residues 85, 96, 115, or 212 affects the quantum yield and kinetics of proton release and uptake by bacteriorhodopsin. Proc Natl Acad Sci U S A. 1989 Jan;86(2):529–533. doi: 10.1073/pnas.86.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marti T., Otto H., Mogi T., Rösselet S. J., Heyn M. P., Khorana H. G. Bacteriorhodopsin mutants containing single substitutions of serine or threonine residues are all active in proton translocation. J Biol Chem. 1991 Apr 15;266(11):6919–6927. [PubMed] [Google Scholar]
- Mathies R. A., Lin S. W., Ames J. B., Pollard W. T. From femtoseconds to biology: mechanism of bacteriorhodopsin's light-driven proton pump. Annu Rev Biophys Biophys Chem. 1991;20:491–518. doi: 10.1146/annurev.bb.20.060191.002423. [DOI] [PubMed] [Google Scholar]
- Mogi T., Marti T., Khorana H. G. Structure-function studies on bacteriorhodopsin. IX. Substitutions of tryptophan residues affect protein-retinal interactions in bacteriorhodopsin. J Biol Chem. 1989 Aug 25;264(24):14197–14201. [PubMed] [Google Scholar]
- Mogi T., Stern L. J., Chao B. H., Khorana H. G. Structure-function studies on bacteriorhodopsin. VIII. Substitutions of the membrane-embedded prolines 50, 91, and 186: the effects are determined by the substituting amino acids. J Biol Chem. 1989 Aug 25;264(24):14192–14196. [PubMed] [Google Scholar]
- Mogi T., Stern L. J., Hackett N. R., Khorana H. G. Bacteriorhodopsin mutants containing single tyrosine to phenylalanine substitutions are all active in proton translocation. Proc Natl Acad Sci U S A. 1987 Aug;84(16):5595–5599. doi: 10.1073/pnas.84.16.5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogi T., Stern L. J., Marti T., Chao B. H., Khorana H. G. Aspartic acid substitutions affect proton translocation by bacteriorhodopsin. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4148–4152. doi: 10.1073/pnas.85.12.4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassal M., Mogi T., Karnik S. S., Khorana H. G. Structure-function studies on bacteriorhodopsin. III. Total synthesis of a gene for bacterio-opsin and its expression in Escherichia coli. J Biol Chem. 1987 Jul 5;262(19):9264–9270. [PubMed] [Google Scholar]
- Needleman R., Chang M., Ni B., Váró G., Fornés J., White S. H., Lanyi J. K. Properties of Asp212----Asn bacteriorhodopsin suggest that Asp212 and Asp85 both participate in a counterion and proton acceptor complex near the Schiff base. J Biol Chem. 1991 Jun 25;266(18):11478–11484. [PubMed] [Google Scholar]
- Ni B. F., Chang M., Duschl A., Lanyi J., Needleman R. An efficient system for the synthesis of bacteriorhodopsin in Halobacterium halobium. Gene. 1990 May 31;90(1):169–172. doi: 10.1016/0378-1119(90)90456-2. [DOI] [PubMed] [Google Scholar]
- Otto H., Marti T., Holz M., Mogi T., Lindau M., Khorana H. G., Heyn M. P. Aspartic acid-96 is the internal proton donor in the reprotonation of the Schiff base of bacteriorhodopsin. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9228–9232. doi: 10.1073/pnas.86.23.9228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto H., Marti T., Holz M., Mogi T., Stern L. J., Engel F., Khorana H. G., Heyn M. P. Substitution of amino acids Asp-85, Asp-212, and Arg-82 in bacteriorhodopsin affects the proton release phase of the pump and the pK of the Schiff base. Proc Natl Acad Sci U S A. 1990 Feb;87(3):1018–1022. doi: 10.1073/pnas.87.3.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovchinnikov Y. A., Abdulaev N. G., Feigina M. Y., Kiselev A. V., Lobanov N. A. The structural basis of the functioning of bacteriorhodopsin: an overview. FEBS Lett. 1979 Apr 15;100(2):219–224. doi: 10.1016/0014-5793(79)80338-5. [DOI] [PubMed] [Google Scholar]
- Rothschild K. J. FTIR difference spectroscopy of bacteriorhodopsin: toward a molecular model. J Bioenerg Biomembr. 1992 Apr;24(2):147–167. doi: 10.1007/BF00762674. [DOI] [PubMed] [Google Scholar]
- Soppa J., Oesterhelt D. Bacteriorhodopsin mutants of Halobacterium sp. GRB. I. The 5-bromo-2'-deoxyuridine selection as a method to isolate point mutants in halobacteria. J Biol Chem. 1989 Aug 5;264(22):13043–13048. [PubMed] [Google Scholar]
- Soppa J., Otomo J., Straub J., Tittor J., Meessen S., Oesterhelt D. Bacteriorhodopsin mutants of Halobacterium sp. GRB. II. Characterization of mutants. J Biol Chem. 1989 Aug 5;264(22):13049–13056. [PubMed] [Google Scholar]
- Stern L. J., Ahl P. L., Marti T., Mogi T., Duñach M., Berkowitz S., Rothschild K. J., Khorana H. G. Substitution of membrane-embedded aspartic acids in bacteriorhodopsin causes specific changes in different steps of the photochemical cycle. Biochemistry. 1989 Dec 26;28(26):10035–10042. doi: 10.1021/bi00452a023. [DOI] [PubMed] [Google Scholar]
- Stern L. J., Khorana H. G. Structure-function studies on bacteriorhodopsin. X. Individual substitutions of arginine residues by glutamine affect chromophore formation, photocycle, and proton translocation. J Biol Chem. 1989 Aug 25;264(24):14202–14208. [PubMed] [Google Scholar]
- Stoeckenius W., Lozier R. H., Bogomolni R. A. Bacteriorhodopsin and the purple membrane of halobacteria. Biochim Biophys Acta. 1979 Mar 14;505(3-4):215–278. doi: 10.1016/0304-4173(79)90006-5. [DOI] [PubMed] [Google Scholar]
- Subramaniam S., Greenhalgh D. A., Khorana H. G. Aspartic acid 85 in bacteriorhodopsin functions both as proton acceptor and negative counterion to the Schiff base. J Biol Chem. 1992 Dec 25;267(36):25730–25733. [PubMed] [Google Scholar]
- Subramaniam S., Greenhalgh D. A., Rath P., Rothschild K. J., Khorana H. G. Replacement of leucine-93 by alanine or threonine slows down the decay of the N and O intermediates in the photocycle of bacteriorhodopsin: implications for proton uptake and 13-cis-retinal----all-trans-retinal reisomerization. Proc Natl Acad Sci U S A. 1991 Aug 1;88(15):6873–6877. doi: 10.1073/pnas.88.15.6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam S., Marti T., Khorana H. G. Protonation state of Asp (Glu)-85 regulates the purple-to-blue transition in bacteriorhodopsin mutants Arg-82----Ala and Asp-85----Glu: the blue form is inactive in proton translocation. Proc Natl Acad Sci U S A. 1990 Feb;87(3):1013–1017. doi: 10.1073/pnas.87.3.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uegaki K., Sugiyama Y., Mukohata Y. Archaerhodopsin-2, from Halobacterium sp. aus-2 further reveals essential amino acid residues for light-driven proton pumps. Arch Biochem Biophys. 1991 Apr;286(1):107–110. doi: 10.1016/0003-9861(91)90014-a. [DOI] [PubMed] [Google Scholar]
- Váró G., Zimányi L., Chang M., Ni B., Needleman R., Lanyi J. K. A residue substitution near the beta-ionone ring of the retinal affects the M substates of bacteriorhodopsin. Biophys J. 1992 Mar;61(3):820–826. doi: 10.1016/S0006-3495(92)81887-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L., Herzfeld J. NMR studies of retinal proteins. J Bioenerg Biomembr. 1992 Apr;24(2):139–146. doi: 10.1007/BF00762673. [DOI] [PubMed] [Google Scholar]