Abstract
Four species representing three genera of halophilic archaebacteria were examined for the presence of genomic sequences that encode proteins of the superoxide dismutase family. Three species, Halobacterium cutirubrum, Halobacterium sp. strain GRB, and Haloferax volcanii, contain duplicated (paralogous) genes of the sod family; a fourth species, Haloarcula marismortui, contains only a single gene. These seven genes were cloned and sequenced, and their transcripts were characterized by Northern (RNA) hybridization, S1 nuclease protection, and primer extension. The expression of one of the two genes in H. cutirubrum, Halobacterium sp. strain GRB, and Haloferax volcanii was shown to be elevated in the presence of paraquat, a generator of superoxide radicals. The other genes, including the single gene from Haloarcula marismortui, exhibited no elevated expression in the presence of paraquat. The 5' and 3' flanking regions of all the genes contain recognizable promoter and terminator elements that are appropriately positioned relative to the 5' and 3' transcript end sites. Between genera, the orthologous paraquat-responsive genes exhibit no sequence similarity in either their 5' or 3' flanking regions, whereas the orthologous nonresponsive genes exhibit limited sequence similarity but only in the 5' flanking region. Within the coding region, the two paralogous genes of Haloferax volcanii are virtually identical (99.5%) despite the absence of similarity in the flanking regions. In contrast, the paralogous genes of H. cutirubrum and Halobacterium sp. strain GRB are only about 87% identical. In the alignment of all seven sequences, there are nine codon positions where both the TCN and AGY serine codons are utilized; some or all of these may well be examples of convergent evolution.
Full text
PDF






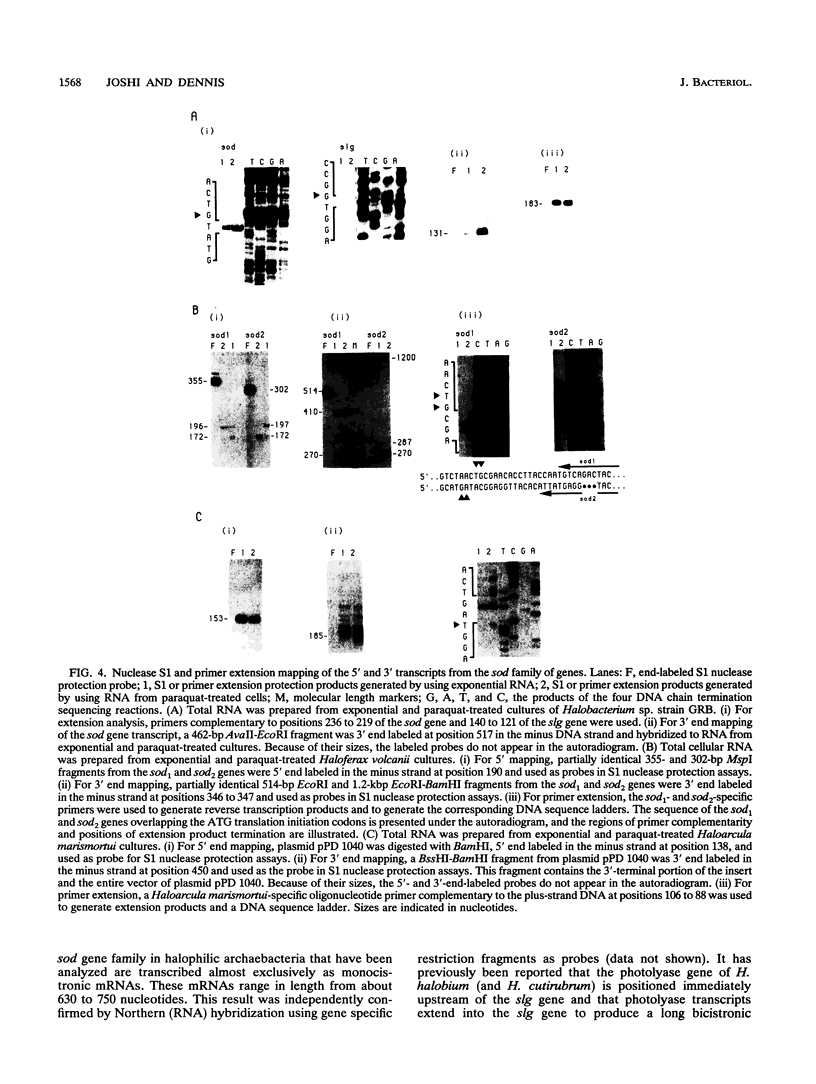



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chant J., Dennis P. Archaebacteria: transcription and processing of ribosomal RNA sequences in Halobacterium cutirubrum. EMBO J. 1986 May;5(5):1091–1097. doi: 10.1002/j.1460-2075.1986.tb04327.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlebois R. L., Schalkwyk L. C., Hofman J. D., Doolittle W. F. Detailed physical map and set of overlapping clones covering the genome of the archaebacterium Haloferax volcanii DS2. J Mol Biol. 1991 Dec 5;222(3):509–524. doi: 10.1016/0022-2836(91)90493-p. [DOI] [PubMed] [Google Scholar]
- Daniels C. J., McKee A. H., Doolittle W. F. Archaebacterial heat-shock proteins. EMBO J. 1984 Apr;3(4):745–749. doi: 10.1002/j.1460-2075.1984.tb01878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demple B., Amábile-Cuevas C. F. Redox redux: the control of oxidative stress responses. Cell. 1991 Nov 29;67(5):837–839. doi: 10.1016/0092-8674(91)90355-3. [DOI] [PubMed] [Google Scholar]
- Dennis P. P. Multiple promoters for the transcription of the ribosomal RNA gene cluster in Halobacterium cutirubrum. J Mol Biol. 1985 Nov 20;186(2):457–461. doi: 10.1016/0022-2836(85)90117-2. [DOI] [PubMed] [Google Scholar]
- Dente L., Cortese R. pEMBL: a new family of single-stranded plasmids for sequencing DNA. Methods Enzymol. 1987;155:111–119. doi: 10.1016/0076-6879(87)55011-x. [DOI] [PubMed] [Google Scholar]
- Farr S. B., Kogoma T. Oxidative stress responses in Escherichia coli and Salmonella typhimurium. Microbiol Rev. 1991 Dec;55(4):561–585. doi: 10.1128/mr.55.4.561-585.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fee J. A. Regulation of sod genes in Escherichia coli: relevance to superoxide dismutase function. Mol Microbiol. 1991 Nov;5(11):2599–2610. doi: 10.1111/j.1365-2958.1991.tb01968.x. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Hassan H. M., Fridovich I. Regulation of the synthesis of superoxide dismutase in Escherichia coli. Induction by methyl viologen. J Biol Chem. 1977 Nov 10;252(21):7667–7672. [PubMed] [Google Scholar]
- Hsia H. C., Lebkowski J. S., Leong P. M., Calos M. P., Miller J. H. Comparison of ultraviolet irradiation-induced mutagenesis of the lacI gene in Escherichia coli and in human 293 cells. J Mol Biol. 1989 Jan 5;205(1):103–113. doi: 10.1016/0022-2836(89)90368-9. [DOI] [PubMed] [Google Scholar]
- Joshi P., Dennis P. P. Structure, function, and evolution of the family of superoxide dismutase proteins from halophilic archaebacteria. J Bacteriol. 1993 Mar;175(6):1572–1579. doi: 10.1128/jb.175.6.1572-1579.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. How do eucaryotic ribosomes select initiation regions in messenger RNA? Cell. 1978 Dec;15(4):1109–1123. doi: 10.1016/0092-8674(78)90039-9. [DOI] [PubMed] [Google Scholar]
- Marklund S., Marklund G. Involvement of the superoxide anion radical in the autoxidation of pyrogallol and a convenient assay for superoxide dismutase. Eur J Biochem. 1974 Sep 16;47(3):469–474. doi: 10.1111/j.1432-1033.1974.tb03714.x. [DOI] [PubMed] [Google Scholar]
- May B. P., Dennis P. P. Evolution and regulation of the gene encoding superoxide dismutase from the archaebacterium Halobacterium cutirubrum. J Biol Chem. 1989 Jul 25;264(21):12253–12258. [PubMed] [Google Scholar]
- May B. P., Dennis P. P. Superoxide dismutase from the extremely halophilic archaebacterium Halobacterium cutirubrum. J Bacteriol. 1987 Apr;169(4):1417–1422. doi: 10.1128/jb.169.4.1417-1422.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May B. P., Dennis P. P. Unusual evolution of a superoxide dismutase-like gene from the extremely halophilic archaebacterium Halobacterium cutirubrum. J Bacteriol. 1990 Jul;172(7):3725–3729. doi: 10.1128/jb.172.7.3725-3729.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May B. P., Tam P., Dennis P. P. The expression of the superoxide dismutase gene in Halobacterium cutirubrum and Halobacterium volcanii. Can J Microbiol. 1989 Jan;35(1):171–175. doi: 10.1139/m89-026. [DOI] [PubMed] [Google Scholar]
- McCarthy R. A., Warrington E. K. Evidence for modality-specific meaning systems in the brain. Nature. 1988 Aug 4;334(6181):428–430. doi: 10.1038/334428a0. [DOI] [PubMed] [Google Scholar]
- McCord J. M., Fridovich I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein). J Biol Chem. 1969 Nov 25;244(22):6049–6055. [PubMed] [Google Scholar]
- Mullakhanbhai M. F., Larsen H. Halobacterium volcanii spec. nov., a Dead Sea halobacterium with a moderate salt requirement. Arch Microbiol. 1975 Aug 28;104(3):207–214. doi: 10.1007/BF00447326. [DOI] [PubMed] [Google Scholar]
- Newman A. Specific accessory sequences in Saccharomyces cerevisiae introns control assembly of pre-mRNAs into spliceosomes. EMBO J. 1987 Dec 1;6(12):3833–3839. doi: 10.1002/j.1460-2075.1987.tb02720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oren A., Lau P. P., Fox G. E. The taxonomic status of "Halobacterium marismortui" from the Dead Sea: a comparison with Halobacterium vallismortis. Syst Appl Microbiol. 1988;10(3):251–258. doi: 10.1016/S0723-2020(88)80009-2. [DOI] [PubMed] [Google Scholar]
- Peterson G. L. A simplification of the protein assay method of Lowry et al. which is more generally applicable. Anal Biochem. 1977 Dec;83(2):346–356. doi: 10.1016/0003-2697(77)90043-4. [DOI] [PubMed] [Google Scholar]
- Reiter W. D., Hüdepohl U., Zillig W. Mutational analysis of an archaebacterial promoter: essential role of a TATA box for transcription efficiency and start-site selection in vitro. Proc Natl Acad Sci U S A. 1990 Dec;87(24):9509–9513. doi: 10.1073/pnas.87.24.9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salin M. L., Duke M. V., Oesterhelt D., Ma D. P. Cloning and determination of the nucleotide sequence of the Mn-containing superoxide dismutase gene from Halobacterium halobium. Gene. 1988 Oct 15;70(1):153–159. doi: 10.1016/0378-1119(88)90113-8. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel H., Zillig W., Pfäffle M., Schnabel R., Michel H., Delius H. Halobacterium halobium phage øH. EMBO J. 1982;1(1):87–92. doi: 10.1002/j.1460-2075.1982.tb01129.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Takao M., Kobayashi T., Oikawa A., Yasui A. Tandem arrangement of photolyase and superoxide dismutase genes in Halobacterium halobium. J Bacteriol. 1989 Nov;171(11):6323–6329. doi: 10.1128/jb.171.11.6323-6329.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda Y., Avila H. Structure and gene expression of the E. coli Mn-superoxide dismutase gene. Nucleic Acids Res. 1986 Jun 11;14(11):4577–4589. doi: 10.1093/nar/14.11.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Scholl R., Browse J., Somerville C. Double stranded DNA sequencing as a choice for DNA sequencing. Nucleic Acids Res. 1988 Feb 11;16(3):1220–1220. doi: 10.1093/nar/16.3.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]




