Abstract
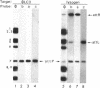
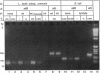
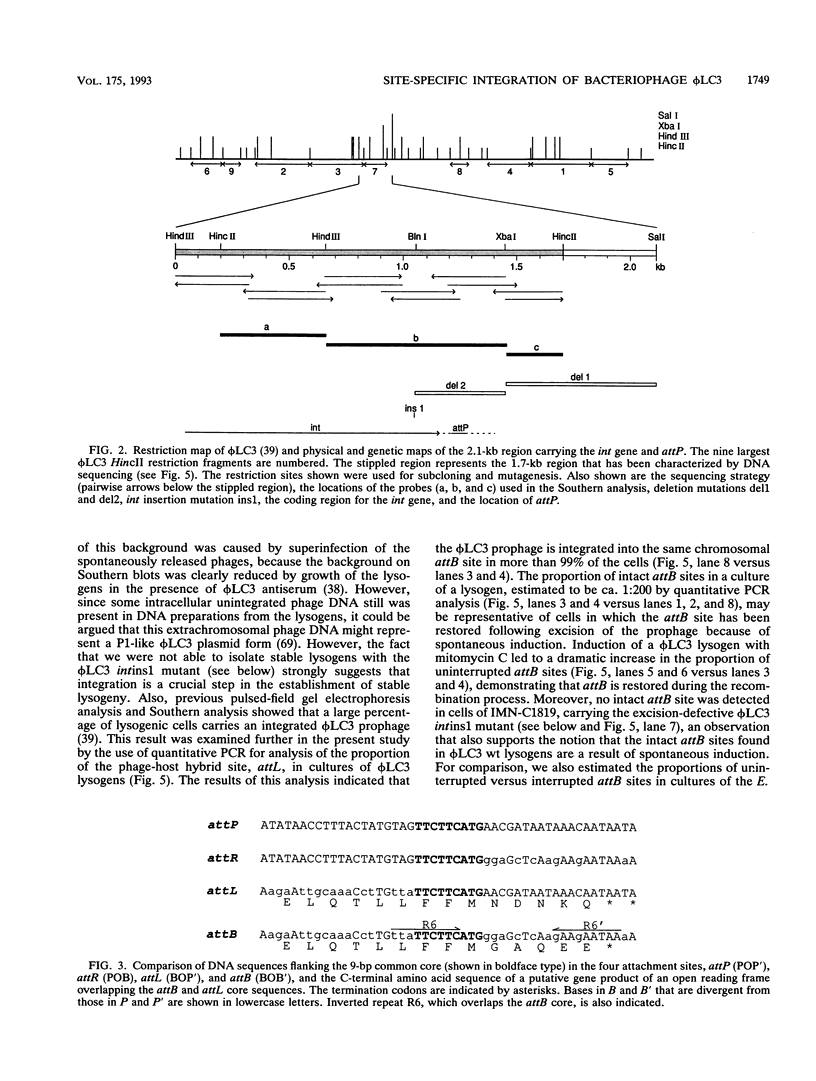
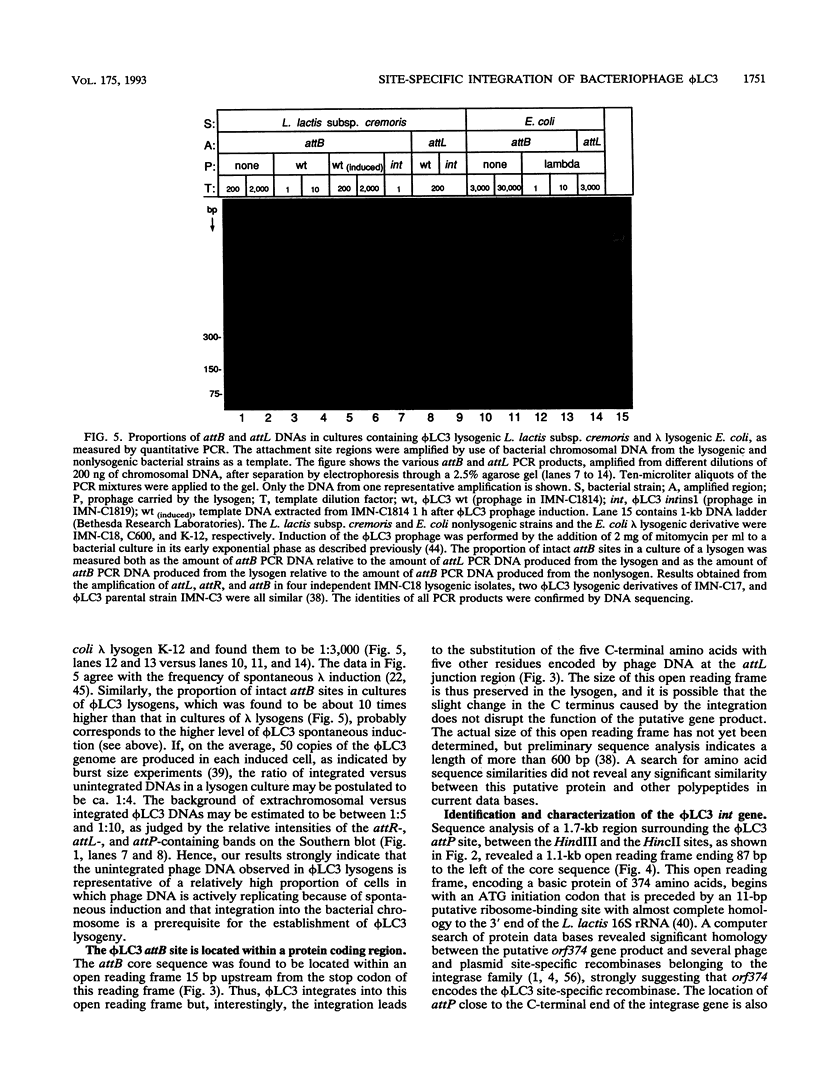
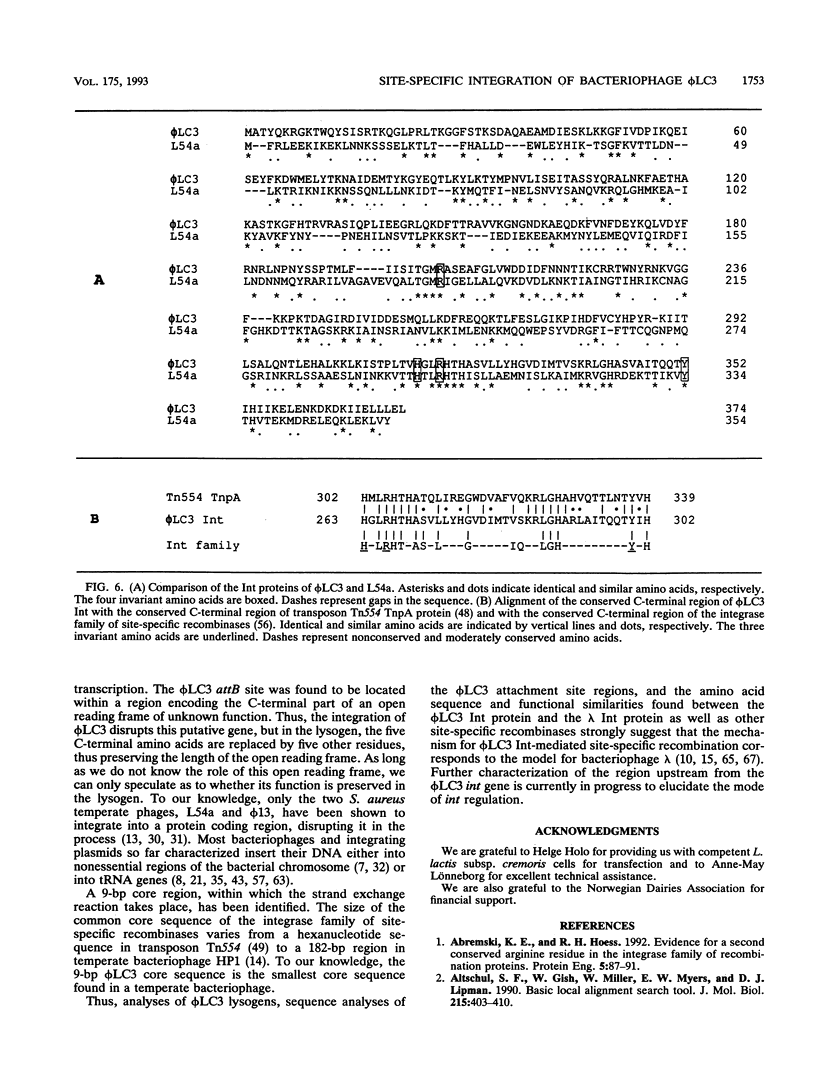
The genetic elements required for the integration of the temperate lactococcal bacteriophage phi LC3 into the chromosome of its bacterial host, Lactococcus lactis subsp. cremoris, were identified and characterized. The phi LC3 phage attachment site, attP, was mapped and sequenced. DNA sequence analysis of attP and of the bacterial attachment site, attB, as well as the two phage-host junctions, attR and attL, in the chromosome of a phi LC3 lysogen, identified a 9-bp common core region, 5'-TTCTTCATG'-3, within which the strand exchange reaction takes place during integration. The attB core sequence is located within the C-terminal part of an open reading frame of unknown function. The phi LC3 integrase gene (int), encoding the phi LC3 site-specific recombinase, was identified and is located adjacent to attP. The phi LC3 Int protein, as deduced from the nucleotide sequence, is a basic protein of 374 amino acids that shares significant sequence similarity with other site-specific recombinases of the integrase family. Phage phi LC3 int- and int-attP-defective mutants, conferring an abortive lysogenic phenotype, were constructed.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abremski K. E., Hoess R. H. Evidence for a second conserved arginine residue in the integrase family of recombination proteins. Protein Eng. 1992 Jan;5(1):87–91. doi: 10.1093/protein/5.1.87. [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Appleyard R K. Segregation of New Lysogenic Types during Growth of a Doubly Lysogenic Strain Derived from Escherichia Coli K12. Genetics. 1954 Jul;39(4):440–452. doi: 10.1093/genetics/39.4.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argos P., Landy A., Abremski K., Egan J. B., Haggard-Ljungquist E., Hoess R. H., Kahn M. L., Kalionis B., Narayana S. V., Pierson L. S., 3rd The integrase family of site-specific recombinases: regional similarities and global diversity. EMBO J. 1986 Feb;5(2):433–440. doi: 10.1002/j.1460-2075.1986.tb04229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball C. A., Johnson R. C. Efficient excision of phage lambda from the Escherichia coli chromosome requires the Fis protein. J Bacteriol. 1991 Jul;173(13):4027–4031. doi: 10.1128/jb.173.13.4027-4031.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball C. A., Johnson R. C. Multiple effects of Fis on integration and the control of lysogeny in phage lambda. J Bacteriol. 1991 Jul;173(13):4032–4038. doi: 10.1128/jb.173.13.4032-4038.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreiro V., Haggård-Ljungquist E. Attachment sites for bacteriophage P2 on the Escherichia coli chromosome: DNA sequences, localization on the physical map, and detection of a P2-like remnant in E. coli K-12 derivatives. J Bacteriol. 1992 Jun;174(12):4086–4093. doi: 10.1128/jb.174.12.4086-4093.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D. P., Idler K. B., Katz L. Characterization of the genetic elements required for site-specific integration of plasmid pSE211 in Saccharopolyspora erythraea. J Bacteriol. 1990 Apr;172(4):1877–1888. doi: 10.1128/jb.172.4.1877-1888.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calendar R., Ljungquist E., Deho G., Usher D. C., Goldstein R., Youderian P., Sironi G., Six E. W. Lysogenization by satellite phage P4. Virology. 1981 Aug;113(1):20–38. doi: 10.1016/0042-6822(81)90133-1. [DOI] [PubMed] [Google Scholar]
- Casanova J. L., Pannetier C., Jaulin C., Kourilsky P. Optimal conditions for directly sequencing double-stranded PCR products with sequenase. Nucleic Acids Res. 1990 Jul 11;18(13):4028–4028. doi: 10.1093/nar/18.13.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman D., Knights J., Russell R., Shanley D., Birkbeck T. H., Dougan G., Charles I. Insertional inactivation of the Staphylococcus aureus beta-toxin by bacteriophage phi 13 occurs by site- and orientation-specific integration of the phi 13 genome. Mol Microbiol. 1991 Apr;5(4):933–939. doi: 10.1111/j.1365-2958.1991.tb00768.x. [DOI] [PubMed] [Google Scholar]
- Goodman S. D., Scocca J. J. Nucleotide sequence and expression of the gene for the site-specific integration protein from bacteriophage HP1 of Haemophilus influenzae. J Bacteriol. 1989 Aug;171(8):4232–4240. doi: 10.1128/jb.171.8.4232-4240.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman M. E., Yarmolinsky M. B. Integration-negative mutants of bacteriophage lambda. J Mol Biol. 1968 Feb 14;31(3):487–505. doi: 10.1016/0022-2836(68)90423-3. [DOI] [PubMed] [Google Scholar]
- Hahn D. R., McHenney M. A., Baltz R. H. Properties of the streptomycete temperate bacteriophage FP43. J Bacteriol. 1991 Jun;173(12):3770–3775. doi: 10.1128/jb.173.12.3770-3775.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall R. M., Vockler C. The region of the IncN plasmid R46 coding for resistance to beta-lactam antibiotics, streptomycin/spectinomycin and sulphonamides is closely related to antibiotic resistance segments found in IncW plasmids and in Tn21-like transposons. Nucleic Acids Res. 1987 Sep 25;15(18):7491–7501. doi: 10.1093/nar/15.18.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hartley J. L., Donelson J. E. Nucleotide sequence of the yeast plasmid. Nature. 1980 Aug 28;286(5776):860–865. doi: 10.1038/286860a0. [DOI] [PubMed] [Google Scholar]
- Hauser M. A., Scocca J. J. Location of the host attachment site for phage HPl within a cluster of Haemophilus influenzae tRNA genes. Nucleic Acids Res. 1990 Sep 11;18(17):5305–5305. doi: 10.1093/nar/18.17.5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins D. G., Bleasby A. J., Fuchs R. CLUSTAL V: improved software for multiple sequence alignment. Comput Appl Biosci. 1992 Apr;8(2):189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- Hoess R. H., Foeller C., Bidwell K., Landy A. Site-specific recombination functions of bacteriophage lambda: DNA sequence of regulatory regions and overlapping structural genes for Int and Xis. Proc Natl Acad Sci U S A. 1980 May;77(5):2482–2486. doi: 10.1073/pnas.77.5.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holo H., Nes I. F. High-Frequency Transformation, by Electroporation, of Lactococcus lactis subsp. cremoris Grown with Glycine in Osmotically Stabilized Media. Appl Environ Microbiol. 1989 Dec;55(12):3119–3123. doi: 10.1128/aem.55.12.3119-3123.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm P. Two regulatory fim genes, fimB and fimE, control the phase variation of type 1 fimbriae in Escherichia coli. EMBO J. 1986 Jun;5(6):1389–1393. doi: 10.1002/j.1460-2075.1986.tb04372.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A., Kusukawa A., Komano T. Nucleotide sequence of the rci gene encoding shufflon-specific DNA recombinase in the IncI1 plasmid R64: homology to the site-specific recombinases of integrase family. Mol Gen Genet. 1988 Jul;213(1):30–35. doi: 10.1007/BF00333394. [DOI] [PubMed] [Google Scholar]
- Landy A., Ross W. Viral integration and excision: structure of the lambda att sites. Science. 1977 Sep 16;197(4309):1147–1160. doi: 10.1126/science.331474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. Y., Buranen S. L. Extent of the DNA sequence required in integration of staphylococcal bacteriophage L54a. J Bacteriol. 1989 Mar;171(3):1652–1657. doi: 10.1128/jb.171.3.1652-1657.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. Y., Iandolo J. J. Integration of staphylococcal phage L54a occurs by site-specific recombination: structural analysis of the attachment sites. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5474–5478. doi: 10.1073/pnas.83.15.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. Y., Iandolo J. J. Lysogenic conversion of staphylococcal lipase is caused by insertion of the bacteriophage L54a genome into the lipase structural gene. J Bacteriol. 1986 May;166(2):385–391. doi: 10.1128/jb.166.2.385-391.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. Y., Iandolo J. J. Mechanism of bacteriophage conversion of lipase activity in Staphylococcus aureus. J Bacteriol. 1985 Oct;164(1):288–293. doi: 10.1128/jb.164.1.288-293.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. Y., Iandolo J. J. Structural analysis of staphylococcal bacteriophage phi 11 attachment sites. J Bacteriol. 1988 May;170(5):2409–2411. doi: 10.1128/jb.170.5.2409-2411.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. H., Pascopella L., Jacobs W. R., Jr, Hatfull G. F. Site-specific integration of mycobacteriophage L5: integration-proficient vectors for Mycobacterium smegmatis, Mycobacterium tuberculosis, and bacille Calmette-Guérin. Proc Natl Acad Sci U S A. 1991 Apr 15;88(8):3111–3115. doi: 10.1073/pnas.88.8.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leenhouts K. J., Kok J., Venema G. Stability of Integrated Plasmids in the Chromosome of Lactococcus lactis. Appl Environ Microbiol. 1990 Sep;56(9):2726–2735. doi: 10.1128/aem.56.9.2726-2735.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong J. M., Nunes-Düby S. E., Oser A. B., Lesser C. F., Youderian P., Susskind M. M., Landy A. Structural and regulatory divergence among site-specific recombination genes of lambdoid phage. J Mol Biol. 1986 Jun 20;189(4):603–616. doi: 10.1016/0022-2836(86)90491-2. [DOI] [PubMed] [Google Scholar]
- Lillehaug D., Lindqvist B., Birkeland N. K. Characterization of phiLC3, a Lactococcus lactis subsp. cremoris temperature bacteriophage with cohesive single-stranded DNA ends. Appl Environ Microbiol. 1991 Nov;57(11):3206–3211. doi: 10.1128/aem.57.11.3206-3211.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig W., Seewaldt E., Kilpper-Bälz R., Schleifer K. H., Magrum L., Woese C. R., Fox G. E., Stackebrandt E. The phylogenetic position of Streptococcus and Enterococcus. J Gen Microbiol. 1985 Mar;131(3):543–551. doi: 10.1099/00221287-131-3-543. [DOI] [PubMed] [Google Scholar]
- Mahillon J., Lereclus D. Structural and functional analysis of Tn4430: identification of an integrase-like protein involved in the co-integrate-resolution process. EMBO J. 1988 May;7(5):1515–1526. doi: 10.1002/j.1460-2075.1988.tb02971.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazodier P., Thompson C., Boccard F. The chromosomal integration site of the Streptomyces element pSAM2 overlaps a putative tRNA gene conserved among actinomycetes. Mol Gen Genet. 1990 Jul;222(2-3):431–434. doi: 10.1007/BF00633850. [DOI] [PubMed] [Google Scholar]
- Melechen N. E., Go G., Lozeron H. A. Effect of cI repressor level on thymineless and spontaneous induction; specificity of lambda RNA transcription. Mol Gen Genet. 1978 Jul 11;163(2):213–221. doi: 10.1007/BF00267412. [DOI] [PubMed] [Google Scholar]
- Moitoso de Vargas L., Landy A. A switch in the formation of alternative DNA loops modulates lambda site-specific recombination. Proc Natl Acad Sci U S A. 1991 Jan 15;88(2):588–592. doi: 10.1073/pnas.88.2.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy E., Huwyler L., de Freire Bastos M. do C. Transposon Tn554: complete nucleotide sequence and isolation of transposition-defective and antibiotic-sensitive mutants. EMBO J. 1985 Dec 1;4(12):3357–3365. doi: 10.1002/j.1460-2075.1985.tb04089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy E., Löfdahl S. Transposition of Tn554 does not generate a target duplication. Nature. 1984 Jan 19;307(5948):292–294. doi: 10.1038/307292a0. [DOI] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Ouellette M., Roy P. H. Homology of ORFs from Tn2603 and from R46 to site-specific recombinases. Nucleic Acids Res. 1987 Dec 10;15(23):10055–10055. doi: 10.1093/nar/15.23.10055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pargellis C. A., Nunes-Düby S. E., de Vargas L. M., Landy A. Suicide recombination substrates yield covalent lambda integrase-DNA complexes and lead to identification of the active site tyrosine. J Biol Chem. 1988 Jun 5;263(16):7678–7685. [PubMed] [Google Scholar]
- Parker J. D., Rabinovitch P. S., Burmer G. C. Targeted gene walking polymerase chain reaction. Nucleic Acids Res. 1991 Jun 11;19(11):3055–3060. doi: 10.1093/nar/19.11.3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyart-Salmeron C., Trieu-Cuot P., Carlier C., Courvalin P. Molecular characterization of two proteins involved in the excision of the conjugative transposon Tn1545: homologies with other site-specific recombinases. EMBO J. 1989 Aug;8(8):2425–2433. doi: 10.1002/j.1460-2075.1989.tb08373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter W. D., Palm P., Yeats S. Transfer RNA genes frequently serve as integration sites for prokaryotic genetic elements. Nucleic Acids Res. 1989 Mar 11;17(5):1907–1914. doi: 10.1093/nar/17.5.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar G., Kapelner S., Sommer S. S. Formamide can dramatically improve the specificity of PCR. Nucleic Acids Res. 1990 Dec 25;18(24):7465–7465. doi: 10.1093/nar/18.24.7465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirai M., Nara H., Sato A., Aida T., Takahashi H. Site-specific integration of the actinophage R4 genome into the chromosome of Streptomyces parvulus upon lysogenization. J Bacteriol. 1991 Jul;173(13):4237–4239. doi: 10.1128/jb.173.13.4237-4239.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Inouye M., Inouye S. Association of a retroelement with a P4-like cryptic prophage (retronphage phi R73) integrated into the selenocystyl tRNA gene of Escherichia coli. J Bacteriol. 1991 Jul;173(13):4171–4181. doi: 10.1128/jb.173.13.4171-4181.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzaghi B. E., Sandine W. E. Improved medium for lactic streptococci and their bacteriophages. Appl Microbiol. 1975 Jun;29(6):807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldman A. S., Fitzmaurice W. P., Scocca J. J. Integration of the bacteriophage HP1c1 genome into the Haemophilus influenzae Rd chromosome in the lysogenic state. J Bacteriol. 1986 Jan;165(1):297–300. doi: 10.1128/jb.165.1.297-300.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Ye Z. H., Buranen S. L., Lee C. Y. Sequence analysis and comparison of int and xis genes from staphylococcal bacteriophages L54a and phi 11. J Bacteriol. 1990 May;172(5):2568–2575. doi: 10.1128/jb.172.5.2568-2575.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z. H., Lee C. Y. Nucleotide sequence and genetic characterization of staphylococcal bacteriophage L54a int and xis genes. J Bacteriol. 1989 Aug;171(8):4146–4153. doi: 10.1128/jb.171.8.4146-4153.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu A., Bertani L. E., Haggård-Ljungquist E. Control of prophage integration and excision in bacteriophage P2: nucleotide sequences of the int gene and att sites. Gene. 1989 Aug 1;80(1):1–11. doi: 10.1016/0378-1119(89)90244-8. [DOI] [PubMed] [Google Scholar]
- Zimmerman L. J., Fuscoe J. C. Direct DNA sequencing of PCR products. Environ Mol Mutagen. 1991;18(4):274–276. doi: 10.1002/em.2850180413. [DOI] [PubMed] [Google Scholar]