Abstract
4-Hydroxyequilenin (4-OHEN)-dC is a major, potentially mutagenic DNA adduct induced by equine estrogens used for hormone replacement therapy. To study the miscoding property of 4-OHEN-dC and the involvement of Y-family human DNA polymerases (pols) η, κ and ι in that process, we incorporated 4-OHEN-dC into oligodeoxynucleotides and used them as templates in primer extension reactions catalyzed by pol η, κ and ι. Pol η inserted dAMP opposite 4-OHEN-dC, accompanied by lesser amounts of dCMP and dTMP incorporation and base deletion. Pol κ promoted base deletions as well as direct incorporation of dAMP and dCMP. Pol ι worked in conjunction with pol κ, but not with pol η, at a replication fork stalled by the adduct, resulting in increased dTMP incorporation. Our results provide a direct evidence that Y-family DNA pols can switch with one another during synthesis past the lesion. No direct incorporation of dGMP, the correct base, was observed with Y-family enzymes. The miscoding potency of 4-OHEN-dC may be associated with the development of reproductive cancers observed in women receiving hormone replacement therapy.
Keywords: DNA adduct, DNA polymerase, equilenin, equilin, hormone replacement therapy
Introduction
Hormone replacement therapy is widely used for postmenopausal women to decrease menopausal symptoms and to protect against osteoporosis. However, hormone replacement therapy is associated with a significantly increased risk of breast, ovarian and endometrial cancers.1-3 Premarin (Wyeth-Ayerst), composed of approximately 30% (w/w) equilin, 10% (w/w) equilenin and other estrogens, is frequently used for this purpose.4 Like estrogens, equilenin and equilin are metabolized to 4-hydroxyequilenin (4-OHEN) and 4-hydroxyequilin (4-OHEQ), respectively.4 4-OHEN is rapidly autoxidized to an o-quinone, which reacts readily with DNA, resulting in the formation of several diastereoisomers of unique 4-OHEN-dC (Figure 1), 4-OHEN-dA and 4-OHEN-dG adducts.4-6 4-OHEQ is also autoxidized to an o-quinone that isomerizes to 4-OHEN-o-quinone; therefore, 4-OHEQ produces DNA adducts identical with those observed for 4-OHEN.7 Equine estrogen-derived DNA adducts have been detected in breast tumor and adjacent normal tissues of several patients receiving hormone replacement therapy, and in paraffin-embedded breast tumor tissues.8
Figure 1.
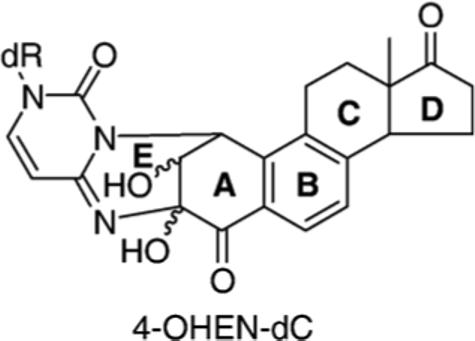
Structures of diastereoisomers of the 4-OHEN-dC adduct.
Many types of DNA adducts block the processivity of the replication machinery during DNA synthesis. Cells utilize translesion synthesis (TLS) DNA polymerases (pols) to extend the nascent strand when a replication fork is stalled.9-11 The Y-family DNA pols, including pol η, κ and ι, are especially associated with translesion synthesis past DNA adducts.12,13 In addition, switching of TLS pols, including Y-family enzymes, at DNA adducts has recently been proposed9,11 on the basis of the results from the TLS bypass assay14-16 and/or steady-state kinetic studies performed with reactions containing a single dNTP.17-20 Interestingly, Y-family DNA pols are highly expressed in human testis, ovary and uterus, where steroid hormones are produced.21-24 Therefore, the presence of equine estrogen-derived DNA adducts and high levels of Y-family DNA pols together in reproductive organs may result in a significant level of mutagenesis during translesion synthesis, leading to an increased risk of breast, ovary and endometrial cancers.
Oligodeoxynucleotides containing a single 4-OHEN-dC, a major DNA adduct derived from 4-OHEN, were prepared by a post-synthetic method and used as templates for steady-state kinetic studies.25 Our previous results indicated that DNA pol η and κ bypassed the 4-OHEN-dC by inserting dAMP and dCMP and extended past the lesion. Although the relative bypass frequencies of nucleotide past the lesion can be obtained using the kinetic study, this assay cannot provide detailed miscoding specificity and frequency occurring at the lesion site. To explore the miscoding property of major diastereoisomers (fr-3 and fr-4) of 4-OHEN-dC,4 we prepared a different 4-OHEN-modified template that can be used for quantitative miscoding analysis using two-phase PAGE.26,27 Miscoding events occurring at the 4-OHEN-dC adduct during DNA synthesis catalyzed by human DNA pol η, κ,or ι were determined in the presence of four dNTPs and proliferating cell nuclear antigen (PCNA). Both pol η and pol κ generate a high frequency of mutagenic events during translesion synthesis past the 4-OHEN-dC lesion. We observed also that pol ι can work in conjunction with pol κ, but not with pol η, at a replication fork stalled by a 4-OHEN-dC adduct during DNA synthesis. This is direct evidence that Y-family DNA pols can switch with one another during translesion synthesis.
Results
Preparation of 38-mer oligomer templates containing a single 4-OHEN-dC adduct
To quantify the miscoding frequency and specificity of 4-OHEN-dC adduct using a two-phase PAGE system,26,27 the sequence; 5'CATGCTGATGAATTCCTTCXCTTCTTTCCTCTCCCTTT, where X is 4-OHEN-dC) is required as a template (Figure 2). When a 7-mer oligomer (5'TTCCCTT) containing three C bases was reacted with 4-OHEN, eight products (peaks 2–9) containing one or more 4-OHEN-dC adduct were produced and resolved from the unmodified oligomer (peak 1) using a μBondapak C18 HPLC column (Figure 3). With the XTerra MS C18 HPLC column, peaks 2 and 3 and peaks 6 and 7 were completely resolved (data not shown). 4-OHEN-dC absorbs UV wavelengths greater than 300 nm, and the absorbance is linearly related to the number of 4-OHEN-dC-modifications; therefore, on the basis of the 320 nm/260 nm absorbance ratio, six products (peaks 2–7) are expected to contain a single 4-OHEN-dC modification, and two products (peaks 8 and 9) should contain two 4-OHEN-dC adducts.25 The 4-OHEN-dC-modified 7-mers were ligated to two unmodified oligomers27 for preparation of the 38-mer templates (Figure 2). The adduct position in the three C bases was determined according to the site of termination of primer extension reactions catalyzed by DNA pol η (Figure 4). The primer extension reactions on the templates produced from peaks 2 and 5 were blocked at position 13, indicating that peaks 2 and 5 contain a single stereoisomer of 4-OHEN-dC positioned in the middle of the three C bases (5'TTCC4-OHENCTT). Similarly, peaks 3 and 6, showing blockage at position 12, were predicted to be 5'TTCCC4-OHENTT; and peaks 4 and 7, showing the blockage at position 14, were predicted to be 5'TTC4-OHENCCTT. The molecular mass of the products was confirmed using MALDI-TOF mass spectrometry.25 The parent ions of 4-OHEN-dC-modified 7-mers (peaks 2 and 5) exhibited peaks at m/z 2320, identifying the molecular mass as 2319 Da, while the molecular mass of the unmodified 7-mer exhibited an ion peak at m/z 2024, identifying the molecular mass as 2023 Da (data not shown). The difference in molecular mass between the modified and the unmodified oligomer was 296 Da, which is consistent with the molecular mass of the 4-hydro-xyequilenin moiety in the unique dC adduct (Figure 1). Thus, the reaction products were identified as 7-mers containing a single diastereoisomer of 4-OHEN-dC positioned in the middle of the three C bases. Enzymatic digestion analysis25 showed that peaks 2 and 5 contain fr-4 and fr-3 isomers of monomeric 4-OHEN-dC, respectively. As before, the oligomers containing fr-4 and fr-3 are referred as Pk-3 and Pk-4, respectively.25
Figure 2.
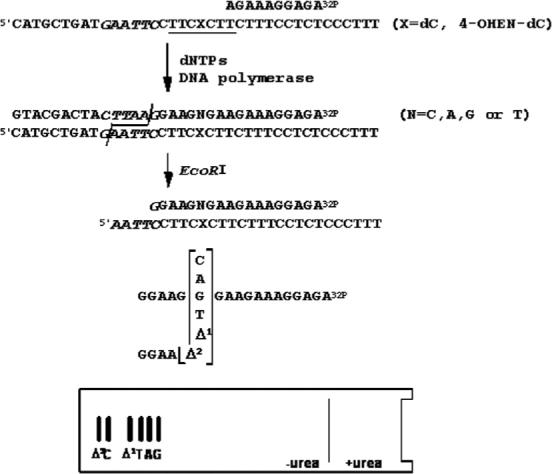
Diagram of the method used to determine miscoding specificity. Unmodified or 4-OHEN-dC-modified 38-mer templates were annealed to a 32P-labeled 10-mer primer. Primer extension reactions catalyzed by Y-family enzymes (pol η, κΔC or ι) were conducted in the presence of four dNTPs. Fully extended products formed during DNA synthesis were recovered from the polyacrylamide gel, annealed with a complementary 38-mer, cleaved with EcoRI, and subjected to two-phase PAGE as described in Materials and Methods. To determine miscoding specificity, the mobility of the reaction products was compared with those of 18-mer standards containing dC, dA, dG, or dT opposite the lesion and one base (Δ1) or two-base (Δ2) deletions.
Figure 3.
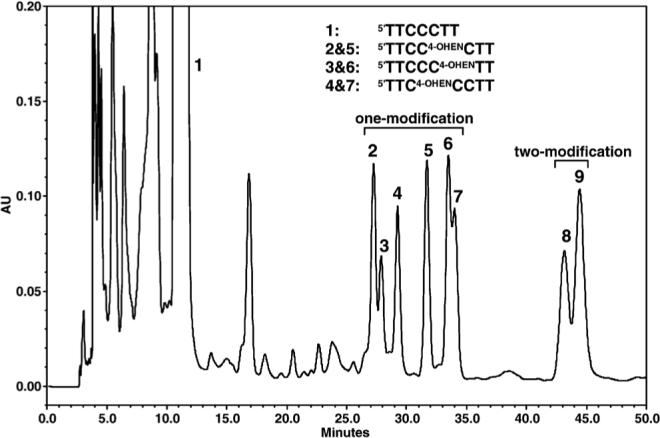
HPLC separation of oligodeoxynucleotides containing a single 4-OHEN-dC adduct. A 200 μg sample of 5'TTCCCTT was reacted at 37 °C for 3 h with 4-OHEN (1 mg/50 μl acetone) in 1 ml of 25 mM potassium phosphate buffer (pH 7.4). After centrifugation, the supernatant was evaporated to dryness and subjected to HPLC. The 7-mer oligomers containing diastereoisoforms of one and more 4-OHEN-dC adducts were isolated on a reverse-phase μBondapak C18 column (0.39 cm × 30 cm, Waters), using a linear gradient of 0.05 M triethylammonium acetate (pH 7.0) in 10%–20% (v/v) acetonitrile, an elution time of 60 min, and a flow-rate of 1.0 ml/min.
Figure 4.
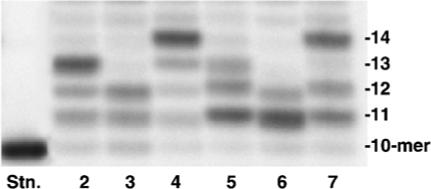
Blockage of primer extension reactions catalyzed by pol η at the 4-OHEN-dC adduct site. Six products containing a single diastereoisomer of 4-OHEN-dC obtained as in Figure 3 were used for preparing their 38-mer oligomer templates as described in Materials and Methods. Using 150 fmol of the modified templates primed with 100 fmol of 32P-labeled 10-mer, primer extension reactions were carried out at 25 °C for 5 min in a buffer containing four dNTPs (100 μM each) and 10 fmol of pol η. One-third of the reaction mixture was subjected to denaturing PAGE (20% (w/v) polyacrylamide) in a 35 cm × 42 cm × 0.04 cm slab gel.
Primer extension reactions catalyzed by pol η, κ, or ι on 4-OHEN-dC-modified DNA template
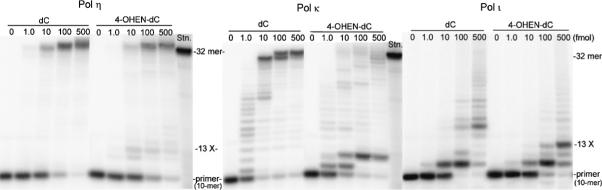
Using unmodified and 4-OHEN-dC-modified 38-mer (Pk-3) templates primed with 32P-labeled 10-mer primer, primer extension reactions catalyzed by either pol η, a truncated form of pol κ (pol κΔC) or pol ι were conducted in the presence of four dNTPs and PCNA (Figure 5). Pol η or κΔC readily extended the primer to form the fully extended product on an unmodified template. Using 4-OHEN-dC-modified template, primer extension reactions catalyzed by pol η were slightly retarded opposite the lesion, and then bypassed the lesion to form the fully-extended product (Figure 5). With pol kΔC, primer extension reactions were retarded more strongly one base before the lesion, and fewer primer extensions bypassed the lesion to form the fully extended product (Figure 5). When the amount of pol η or κΔC was increased, products longer than 32 bases were produced on both the unmodified and 4-OHEN-dC-modified templates. These are blunt-end extension products, as reported earlier for Escherichia coli and mammalian DNA pols.28,29 In contrast, when pol ι was used, primer extensions occurred rarely, even on the unmodified template with a large amount (500 fmol) of this enzyme; no fully-extended product was produced (Figure 5). On the 4-OHEN-dC-modified template, the primer extension reactions were strongly blocked both before and opposite the lesion; only a small amount of further extension was detected.
Figure 5.
Primer extension reactions catalyzed by pol η, κ,or ι on 4-OHEN-dC-modified DNA templates. Using 4-OHEN-dC-modified or unmodified 38-mer template (150 fmol) primed with 100 fmol of 32P-labeled 10-mer, primer extension reactions were carried out at 25 °C for 30 min in a buffer containing four dNTPs (100 μM each) and 0, 1.0 fmol, 10 fmol, 100 fmol or 500 fmol of pol η, pol κΔC, and pol ι, as described in Materials and Methods. One-third of the reaction mixture was subjected to denaturing PAGE as described for Figure 4. A 32-mer (5'AGAGGAAAGTAGCGAAGGAATTCATCAGCATG) was used as a marker of fully extended product. 13X represents the adducted position.
Miscoding properties of 4-OHEN-dC catalyzed by pol η or pol κ
Translesion synthesis catalyzed by pol η or κΔC was conducted in the presence of four dNTPs and PCNA on the unmodified templates and 4-OHEN-dC-modified templates (Pk-3 or Pk-4). The fully extended products (>26 bases long) were recovered from PAGE gels and used for the miscoding analysis using a two-phase PAGE system (Figure 6). A standard mixture of six 32P-labeled oligodeoxynucleotides containing dC, dA, dG, or dT opposite the lesion or one-base and two-base deletions can be resolved by this method (Figure 2). The percentage of miscoding was normalized to the amount of the starting 32P-labeled primer. When products obtained from the primer extension reaction on the unmodified template were analyzed, the expected incorporation of dGMP (75% for pol η and 88% for pol κΔC), the correct base, was observed opposite dC (Figure 6 and Table 1). With pol η for Pk-3, preferential incorporation of dAMP (36.6%) was observed opposite the 4-OHEN-dC, accompanied by smaller amounts of dCMP (12.3%) and dTMP (9.4%) incorporation. Deletions of one base (20.5%) and two bases (4.5%) were also observed. Similar miscoding specificities and frequencies were observed on the Pk-4 template (Figure 6).
Figure 6.
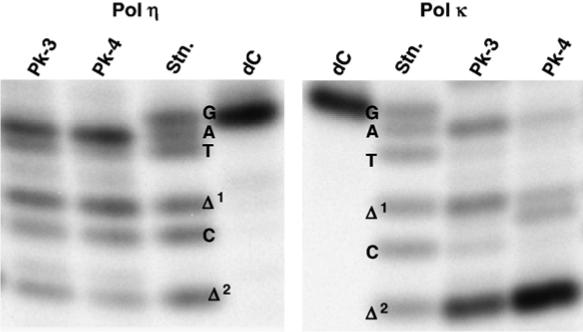
Miscoding specificities of 4-OHEN-dC lesion in reactions catalyzed by pol η or κ. Using 150 fmol of 4-OHEN-dC-modified and unmodified 38-mer template primed with 100 fmol of a 32P-labeled 12-mer primer, primer extension reactions were conducted at 25 °C for 30 min in a buffer containing four dNTPs (100 μM each) and either 100 fmol of pol η or 100 fmol of pol κΔC, as described in Materials and Methods. The extended reaction products (>26 bases long) produced on the unmodified and 4-OHEN-dC-modified templates was extracted following PAGE. The recovered oligodeoxynucleotides were annealed to an unmodified 38-mer and cleaved with EcoRI restriction enzyme, as described in Materials and Methods. Half of the reaction product from the unmodified template and the entire product from the 4-OHEN-dC-modified template were subjected to two-phased electrophoresis in a 20% (w/v) polyacrylamide 15 cm× 72 cm × 0.04 cm slab gel. Mobilities of the reaction products were compared with those of 18-mer standards (Figure 2) containing dC, dA, dG, or dT opposite the lesion and one-base (Δ1)ortwo-base(Δ2) deletions.
Table 1.
Base incorporation and deletions opposite 4-OHEN-dC in reactions catalyzed by Pol η and Pol κΔC
| Miscoding event (%)a |
|||||||
|---|---|---|---|---|---|---|---|
| DNA adduct | C | A | G | T | Δ1 | Δ2 | |
| Pol η | |||||||
| dC | ndb | nd | 75.0±4.5 | nd | nd | nd | |
| 4-OHEN-dC (Pk-3) | 12.3±1.2 | 36.6±4.0 | nd | 9.4±1.0 | 20.5±1.5 | 4.5±0.3 | |
| 4-OHEN-dC (Pk-4) | 12.7±2.1 | 40.2±3.5 | nd | 6.8±1.1 | 22.1±1.8 | 2.6±0.5 | |
| Pol κΔC | |||||||
| dC | nd | nd | 88.0±6.2 | nd | nd | nd | |
| 4-OHEN-dC (Pk-3) | 3.4±0.3 | 16.7±2.1 | nd | 1.0±0.1 | 14.5±2.3 | 42.0±3.5 | |
| 4-OHEN-dC (Pk-4) | 1.6±0.2 | 5.0±0.4 | nd | 0.5±0.1 | 5.5±1.3 | 74.4±7.1 | |
The percentage of nucleotide incorporation (C, A, G, and T) opposite the lesion and deletions (Δ1 and Δ2) were calculated on the basis of the amount of starting primer extension. Data are expressed as the mean±SD obtained from three independent experiments.
Not detectable.
Pol κΔC preferentially promoted deletions (Figure 6 and Table 1); with Pk-3 template, both two-base (42.0%) and one-base (14.5%) deletions were detected. Direct incorporation of dAMP (16.7%), dCMP (3.4%), and dTMP (1.0%) opposite the lesion was also observed. Similar miscoding specificity was observed with Pk-4 template; however, the frequency of two-base deletions was almost twice as high as that of Pk-3 and the frequencies of dAMP and dCMP incorporation were at least 2.5-fold lower than that observed with Pk-3. Surprisingly, with pol κΔC and pol η, no incorporation of dGMP, the correct base, was detected opposite the 4-OHEN-dC adduct in Pk-3 and Pk-4 templates.
Formation of deletions during translesion synthesis by pol κ
To explore the mechanism of forming deletions during translesion synthesis catalyzed by pol κΔC, 4-OHEN-dC adduct in the 38-mer template (Pk-3) was paired with dNMP using 32P-labeled 13N-mer and primer extension reactions catalyzed by pol κΔCwere conducted in the presence of four dNTPs and PCNA. The fully-extended products (>26 bases long) recovered from PAGE gels were used for analysis of miscoding using a two-phase PAGE system (Figure 7). When a 13G-primer was used, only a one-base deletion was formed at the 4-OHEN-dC adduct site; no dGMP incorporation opposite the lesion was observed. With a 13A-primer, a two-base deletion was primarily observed, accompanied by a smaller amount of dAMP incorporation opposite the lesion. With 13C and 13T-primers, incorporation of dCMP and dTMP, respectively, occurred. Thus, when dGMP and dAMP are inserted opposite the 4-OHEN-dC, one-base and two-base deletions, respectively, can be formed during the translesion synthesis.
Figure 7.
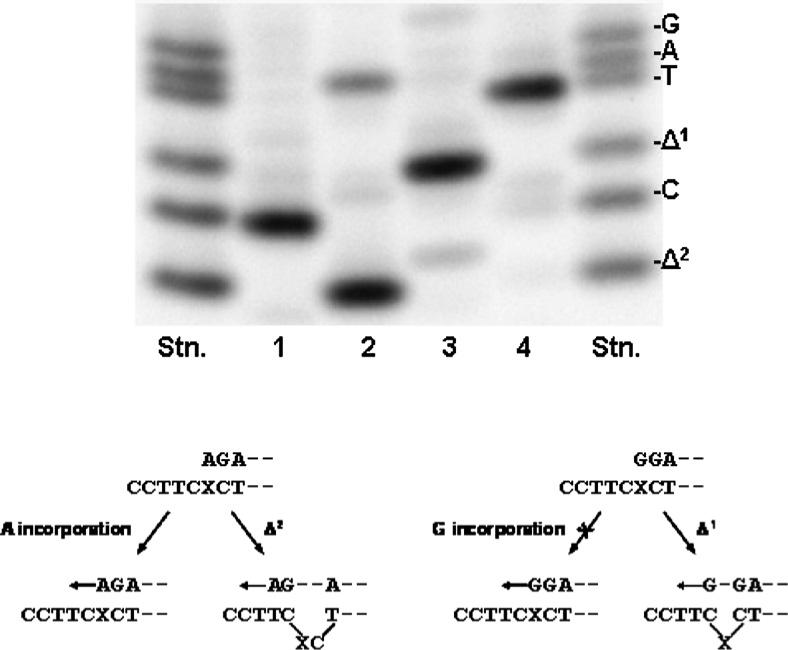
Mechanism of forming deletions and base substitutions. Using 150 fmol of the 4-OHEN-dC modified 38-mer template primed with 100 fmol of a 32P-labeled 13N-mer primer (N = C (1), A (2), G (3), T (4)), primer extension reactions were conducted at 25 °C for 30 min in a buffer containing four dNTPs (100 μM each) and 100 fmol of pol κΔC. The fully extended reaction products (>26 bases long) extracted from PAGE were used for analysis of base substitutions and deletions occurring at the 4-OHEN-dC lesion, as described in the legend to Figure 6.
Kinetic study on 4-OHEN-dC-modified template with pol ι
Steady-state kinetic studies were performed to explore the miscoding properties of 4-OHEN-dC adducts induced by pol ι, as described previously for pol η and κΔC.25 Using unmodified and 4-OHEN-dC-modified 25-mer templates (5'TTTGTXTTTTCTTCTTTCCTCTCCC, where X is dC or 4-OHEN-dC) primed with a 32P-labeled 12-mer (5'GAAAGAAGAAAA), the frequencies of dNTP insertion (Fins) and chain extension (Fext) with pol ι were measured in a buffer containing a single dNTP opposite the 4-OHEN-dC within the linear range of the reaction (Table 2). Fins for dTTP was 2.2-fold higher than that for dGTP, the correct base, and 9.5-fold higher than that for dATP. Fins for dCTP could not be determined because of the poor insertion. Thus, pol ι preferentially incorporated dTMP opposite the 4-OHEN-dC. On the other hand, Fext for the dN·4-OHEN-dC pair could not be determined due to the poor chain extension from the adduct site.
Table 2.
Kinetic parameters for nucleotide insertion reactions catalyzed by pol ι
 | |||
|---|---|---|---|
| N:X | Km (M) | Vmax (% min−1) | Fins |
| X=dC | |||
| G:X | 0.28±0.01 | 6.09±1.84 | 1.0 |
| X=4-OHEN-dC | |||
| C:X | n.d. | n.d. | n.d. |
| A:X | 61.8±4.46 | 0.94±0.11 | 6.91 × 10−4 |
| G:X | 20.8±2.26 | 1.39±0.12 | 3.07 × 10−3 |
| T:X | 13.0±2.57 | 1.84±0.14 | 6.58 × 10−3 |
The kinetics of nucleotide insertion and chain extension reactions were determined as described in Materials and Methods. Frequencies of nucleotide insertion (Fins) were estimated by the equation: F = (Vmax/Km)[wrong pair]/(Vmax/Km)[correct pair]. Data are expressed as the mean±S.D. n.d., not detectable.
Primer extension reactions catalyzed by the combination of pol η+ι or pol κΔC+ι
Primer extension reactions catalyzed by a combination of pol η and pol ι (pol η+ι) or a combination of pol κΔC and pol ι (pol κΔC+ι) were conducted in the presence of four dNTPs and PCNA on unmodified or 4-OHEN-dC-modified 38-mer template (Pk-3) primed with 32P-labeled 12-mer. Using pol η+ι, the primer extension readily occurred to form fully extended products on both unmodified and 4-OHEN-dC modified templates (Figure 8). With pol κΔC alone, the primer extension reactions were blocked before the lesion. The amount of the one-base extended product observed opposite the lesion (position 13X) was increased in parallel with increasing amounts of pol ι, which was capable of inserting a base opposite the 4-OHEN-dC lesion.
Figure 8.
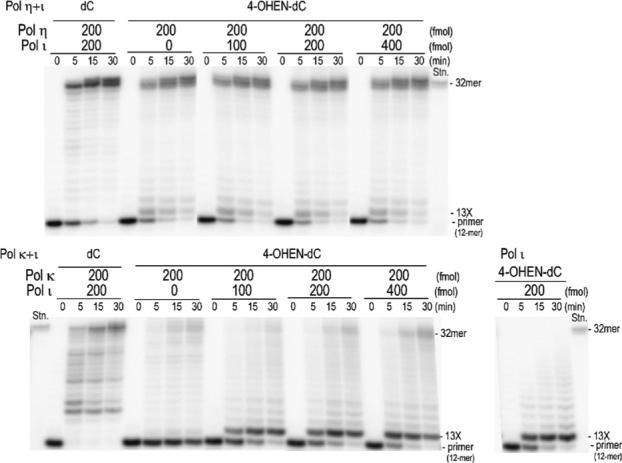
Primer extension reactions catalyzed by the combination of pol η+ι or pol κ+ι on 4-OHEN-dC-modified DNA templates. Using 150 fmol of 4-OHEN-dC-modified or unmodified 38-mer template primed with 100 fmol of 32P-labeled 12-mer, primer extension reactions were carried out at 25 °C for 30 min in a buffer containing four dNTPs (100 μM each) and the combination of pol η+ι or pol κΔC+ι. One-third of the reaction mixture was subjected to denaturing PAGE in a 20% (w/v) polyacrylamide 35 × cm 42 × cm 0.04 cm slab gel. A 32-mer (5'AGAGGAAAGTAGCGAAGGAATTCATCAGCATG) was used as a marker of fully extended product. 13X represents the adducted position.
This indicates that pol ι inserts a base opposite the lesion, but stalls during the primer extension. In addition, the fully extended products (14–32-mers) were also increased in parallel with increasing amounts of this enzyme.
Miscoding specificities of 4-OHEN-dC lesion in reactions catalyzed by the combination of pol η+ι or pol κΔC+ι
The fully extended products (>26 bases long) were subjected to two-phased PAGE to quantify base substitutions and deletions formed opposite the 4-OHEN-dC. When pol ι was added to the primer extension reaction catalyzed by pol κΔC, the amount of dTMP incorporation opposite the 4-OHEN-dC was increased in parallel with the increase in amount of pol ι. In addition, both the amount of dAMP incorporation opposite the lesion and amounts of the one-base and two-base deletions were decreased (Figure 9(b)). In contrast, the miscoding property observed with pol η was not affected by addition of pol ι (Figure 9(a)). No incorporation of dGMP, the correct base, opposite the lesion was detected using a combination of pol κΔC and ι or a combination of pol η and ι.
Figure 9.
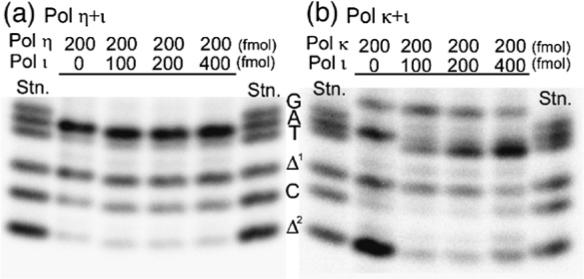
Miscoding specificities of 4-OHEN-dC lesion in reactions catalyzed by the combination of pol η+ι or pol κ+ι. Using 150 fmol of a 4-OHEN-dC-modified and unmodified 38-mer template primed with 100 fmol of a 32P-labeled 12-mer primer, we conducted primer extension reactions at 25 °C for 30 min in a buffer containing four dNTPs (100 μM each) and either (a) pol η+ι (0–400 fmol for pol ι, 200 fmol for pol η) or (b) pol κΔC+ι (0–400 fmol for pol ι, 200 fmol for pol κΔC), as described in Materials and Methods. The extended reaction products (>26 bases long) produced on the unmodified and 4-OHEN-dC-modified templates were extracted following PAGE. The recovered oligodeoxynucleotides were annealed to an unmodified 38-mer and cleaved with EcoRI restriction enzyme, as described in Materials and Methods. Half of the reaction product from the unmodified template and the entire product from the 4-OHEN-dC-modified template were subjected to two-phased electrophoresis in a 20% (w/v) polyacrylamide 15 cm × 72 cm × 0.04 cm slab gel.
Discussion
We have previously observed that 4-OHEN-dC, a major equine estrogen-DNA adduct, strongly blocked the primer extension reactions catalyzed by replicative human DNA pol α or δ;25 no nucleotide was inserted opposite the lesion even using a large amount of the enzyme. Since Y-family DNA pols (pol η, κ or ι) are involved in translesion synthesis past bulky DNA adducts,12,13 and are highly expressed in reproductive organs,21-24 we used these DNA pols to explore miscoding events occurring at a major diastereoisomer (fr-3 or fr-4) of 4-OHEN-dC during translesion synthesis. The relative bypass frequency of nucleotide past the DNA adducts have been estimated using steady-state kinetic studies employing primer extension reactions catalyzed by DNA pol in the presence of only a single dNTP where there is no competition between nucleotides.25 In the present study, a two-phased PAGE system established previously in our laboratory was utilized to analyze the fully extended products past the DNA adduct in the presence of all four dNTPs and PCNA for quantification of the miscoding frequency and specificity occurring at the lesion site.26,27 Using this technique, pol η preferentially inserted dAMP opposite 4-OHEN-dC, accompanied by smaller amounts of dCMP and dTMP incorporation and base deletions. Although pol κΔC promoted one-base and two-base deletions preferentially, direct incorporation of dAMP and, to a lesser extent, dCMP and dTMP was also detected. Thus, the miscoding properties vary depending on the DNA polymerase used.
Surprisingly, with both pol η and pol κΔC, no direct incorporation of dGMP, the correct base, was observed opposite the 4-OHEN-dC adduct. It was proved also that no dGMP incorporation opposite the adduct was formed even when the dG•4-OHEN-dC pair was extended using pol κΔC(Figure 7). Although conformational studies were performed for the dG·4-OHEN-dC pair in duplex DNA,30 we did not observe such correct base-pairing during the translesion synthesis catalyzed by pol η and κ. These results indicate that the major isomers of 4-OHEN-dC are highly miscoding lesions, generating primarily C→T transitions, along with C→G and C→Atransversions.
Pk-3- and Pk-4-oligomers contain the fr-4 and fr-3 diastereoisomers of 4-OHEN-dC, respectively. The absolute structures of these isomers have not been determined despite attempts using several NMR techniques;4 a crystallographic technique may be required to establish it. Pol η promoted primarily dAMP incorporation opposite each lesion, along with dCMP incorporation. The miscoding events occurring at fr-4 of 4-OHEN-dC (Pk-3 template) was identical with that observed with fr-3 (Pk-4 template). The observation was supported by steady-state kinetic studies performed previously with a different template sequence.25 Although, with pol κΔC, the miscoding specificity observed with Pk-3 template was similar to that of Pk-4 template, the frequency of two-base deletions was increased and the frequencies of dAMP and dCMP incorporation were decreased. Geacintov and his colleagues indicated that 4-OHEN-dC isomers with positive circular dichroism absorbance (CD) were oriented toward the 5' terminus in duplex DNA while the isomers with negative CD were oriented toward the 3' terminus in duplex DNA.6,31 Since fr-3 and fr-4 isolated in our study have negative and positive CD, respectively,25 fr-3 in Pk-4-oligomer orients in the 3'-direction and fr-4 in Pk-3-oligomer orients in the 5'-direction in duplex DNA. Such adduct stereochemistry may affect the miscoding property generated by pol κΔC; but not by pol η.
With a template having a different sequence context, steady-state kinetic studies performed using pol κΔC showed that the relative bypass frequency past the dC·4-OHEN-dC pair was more efficient than that for the dA·4-OHEN-dC pair. The result was the opposite of that obtained with two-phase PAGE assay, suggesting that pol κΔC promoted primarily direct dAMP incorporation, along with dCMP incorporation, at the lesion. Such a difference may reflect the sequence context adjacent to the lesion.
Formation of one-base and two-base deletions was observed primarily with pol κΔC during translesion synthesis past the 4-OHEN-dC. To explore the deletion mechanism, we analyzed the fully extended products obtained from the chain extension reactions from the dN•4-OHEN-dC pairs (Figure 7). When the dA•4-OHEN-dC pair was used, the predominant product formed was a two-base deletion. A smaller amount of product with dAMP opposite the lesion was also formed. These results may be explained if the terminal dAMP of the primer preferentially pairs with T two bases 5' to the lesion, forming the two-base deletions; whereas, when dAMP is positioned directly opposite the lesion, only a portion of such primers may be chain-extended (Figure 7). With the dG·4-OHEN-dC pair, only one-base deletions were formed. dGMP positioned opposite the lesion may not be chain-extended; the majority of dGMP may pair with the C immediately 5' to the lesion, forming the one-base deletions (Figure 7). Since dCMP or dTMP positioned opposite the lesion cannot pair with any base 5' to the lesion on the template, no deletion could be formed. Steady-state kinetic studies performed previously25 indicated that pol κΔC inserted dATP and dCMP opposite the 4-OHEN-dC lesion. Therefore, two-base deletions could be formed primarily at the lesion site. As demonstrated for acetylaminofluorene-derived DNA adducts,32,33 the insertion frequency of dNTP opposite the lesion may be a limiting factor in generating frameshift deletions in vitro.
Switching mechanisms between replicative DNA pols and Y-family DNA pols have recently been proposed to explain translesion synthesis past DNA adducts,9,11 based on indirect assay systems employing a simple TLS bypass assay14-16 and/or steady-state kinetic studies.17-20 In addition, expression and localization of fluorescently tagged TLS pols have been employed to predict which TLS pol catalyzes synthesis at stalled DNA replication forks following DNA-damaging treatment.34,35 In the present studies, using a combination of either pol κΔC+ι or pol η+ι for primer extension reactions past the 4-OHEN-dC lesion, the miscoding specificity and frequency were quantified using a two-phase PAGE system (Figures 8 and 9). Although the majority of primer extension reactions catalyzed by pol κΔC alone were blocked one base before the 4-OHEN-dC lesion, the additional pol ι proceeded to insert dTMP opposite the lesion, but then further chain extension was blocked. Since pol ι alone has only a weak ability to extend the primer from the adducted base-pair, the increase of fully extension product may be reflected by the increased dTMP insertion opposite the lesion by the additional pol ι, followed by chain extension by pol κΔC. Among DNA pols, pol ι is the only one that prefers to insert dTMP opposite the 4-OHEN-dC lesion, as supported by our steady-state kinetics study (Table 2). On the other hand, the original incorporation of dAMP and the one-base and two-base deletions observed opposite the 4-OHEN-dC lesion with pol κΔC alone were reduced by addition of pol ι. This indicates that pol ι assists in extending the primer strand on which pol κ is stalled (before the lesion) by incorporating dTMP opposite the lesion (Figure 10). After that, pol κ may again take the place of pol ι to extend the primer, as reported previously.19,20 Our results are direct evidence that Y-family DNA pols can switch with each other during translesion synthesis past the lesion. In contrast, the original miscoding property observed with pol η alone was not affected by addition of pol ι. Although pol η and pol ι associate with each other and have overlapping binding sites on PCNA during DNA replication,9,34 our observation indicates that pol η can work independently from pol ι when bypassing the 4-OHEN-dC lesion.
Figure 10.
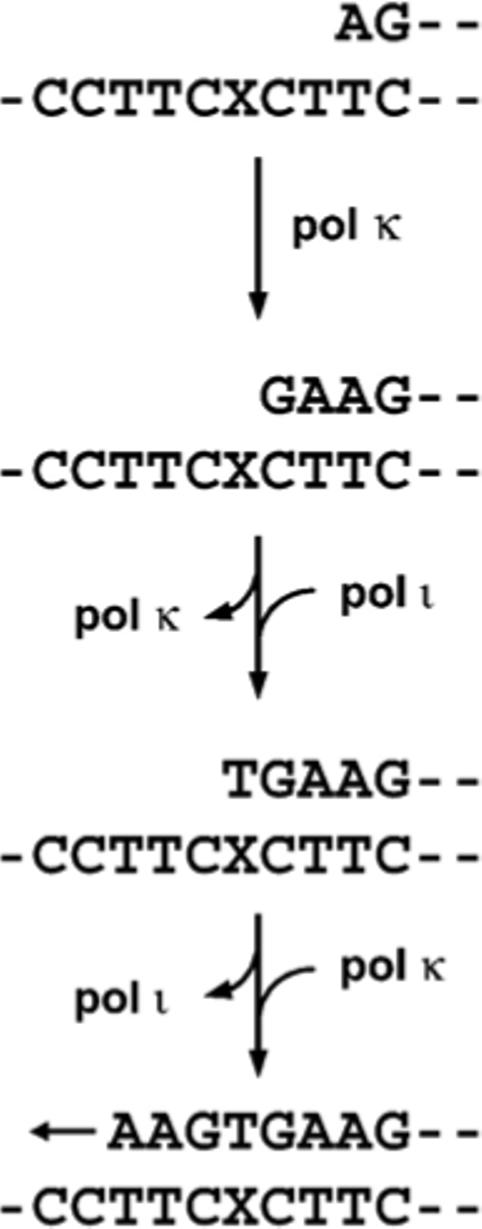
Model for enzymatic switching during translesion synthesis past 4-OHEN-dC.
We have here demonstrated that the two-phase PAGE can be used to determine quantitatively miscoding properties of DNA adduct in the presence of a mixture of DNA pols and co-factors. Using this system, the influence of replicative DNA pols on the miscoding specificity and frequency of the 4-OHEN-dC adduct generated by the Y-family enzymes could be investigated. In addition, when concentration of each DNA polymerases existing in reproductive organs is established, the miscoding properties occurring under physiological condition could be explored. Such important studies will be required in future to understand the translesion mechanism past the DNA adduct.
When 4-OHEN-dC-containing pMY189 shuttle vector plasmid carrying a bacteria suppressor tRNA gene, supF, was propagated in human lung fibroblast cells, C:G → G:C and C:G → A:T mutations were primarily detected, accompanied by fewer C:G → T:A transitions.36 Therefore, the majority of mutations in the cells may have been due to the direct incorporation of dCMP or dTMP, not dAMP, opposite the 4-OHEN-dC lesion. Since pol η, κ, and ι are expressed in human lung tissues, as they are in reproductive organs,21,24 these pols may be associated with the 4-OHEN-induced mutations detected in the fibroblast cells. However, the mutational spectrum observed in the cells is not consistent with our present results showing that direct dAMP incorporation, followed by dCMP incorporation, are the major base-substitutions promoted by pol η or κΔC. Rather, only a small amount of dTMP was incorporated opposite 4-OHEN-dC during translesion synthesis catalyzed by pol η and κΔC(Figure 9). The higher frequency of C:G → A:T mutations observed in the fibroblast cells may be due to the increased dTMP incorporation opposite the 4-OHEN-dC lesion brought about by the association of pol ι and κΔC. Alternatively, oxidative DNA damages, including 8-oxo-7,8-dihydro-2'-deoxyguanosine (8-oxodG), may be induced by reactive oxygen species generated during redox reaction between the 4-OHEN o-quinone and their semiqui-none radicals.37-42 Since 8-oxodG generates G → T transversions mediated primarily by several DNA pols, including pol η and pol κ,18,43-45 C:G → A:T mutations observed in the fibroblast cells may be due to oxidative DNA damage occurring at C:G pairs.
In conclusion, the Y-family DNA pols η, κ, and ι showed unique characteristics for translesion synthesis past the major 4-OHEN-dC adducts. In particular, major 4-OHEN-dC was a highly miscoding lesion for pols η and κΔC, which incorporate primarily dAMP or generate deletions during translesion DNA synthesis. The miscoding frequency and specificity was also influenced by the adduct stereochemistry and sequence context adjacent to the lesion. Pol ι could insert dTMP opposite the lesion, but further chain extension was blocked. On the basis of the different miscoding specificities of each Y-family DNA pol, we provided a direct evidence that pol ι can work in conjunction with pol κ, but not with pol η, during translesion synthesis at a replication fork stalled by a 4-OHEN-dC lesion.
Materials and Methods
General
[γ-32P]ATP (spec. act. >6000 Ci/mmol) and dNTPs were obtained from GE Healthcare Bio-Sciences Corp. (Piscataway, NJ). Bacteriophage T4 polynucleotide kinase and EcoRI restriction endonuclease were purchased from New England BioLabs (Ipswich, MA). Human pol η and pol κΔC were provided by Dr Hanaoka and Dr Ohmori, respectively. Human pol κ and ι were purchased from Enzymax, LCC (Lexington, KY).
Synthesis of 4-OHEN-dC modified oligodeoxynucleotides
A 7-mer oligodeoxynucleotide containing three C bases (5'TTCCCTT, 200 μg) was reacted at 37 °C for 3 h with 4-OHEN (1 mg (3.5 μmol)/50 μl acetone) in 1 ml of 25 mM potassium phosphate buffer (pH 7.4).25 To avoid subjecting insoluble materials to HPLC, the reaction mixture was centrifuged. The supernatant was evaporated to dryness and subjected to HPLC. Several products containing one or more 4-OHEN-dC adducts were isolated on a reverse-phase μBondapak C18 column (0.39 cm × 30 cm, Waters), using a linear gradient composed of 0.05 M triethylammonium acetate (pH 7.0) in 10%–20% (v/v) acetonitrile, an elution time of 60 min, and a flow-rate of 1.0 ml/min. Each isolated product was further purified using an XTerra MS C18 column (0.46 cm × 5.0 cm, Waters) column, with the HPLC conditions used for the μBondapak C18 column. 4-OHEN-dC absorbs UV at >300 nm, and the absorbance is linearly related to the number of 4-OHEN-dC-modifications; therefore, on the basis of the 320 nm/260 nm absorbance ratio, the number of 4-OHEN-dC modifications in each product was estimated. Six products containing a single diastereoisomer of 4-OHEN-dC were used for preparing their 38-mer oligomers as described below. The position of the 4-OHEN-dC adduct was determined by the blockage of primer extension reactions catalyzed by pol η on the modified 38-mer oligomers.
MALDI-TOF analysis
The molecular mass of the products was determined by matrix assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry.25 Samples were run on a Voyager-DE STR MALDI-TOF mass spectrometer system (Applied Biosystems, Framingham, MA) operated in the linear mode. Samples were dissolved in a 50% (v/v) solution of acetonitrile/water containing hydroxypicolinic acid (5 mg/ml) and ammonium citrate at an 8:1 (v/v) ratio and dried on a sample plate. A nitrogen laser operating at 337 nm and a 3 ns pulse rate was employed. The accelerating voltage was set to 20 kV and a delay of 250 ns was used to accelerate ions into the flight tube of the mass spectrometer. The mass scale (m/z 1000–10,000) was calibrated in the positive ion mode with a mixture of standard oligodeoxynucleotides and approximately 100 laser shots were used to produce each spectrum.
Primer extension reactions
Pol η and pol κΔC were prepared as described.46,47 Although pol κΔC has a slightly lower processivity than that of full-length pol κ, the miscoding rate on the undamaged DNA by pol κΔC was similar to that of pol κ.48 The 4-OHEN-dC-modified 7-mers (5'TTCC4-OHENCTT) were ligated to two unmodified oligomers 5'CATGCTGATGAATTCC and 5'CTTTCCTCTCCCTTT)25,26 for preparation of the 38-mer templates 5'CATGCTGATGAATTCCTTCC4-OHENCTTCTTTCCTCTCCCTTT used for the miscoding studies (Figure 2). A 10-mer (5'AGAGGAAAGA), 12-mer (5'AGAGGAAAGAAG), or 13N-mer (5'AGAGGAAAGAAGN, N=C, A, G, or T) primer was labeled at the 5'-terminus by treating with T4 polynucleotide kinase in the presence of [γ-32P]ATP.49 Using the 4-OHEN-dC-modified or unmodified 38-mer 5'CATGCTGATGAATTCCTTCXCTTCTTTCCTCTCC where X is dC or 4-OHEN-dC) template (150 fmol) primed with a 32P-labeled 10-mer, 12-mer, or 13-mer primer (100 fmol), primer extension reactions catalyzed by pol η, κΔC, ι, or the mixture of pol η and ι (pol η+ι)orpol κ and ι (pol κΔC+ι) (the amount of each pol is described in the Figure legends) were carried out at 25 °C for 30 min in 20 μl of a buffer containing four dNTPs (100 μM each) and 1400 fmol of PCNA.25 The primer extension reactions catalyzed by pol η+ι or κΔC+ι were started by addition of primer-annealed DNA template,after the two pols,dNTPs, and PCNA were mixed into a reaction buffer. The reaction buffer for pol η and ι was 40 mM Tris–HCl (pH 8.0), 1 mM MgCl2, 10 mM DTT, 250 μg/ml of BSA, 60 mM KCl, 2.5% (v/v) glycerol. A similar reaction buffer was used for pol κ or κΔC, using 5 mM MgCl2 instead of 1 mM MgCl2. Reactions were stopped by the addition of formamide dye. The sample was subjected to denaturing PAGE in a 20% (w/v) polyacrylamide 35 cm×42 cm×0.04 cm slab gel. The radioactivity of the extended products was measured using STORM and ImageQuaNT software (Molecular Dynamics).
Quantification of miscoding specificity
Using 150 fmol of 4-OHEN-dC-modified and unmodified 38-mer template primed with 100 fmol of a 32P-labeled 12-mer primer or 13N-mer, primer extension reactions catalyzed by pol η, κΔC, η+ι,or κΔC+ι were conducted at 25 °C for 30 min in 20 μl of a buffer containing four dNTPs (100 μM each) and 1400 fmol of PCNA. When pol η+ι or pol κΔC+ι was used, the primer extension reactions were initiated by the addition of primer-annealed DNA template, after the two pols, dNTPs, and PCNA were mixed into a reaction buffer. Reactions were stopped by adding formamide dye and then subjected to denaturing PAGE in a 20% (w/v) polyacrylamide 35 cm × 42 cm × 0.04 cm slab gel Fully extended products (>26 bases long) were recovered from the gel, annealed with the complementary oligomer, cleaved by EcoRI (100 U) at 15 °C overnight, and subjected to two-phased PAGE (15 cm × 72 cm × 0.04 cm) with 7 M urea in the upper phase and no urea in the lower phase (Figure 2).26,27 To quantify base substitutions and deletions, mobilities of the reaction products were compared with those of 18-mer standards containing dC, dA, dG, or dT opposite the lesion and one-base (Δ1)or two-base (Δ2) deletions. The percentage of miscoding was normalized to the amount of the starting 32P-labeled primer.
Steady-state kinetics studies
Kinetic parameters at the 4-OHEN-dC lesion in reactions catalyzed by pol ι were determined in a buffer containing a single dNTP (0–500 μM) as described for pol η and κΔC.25 For insertion kinetics, reaction mixtures containing varying amounts (0.32–3.2 fmol) of pol ι were incubated at 25 °C for 2 min in 10 μl of a reaction buffer containing 150 fmol of 25-mer template (5'TTTGTXTTTTCTTCTTTCCTCTCCC, where X is dC or 4-OHEN-dC) primed with 100 fmol of 32P-labeled 12-mer (5'GAAAGAAGAAAA).25 Reaction mixtures containing 150 fmol of 25-mer template primed with 100 fmol of a 32P-labeled 13N-mer(5'GAAAGAAGAAAAN,whereNisC,A,GorT), with 0–500 μM dATP, and 0.32–3.2 fmol of pol ι were used to measure chain extension kinetics. The reaction samples were subjected to denaturing PAGE in a 20% (w/v) polyacrylamide 35 cm × 42 cm × 0.04 cm slab gel. The Michaelis constants (Km) and maximum rates of reaction (Vmax) were obtained from Hanes-Woolf plots. The frequencies of dNTP insertion (Fins) and chain extension (Fext) were determined relative to the dC:dG base-pair according to the equation:50,51
Acknowledgements
We are grateful to Drs F. Hanaoka and H. Ohmori for providing human pol η and pol κΔC, respectively. We thank Ms S. Yasui for experimental assistance. This research was supported by grants ES012408 from the National Institute of Environmental Health Sciences.
Glossary
Abbreviations used:
- EQ
equilin
- EN
equilenin
- 4-OHEQ
4-hydroxyequilin
- 4-OHEN
4-hydroxyequilenin
- 8-oxodG
8-oxo-7,8-dihydro-2′-deoxyguanosine
- dNTP
2′-deoxynucleoside triphosphate
- pol η
DNA polymerase η
- pol κ
DNA polymerase κ
- pol κΔC
a truncated form of pol κ
- pol ι
DNA polymerase ι
- Fins
frequency of insertion
- Fext
frequency of extension
References
- 1.Colditz GA, Hankinson SE, Hunter DJ, Willett WC, Manson JE, Stampfer MJ, et al. The use of estrogens and progestins and the risk of breast cancer in postmenopausal women. N. Engl. J. Med. 1995;332:1589–1593. doi: 10.1056/NEJM199506153322401. [DOI] [PubMed] [Google Scholar]
- 2.Grodstein F, Stampfer MJ, Colditz GA, Willett WC, Manson JE, Joffe M, et al. Post-menopausal hormone therapy and mortality. N. Engl. J. Med. 1997;336:1769–1775. doi: 10.1056/NEJM199706193362501. [DOI] [PubMed] [Google Scholar]
- 3.Lacey JV, Jr, Mink PJ, Lubin JH, Sherman ME, Troisi R, Hartge P, et al. Menopausal hormone replacement therapy and risk of ovarian cancer. JAMA. 2002;288:334–341. doi: 10.1001/jama.288.3.334. [DOI] [PubMed] [Google Scholar]
- 4.Bolton JL, Pisha E, Zhang F, Qiu S. Role of quinoids in estrogen carcinogenesis. Chem. Res. Toxicol. 1998;11:1113–1127. doi: 10.1021/tx9801007. [DOI] [PubMed] [Google Scholar]
- 5.Shen L, Qiu S, Chen Y, Zhang F, van Breemen RB, Nikolic D, Bolton JL. Alkylation of 2′-deoxynucleosides and DNA by the Premarin metabolite 4-hydroxyequilenin semiquinone radical. Chem. Res. Toxicol. 1998;11:94–101. doi: 10.1021/tx970181r. [DOI] [PubMed] [Google Scholar]
- 6.Ding S, Shapiro R, Geacintov NE, Broyde S. Conformations of stereoisomeric base adducts to 4-hydroxyequilenin. Chem. Res. Toxicol. 2003;16:695–707. doi: 10.1021/tx0340246. [DOI] [PubMed] [Google Scholar]
- 7.Zhang F, Chen Y, Pisha E, Shen L, Xiong Y, van Breemen RB, Bolton JL. The major metabolite of equilin, 4-hydroxyequilin, autoxidizes to an o-quinone which isomerizes to the potent cytotoxin 4-hydroxyequilenin-o-quinone. Chem. Res. Toxicol. 1999;12:204–213. doi: 10.1021/tx980217v. [DOI] [PubMed] [Google Scholar]
- 8.Embrechts J, Lemière F, Van Dongen W, Esmans EL, Buytaert P, Van Marck E, et al. Detection of estrogen DNA-adducts in human breast tumor tissue and healthy tissue by combined nano LC-nano ES tandem mass spectrometry. J. Am. Soc. Mass Spectrom. 2003;14:482–491. doi: 10.1016/S1044-0305(03)00130-2. [DOI] [PubMed] [Google Scholar]
- 9.Plosky BS, Woodgate R. Switching from high-fidelity replicases to low-fidelity lesion-bypass polymerases. Curr. Opin. Genet. Dev. 2004;14:113–119. doi: 10.1016/j.gde.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 10.Lehmann AR. Replication of damaged DNA by translesion synthesis in human cells. FEBS Letters. 2005;579:873–876. doi: 10.1016/j.febslet.2004.11.029. [DOI] [PubMed] [Google Scholar]
- 11.Friedberg EC, Lehmann AR, Fuchs RP. Trading places: how do DNA polymerases switch during translesion DNA synthesis? Mol. Cell. 2005;18:499–505. doi: 10.1016/j.molcel.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 12.Goodman MF, Tippen B. The expanding polymerase universe. Nature Rev. Mol. Cell Biol. 2000;1:101–109. doi: 10.1038/35040051. [DOI] [PubMed] [Google Scholar]
- 13.Kunkel TA, Pavlov YI, Bebenek K. Functions of human DNA polymerases η, κ and ι suggested by their properties, including fidelity with undamaged DNA templates. DNA Repair. 2003;2:135–149. doi: 10.1016/s1568-7864(02)00224-0. [DOI] [PubMed] [Google Scholar]
- 14.Kokoska RJ, McCulloch SD, Kunkel TA. The efficiency and specificity of apurinic/ apyrimidinic site bypass by human DNA polymerase η and Sulfolobus solfataricus Dpo4. J. Biol. Chem. 2003;278:50537–50545. doi: 10.1074/jbc.M308515200. [DOI] [PubMed] [Google Scholar]
- 15.McCulloch SD, Kokoska RJ, Chilkova O, Welch CM, Johansson E, Burgers PM, Kunkel TA. Enzymatic switching for efficient and accurate translesion DNA replication. Nucl. Acids Res. 2004;32:4665–4675. doi: 10.1093/nar/gkh777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McCulloch SD, Kokoska RJ, Masutani C, Iwai S, Hanaoka F, Kunkel TA. Preferential cissyn thymine dimer bypass by DNA polymerase η occurs with biased fidelity. Nature. 2004;428:97–100. doi: 10.1038/nature02352. [DOI] [PubMed] [Google Scholar]
- 17.Johnson RE, Washington MT, Haracska L, Prakash S, Prakash L. Eukaryotic polymerases ι and ζ act sequentially to bypass DNA lesions. Nature. 2000;406:1015–1019. doi: 10.1038/35023030. [DOI] [PubMed] [Google Scholar]
- 18.Haracska L, Prakash L, Prakash S. Role of human DNA polymerase κ as an extender in translesion synthesis. Proc. Natl Acad. Sci. USA. 2002;99:16000–16005. doi: 10.1073/pnas.252524999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Washington MT, Minko IG, Johnson RE, Wolfle WT, Harris TM, Lloyd RS, et al. Efficient and error-free replication past a minor-groove DNA adduct by the sequential action of human DNA polymerases ι and κ. Mol. Cell Biol. 2004;24:5687–5693. doi: 10.1128/MCB.24.13.5687-5693.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wolfle WT, Johnson RE, Minko IG, Lloyd RS, Prakash S, Prakash L. Replication past a trans-4-hydroxynonenal minor-groove adduct by the sequential action of human DNA polymerases ι and κ. Mol. Cell Biol. 2006;26:381–386. doi: 10.1128/MCB.26.1.381-386.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerlach VL, Aravind L, Gotway G, Schultz RA, Koonin EV, Friedberg EC. Human and mouse homologs of Escherichia coli DinB (DNA polymerase IV), members of the UmuC/DinB super-family. Proc. Natl Acad. Sci. USA. 1999;96:11922–11927. doi: 10.1073/pnas.96.21.11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ogi T, Kato T,, Jr, Kato T, Ohmori H. Mutation enhancement by DINB1, a mammalian homologue of the Escherichia coli mutagenesis protein DinB. Genes Cells. 1999;4:607–618. doi: 10.1046/j.1365-2443.1999.00289.x. [DOI] [PubMed] [Google Scholar]
- 23.Velasco-Miguel S, Richardson JA, Gerlach VL, Lai WC, Gao T, Russell LD, et al. Constitutive and regulated expression of the mouse Dinb (Pol κ) gene encoding DNA polymerase κ. DNA Repair. 2003;2:91–106. doi: 10.1016/s1568-7864(02)00189-1. [DOI] [PubMed] [Google Scholar]
- 24.Kawamura K, Bahar R, Seimiya M, Chiyo M, Wada A, Okada S, et al. DNA polymerase θ is preferentially expressed in lymphoid tissues and upregulated in human cancers. Int. J. Cancer. 2004;109:9–16. doi: 10.1002/ijc.11666. [DOI] [PubMed] [Google Scholar]
- 25.Suzuki N, Yasui M, Laxmi YRS, Ohmori H, Hanaoka F, Shibutani S. Translesion synthesis past equine estrogen-derived 2′-deoxycytidine DNA adducts by human DNA polymerases η and κ. Biochemistry. 2004;43:11312–11320. doi: 10.1021/bi049273n. [DOI] [PubMed] [Google Scholar]
- 26.Shibutani S. Quantitation of base substitutions and deletions induced by chemical mutagens during DNA synthesis in vitro. Chem. Res. Toxicol. 1993;6:625–629. doi: 10.1021/tx00035a006. [DOI] [PubMed] [Google Scholar]
- 27.Shibutani S, Suzuki N, Matsumoto Y, Grollman AP. Miscoding properties of 3,N4-etheno-2′-deoxycytidine in reactions catalyzed by mammalian DNA polymerases. Biochemistry. 1996;35:14992–14998. doi: 10.1021/bi961446o. [DOI] [PubMed] [Google Scholar]
- 28.Clark JM, Joyce CM, Beardsley GP. Novel blunt-end addition reactions catalyzed by DNA polymerase I of Escherichia coli. J. Mol. Biol. 1987;198:123–127. doi: 10.1016/0022-2836(87)90462-1. [DOI] [PubMed] [Google Scholar]
- 29.Terashima I, Suzuki N, Dasaradhi L, Tan C-K, Downey KM, Shibutani S. Translesional synthesis on DNA templates containing an estrogen quinine-derived adduct: N2-(2-hydroxyestron-6-yl)-2′-deoxyguanosine and N6-(2-hydroxyestron-6-yl)-2′-deoxyadenosine. Biochemistry. 1998;37:13807–13815. doi: 10.1021/bi981235e. [DOI] [PubMed] [Google Scholar]
- 30.Ding S, Shapiro R, Geacintov NE, Broyde S. Equilenin-derived DNA adducts to cytosine in DNA duplexes: structures and thermodynamics. Biochemistry. 2005;44:14565–14576. doi: 10.1021/bi051090t. [DOI] [PubMed] [Google Scholar]
- 31.Kolbanovskiy A, Kuzmin V, Shastry A, Kolbanovskaya M, Chen D, Chang M, et al. Base selectivity and effects of sequence and DNA secondary structure on the formation of covalent adducts derived from the equine estrogen metabolite 4-hydroxyequilenin. Chem. Res. Toxicol. 2005;18:1737–1747. doi: 10.1021/tx050190x. [DOI] [PubMed] [Google Scholar]
- 32.Shibutani S, Grollman AP. On the mechanism of frameshift (deletion) mutagenesis in vitro. J. Biol. Chem. 1993;268:11703–11710. [PubMed] [Google Scholar]
- 33.Shibutani S, Suzuki N, Grollman AP. Mechanism of frameshift (deletion) generated by acetylaminofluorene-derived DNA adducts in vitro. Biochemistry. 2004;43:15929–15935. doi: 10.1021/bi048087e. [DOI] [PubMed] [Google Scholar]
- 34.Kannouche P, Fernández de Henestrosa AR, Coull B, Vidal AE, Gray C, Zicha D, et al. Localization of DNA polymerases η and ι to the replication machinery is tightly co-ordinated in human cells. EMBO J. 2003;22:1223–1233. doi: 10.1093/emboj/cdf618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase η with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell. 2004;14:491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- 36.Yasui M, Matsui S, Laxmi YRS, Suzuki N, Kim SY, Shibutani S, Matsuda T. Mutagenic events induced by 4-hydroxyequilin in supF shuttle vector plasmid propagated in human cells. Carcino-genesis. 2003;24:911–917. doi: 10.1093/carcin/bgg029. [DOI] [PubMed] [Google Scholar]
- 37.Han X, Liehr JG. Microsome-mediated 8-hydroxylation of guanine bases of DNA by steroid estrogens: correlation of DNA damage by free radicals with metabolic activation to quinones. Carcinogenesis. 1995;16:2571–2574. doi: 10.1093/carcin/16.10.2571. [DOI] [PubMed] [Google Scholar]
- 38.Chen Y, Shen L, Zhang F, Lau SS, van Breemen RB, Nikolic D, Bolton JL. The equine estrogen metabolite 4-hydroxyequilenin causes DNA single-strand breaks and oxidation of DNA bases in vitro. Chem. Res. Toxicol. 1998;11:1105–1111. doi: 10.1021/tx980083l. [DOI] [PubMed] [Google Scholar]
- 39.Chen Y, Liu X, Pisha E, Constantinou AI, Hua Y, Shen L, et al. A metabolite of equine estrogens, 4-hydroxyequilenin, induces DNA damage and apoptosis in breast cancer cell lines. Chem. Res. Toxicol. 2000;13:342–350. doi: 10.1021/tx990186j. [DOI] [PubMed] [Google Scholar]
- 40.Zhang F, Swanson SM, van Breemen RB, Liu X, Yang Y, Gu C, Bolton JL. Equine estrogen metabolite 4-hydroxyequilenin induces DNA damage in the rat mammary tissues: formation of single-strand breaks, apurinic sites, stable adducts, and oxidized bases. Chem. Res. Toxicol. 2001;14:1654–1659. doi: 10.1021/tx010158c. [DOI] [PubMed] [Google Scholar]
- 41.Zhang F, Yao D, Hua Y, van Breemen RB, Bolton JL. Synthesis and reactivity of the catechol metabolites from the equine estrogen, 8,9-dehydroestrone. Chem. Res. Toxicol. 2001;14:754–763. doi: 10.1021/tx010049y. [DOI] [PubMed] [Google Scholar]
- 42.Liu X, Yao J, Pisha E, Yang Y, Hua Y, van Breemen RB, Bolton JL. Oxidative DNA damage induced by equine estrogen metabolites: role of estrogen receptor α. Chem. Res. Toxicol. 2002;15:512–519. doi: 10.1021/tx0101649. [DOI] [PubMed] [Google Scholar]
- 43.Shibutani S, Takeshita M, Grollman AP. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 44.Moriya M. Single strand shuttle phagemid for mutagenesis studies in mammalian cells: 8-oxogua-nine in DNA induces targeted G:C → T:A transversion in simian kidney cells. Proc. Natl Acad. Sci. USA. 1993;90:1122–1126. doi: 10.1073/pnas.90.3.1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tan X, Grollman AP, Shibutani S. Comparison of the mutagenic properties of 8-oxo-7,8-dihydro-2′-deoxyadenosine and 8-oxo-7,8-dihydro-2′-deoxyguanosine DNA lesions in mammalian cells. Carcinogenesis. 1999;20:2287–2292. doi: 10.1093/carcin/20.12.2287. [DOI] [PubMed] [Google Scholar]
- 46.Masutani C, Araki M, Yamada A, Kusumoto R, Nogimori T, Maekawa T, et al. Xeroderma pigmentosum variant (XP-V) correcting protein from HeLa cells has a thymine dimer bypass DNA polymerase activity. EMBO J. 1999;18:3491–3501. doi: 10.1093/emboj/18.12.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ohashi E, Ogi T, Kusumoto R, Iwai S, Masutani C, Hanaoka F, Ohmori H. Error-prone bypass of certain DNA lesions by the human DNA polymerase κ. Genes Dev. 2000;14:1589–1594. [PMC free article] [PubMed] [Google Scholar]
- 48.Ohashi E, Bebenek K, Matsuda T, Feaver WJ, Gerlach VL, Friedberg EC, et al. Fidelity and processivity of DNA synthesis by DNA polymerase κ, the product of the human DINB1 gene. J. Biol. Chem. 2000;275:39678–39684. doi: 10.1074/jbc.M005309200. [DOI] [PubMed] [Google Scholar]
- 49.Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1982. [Google Scholar]
- 50.Mendelman LV, Boosalis MS, Petruska J, Goodman MF. Nearest neighbor influences on DNA polymerase insertion fidelity. J. Biol. Chem. 1989;264:14415–14423. [PubMed] [Google Scholar]
- 51.Mendelman LV, Petruska J, Goodman MF. Base mispair extension kinetics. Comparison of DNA polymerase α and reverse transcriptase. J. Biol. Chem. 1990;265:2338–2346. [PubMed] [Google Scholar]