Abstract
As a cellular adaptative response, hypoxia decreases Na,K-ATPase activity by triggering the endocytosis of its α1 subunit in alveolar epithelial cells. Here, we present evidence that the ubiquitin conjugating system is important in the Na,K-ATPase endocytosis during hypoxia and ubiquitination of Na,K-ATPase α1 subunit occurs at the basolateral membrane. Endocytosis and ubiquitination were prevented when the Ser 18 in the PKC phosphorylation motif of the Na,K-ATPase α1 subunit was mutated to an alanine, suggesting that phosphorylation at Ser-18 is required for ubiquitination. Mutation of the four lysines surrounding Ser 18 to arginine prevented Na,K-ATPase ubiquitination and endocytosis during hypoxia; however, only one of them was sufficient to restore hypoxia-induced endocytosis. We provide evidence that ubiquitination plays an important role in cellular adaptation to hypoxia by regulating Na,K-ATPase α1-subunit endocytosis.
Keywords: Na, K-ATPase, endocytosis, phosphorylation, ubiquitination, hypoxia
Introduction
Adaptation to hypoxia at the cellular level is regulated by a dual mechanism; on one hand hypoxia leads to an increase in the efficiency of energy-producing pathways, mostly through anaerobic glycolysis, and on the other hand it decreases energy-consuming processes such as the Na,K-ATPase [1]. The Na,K-ATPase consists of a catalytic α subunit and a regulatory β subunit. Active Na+ and K+ transport by this protein consumes ∼30 % of cellular ATP consumption [1].
We have previously reported that in alveolar epithelial cells hypoxia inhibits Na,K-ATPase catalytic activity by causing its endocytosis via a clathrin dependent mechanism, which requires the phosphorylation of the Na,K-ATPase α subunit at serine 18 by PKC-ζ [2, 3]. Classically, conjugation of the 76 amino acid polypeptide ubiquitin to cytoplasmatic proteins targets them for degradation by the 26S proteasome [4] but recently, it has been described that the ubiquitin system participates in the endocytic pathway [5-7]. In mammalian cells, ubiquitination has been implicated in the internalization of the epithelial sodium channel [8], the growth hormone receptor [9] and the epidermal growth factor receptor [10] among others. Ubiquitination functions at multiple subcellular locations to effect down-regulation of membrane proteins [11]. It can function during internalization of proteins from the plasma membrane and also in the sorting at the endosomal compartment level [12, 13].
We set out to determine whether ubiquitination plays a role in hypoxia-induced Na,K-ATPase endocytosis. Our data suggest that Na,K-ATPase α1 subunit ubiquitination is necessary for hypoxia-induced endocytosis and that its phosphorylation is a pre-requisite for ubiquitination. We have also identified four lysines residues in the N-terminus of the α1 subunit that when mutated, inhibited the Na,K-ATPase ubiquitination and endocytosis. These data suggest that the ubiquitin pathway is required in the regulation of the Na,K-ATPase trafficking which is of importance in the cell adaptation to hypoxia.
Materials and Methods
Materials
Na,K-ATPase α1 subunit monoclonal antibody (clone 464.6) was purchased from Upstate Biotechnology (Lake Placid, New York, USA). Ouabain was purchased from ICN Biomedicals Inc. (Aurora, Ohio, USA). 1,2-dioleoyl-sn-glycerol (DAG) and L-α-phosphatidyl-L-serine (PS) were purchased from Sigma-Aldrich (St Louis, Missouri, USA). Rat brain PKC and MG132 were purchased from Calbiochem (San Diego, California, USA). Percoll was purchased from Amersham Pharmacia Biotech (Uppsala,Sweden), GFP polyclonal antibody was purchased from Clontech (Palo Alto, California, USA), and A/G PLUS-Agarose, GFP monoclonal and ubiquitin-monoclonal antibodies were obtained from Santa Cruz Biotech (Santa Cruz, California, USA). All other reagents were commercial products of the highest grade available.
Cell culture and transfections
A549 cells (ATCC CCL 185, a human adenocarcinoma cell line), A549 cells expressing the rat Na,K-ATPase α1 subunit isoform (α1-A549), or the α1 subunit tagged with GFP (α1-GFP-A549), a gift from Dr Bertorello (Karolinska Institutet) were grown in DMEM supplemented with 10% fetal bovine serum, 2 mM L-glutamine, 50 μg/ml gentamicin, 100 U/ml penicillin, 100 μg/ml streptomycin, and 3 μM ouabain to suppress the endogenous human Na,K-ATPase α1 subunit. Cells were incubated in a humidified atmosphere of 5% CO2/95% air at 37°C. Construction of the GFP-S18A-Na,K-ATPase α1 subunit (GFP-S18A-α1 ) and the establishment of a stable cell line expressing this construct were performed as described [14]. Hypoxic conditions (1.5% O2, 93.5% N2, and 5% CO2) were achieved in a humidified variable aerobic workstation (INVIVO O2, Ruskinn Technologies, Leeds, UK). The INVIVO O2 contains an oxygen sensor that continuously monitors the chamber oxygen tension.
Lysine to arginine mutants of α1-Na,K-ATPase or α1-GFP-Na,K-ATPase (K4R, GFP-K4R) were generated by substituting the lysines 16, 17, 19 and 20 with arginines utilizing the QuikChange Mutagenesis Kit from Stratagene as suggested by the manufacturer. The following primers were used: forward, GTATCAGAAGGGGACAGGAGGAGCAGGGCGAAGAAGGAAAGGGACATG, and reverse, CATCTCCCTCTTCTTCGCCCTCCTGCTCCTGTCCCCATGTTCTGATAC. The Lys-16 (KRSRR), Lys 17 (RKSKK), Lys 19 (RRSKR) and Lys 20 (RRSRK)-α1 Na,K-ATPase were generated from the K4R by mutagenesis as described above. Constructs were verified by automated DNA sequencing. Cells were transfected with expression constructs using the LipofectAMINE technique (LipofectAMINE, Invitrogen, Carlsbad, California,USA). Two days after transfection the cells were transferred to a medium containing 3 μM ouabain as described [2].
The Chinese hamster ovary cell lines (CHO), CHO-ts20 (thermo sensitive), and CHO-E36 (wild type), a gift from Dr Strous [9, 15], were incubated in a humidified atmosphere of 5% CO2 and were exposed to hypoxia or normoxia in a humidified 1L glass chamber at 30°C or 43°C. Experiments were performed with 80% confluent cells.
Preparation of endosomes and basolateral membranes (BLMs)
To prepare the endosomal fractions, after hypoxia cells were transferred to ice and homogenized in ice-cold homogenization buffer (250mM sucrose and 3 mM imidazole, 2 mM EGTA, 10 mM NaF, 30 mM Na4O7P2, 1 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin, 4 μg/ml aprotinin, pH 7.4). Cells were gently homogenized (15−20 strokes) to minimize damage of the endosomes using a Dounce homogenizer, and the samples were subjected to a brief (5 min) centrifugation (4°C, 3 000 ×g). Endosomes were fractionated on a flotation gradient as described [16] by using essentially the technique of Gorvel et al. [17]. BLMs were prepared using Percoll gradient centrifugation as described [2, 18]. Briefly, cells were scraped in phosphate-buffered saline (PBS) centrifuged, resuspended in homogenization buffer (300 mM mannitol in 12 mM Tris-HCl–HEPES [pH 7.6] plus protease inhibitors as described above), homogenized, and centrifuged twice to discard the nuclear and mitochondrial pellet. Supernatant was centrifuged at 48 000 g for 30 min, and the BLM fraction was recovered after the membrane pellet was centrifuged in a 16% Percoll gradient at 48 000 g for 30 min. Equal amounts of proteins were resolved by 10% SDS-PAGE and analyzed by immunoblotting with specific antibodies.
Cell surface labeling
Cells were labeled for 20 min using 1 mg/ml EZ-link NHS-SS-biotin (Pierce Chemical Co., Rockford, Illinois, USA). After labeling, the cells were rinsed three times with PBS containing 50 mM glycine to quench unreacted biotin and then lysed in modified RIPA (mRIPA; 50 mM Tris-HCl [pH 8], 150 mM NaCl, 1% Nonidet P-40, and 1% sodium deoxycholate, containing protease inhibitors as described above). Aliquots (150 μg of protein) were incubated overnight at 4°C with end-over-end shaking in the presence of streptavidin beads (Pierce Chemical Co.). The beads were thoroughly washed and then resuspended in 30 μl of Laemmli sample buffer solution [2]. Proteins were analyzed by SDS-PAGE and Western blot.
Immunoprecipitation and in vitro phosphorylation of the Na,K-ATPase α subunit
GFP α- A549 cells were incubated at 1.5% or 21% O2 for 20 min. The incubation was terminated by placing the cells on ice, aspirating the media, and adding immunoprecipitation buffer (20 mM Tris-HCl, 2 mM EGTA, 2 mM EDTA, 30 mM Na4P2O7, 30 mM NaF, 1 mM Na3VO4, 1 mM PMSF, 10 μg/ml leupeptin [pH 7.4]). The cells were then scraped from the plates, frozen in liquid nitrogen, thawed, sonicated, frozen again, and centrifuged for 2 min at 14,000 g. After protein determination, 0.2% SDS and 1% Triton X-100 were added to each sample. Equal amounts of protein (700 μg) were then incubated with anti-GFP antibody for 2 hours at 4°C. Protein A/G PLUS-Agarose was added, and the samples were incubated overnight at 4°C. The samples were then washed twice with immunoprecipitation buffer supplemented with 0.2% SDS and 1% Triton X-100 and once with 20 mM Tris-HCl (pH 7.4).
The phosphorylation state of the immunoprecipitated Na,K-ATPase–GFPα1 subunit was assessed in vitro by the "back phosphorylation" method [2]. The standard reaction mixture for in vitro back phosphorylation of the Na,K-ATPase α1 subunit by purified PKC (150 ng per 150 μl, 30 min at 30°C) contained 10 mM MgCl2, 0.25 mM EGTA, 0.4 mM CaCl2, 0.32 mg/ml PS, 0.03 mg/ml DAG, 0.1 mg/ml BSA, and 20 mM Tris-HCl (pH 7.5). The phosphorylation reaction was started by the addition of [γ-32P]ATP (final concentration, 100 μM; 1.3 μCi per sample). The reaction was stopped by placing the tubes on ice and washing the beads twice with 20 mM Tris-HCl (pH 7.4). Samples were analyzed by SDS-PAGE using the Laemmli buffer system. Proteins were transferred to nitrocellulose membranes and autoradiographed.
Statistical analysis
Data are expressed as means ± SD. Statistical analyses were performed using one-way ANOVA followed by a multiple comparison test (Tukey) when the F statistic indicated significance. Results were considered significant when P was less than 0.05.
Results
The ubiquitination system is required for internalization of the Na,K-ATPase α1 subunit
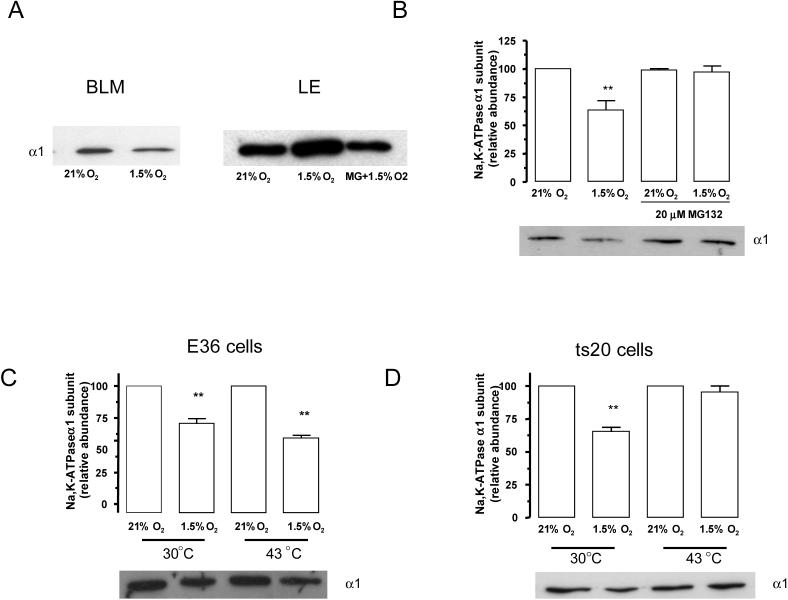
Hypoxia causes Na,K-ATPase to translocate from the plasma membrane to endosomal compartments (Figure 1A). To determine whether the ubiquitin conjugating system is necessary for Na,K-ATPase endocytosis during hypoxia, α1-A549 cells were preincubated in the presence of a proteosomal inhibitor. Approximately 50% of plasma membrane Na,K-ATPase was endocytosed after 60 min of hypoxia, which was prevented by the pre-treatment with MG132 (Figure 1B). MG132 treatment also prevented the translocation of Na,K-ATPase to the endosomal compartments (Figure 1A). To further confirm the role of the ubiquitin conjugating system in the internalization step, we used the CHO-ts20 cell line, which carries a temperature sensitive ubiquitin-activating enzyme (E1) [19]. This enzyme is fully functional when cells are incubated at 30° C, but becomes inactive when cells are incubated at the non-permissive temperature of 43°C. In the parental cell line (CHO-E36) Na,K-ATPase membrane protein abundance decreased during hypoxia at both the permissive and non-permissive temperature (Figure 1C). Conversely, pre-treatment of CHO-ts20 at 43° C prevented the decrease in Na,K-ATPase protein abundance at the plasma membrane (Figure 1D). These data suggest that the ubiquitination machinery is required for internalization of the Na,K-ATPase α1 subunit..
Figure 1.
The ubiquitin conjugation system is necessary for Na,K-ATPase endocytosis during hypoxia. A, A549 cells were exposed to 21 or 1.5% O2 for 60 min at 37° C. Na,K-ATPase α1 subunit abundance in basolateral (BLM), and late endosomes (LE) was determined. Equal amounts of protein (5 μg) were loaded in each lane. A representative Western blot is shown, n=3. B, A549 cells were incubated in the presence or absence MG132 (20 μM) and then exposed to 1.5% O2 for 60 min. Na,K-ATPase endocytosis was determined by biotin labeling of surface proteins followed by streptavidin pull-down and Western blot analysis using a specific anti α1 subunit antibody. Top, quantitative densitometric scan of three experiments (mean ± SEM), **p<0.01, bottom representative Western blot of the α1 subunit abundance. C and D, E36 or CHO-ts20 cells were incubated at 21% O2 or 1.5% O2 for 60 min at either 30°C or 43°C. Na,K-ATPase endocytosis was studied as describe for B. Top, quantitative densitometric scan of three experiments (mean ± SEM), **p<0.01, bottom representative Western blot of the α1 subunit abundance.
Na,K-ATPase is ubiquitinated at t he plasma membrane
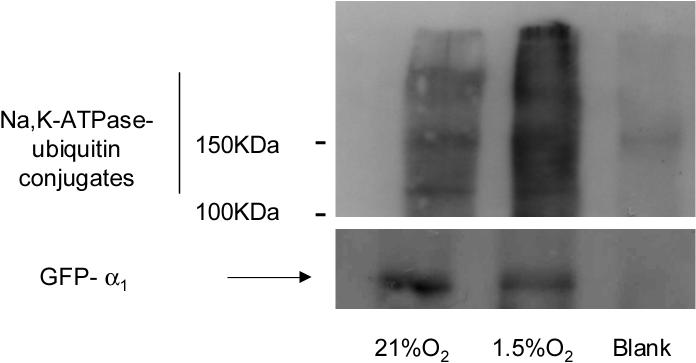
If Na,K-ATPase ubiquitination plays a role in the regulation of its basolateral internalization, the most likely site of ubiquitination is at the basolateral plasma membrane. We assessed the Na,K-ATPase-α1 subunit ubiquitination status in α1-GFP-A549 cells pre-treated with MG-132 and exposed to normoxia or hypoxia. BLMs were isolated and solubilized in homogenization buffer containing 1% Triton X100 then the GFP-α1-Na,K-ATPase was immunoprecipitated from this fraction. In hypoxia exposed cells we observed the appearance of Na,K-ATPase-ubiquitin conjugates, indicating that ubiquitination occurs at basolateral membrane (Figure 2).
Figure 2.
Hypoxia increases Na,K-ATPase (NKA)-ubiquitin conjugates at the plasma membrane. GFP-α1-A549 cells were incubated at 21% O2 in the presence or absence MG132 (20 μM) and then exposed to 1.5% O2 for 60 min. At the end of the incubation, BLMs were isolated; membrane proteins were solubilized with 1% Triton-X100. Equal amount of proteins (20−30 μg) were immunoprecipitated with an anti-GFP antibody. Immunoprecipitates were immunoblotted against ubiquitin. The membranes were then stripped and reblotted against GFP as a loading control. A representative Western blot is shown, n=3.
Phosphorylation at Ser-18-α1 Na,K-ATPase is necessary for ubiquitination to occur
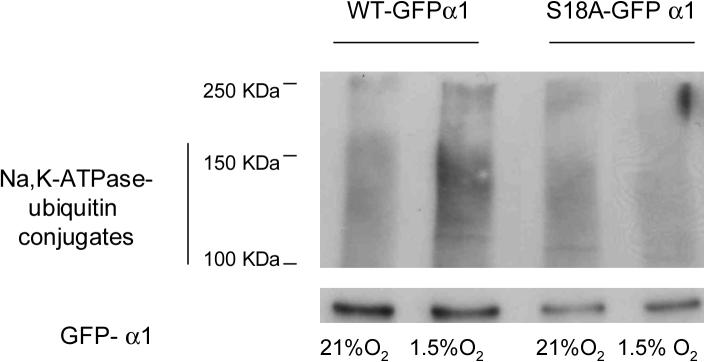
A crucial step in the targeting of plasma membrane proteins for internalization involves the recognition of the cargo molecule by the endocytic system [20]. It has been previously reported that phosphorylation at Ser 18 in the Na,K-ATPase-α1 subunit is necessary for endocytosis [2, 21]. Here, we used point mutation analysis to investigate the role of this serine in the ubiquitination process. When the GFP-WT and GFP-S18A-α1 Na,K-ATPase molecules were immunoprecipitated and then immunobloted against ubiquitin we observed that the cells lysates from the GFP-S18A-α1 cells had little or no ubiquitin conjugates during hypoxia (Figure 3), suggesting that phosphorylation of the Na,K-ATPase is required for ubiquitination.
Figure 3.
Hypoxia induction of Na,K-ATPase-ubiquitin conjugates is dependent on Ser-18 phosphorylation. WT or S18A GFP-α1-A549 were incubated at 21% O2 in the presence of MG132 (20 μM) and then exposed to 1.5% O2 for 60 min Cells were processed as described for Fig. 2. A representative western blot is shown, n=3.
Characterization of the anchoring lysines in Na,K-ATPase α1 subunit
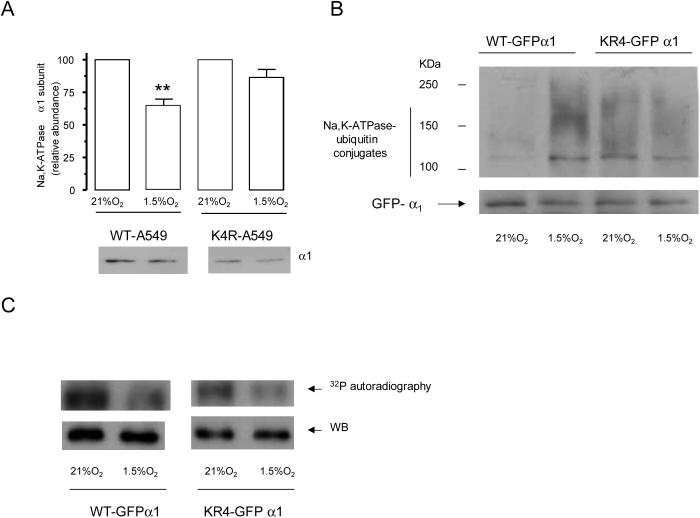
It has been previously described that ubiquitination occurs in the vicinity of phosphorylation sites [22, 23]. We performed site directed mutagenesis to substitute with arginine the four lysine residues adjacent to the Ser 18 in the Na,K-ATPase α1-subunit molecule (KK18SKK→RR18SRR). As shown in Figure 4A, hypoxia-induced endocytosis was prevented in the cells transfected with K4R-α1-Na,K-ATPase. Also, substitution of the lysines in K4R-α1-Na,K-ATPase resulted in the loss of ubiquitinated conjugates as analyzed by immunoprecipitation after hypoxia exposure (Figure 4B). These results suggest that the lysine residues surrounding Ser 18 serve as ubiquitin binding sites. PKC typically phosphorylates serine or threonine residues in basic sequences [24] and since we mutated the lysines surrounding Ser-18, we investigated whether the decrease in ubiquitination was due to the inability of PKC to phosphorylate the K4R-α1-Na,K-ATPase molecule. As shown in Figure 4C, the phosphorylation status was not altered in the mutated molecules as compared to the wild type α1 subunit, suggesting that a defective phosphorylation is not the cause for impaired ubiquitination.
Figure 4.
Mutations of ubiquitination sites prevent Na,K-ATPase endocytosis during hypoxia. A, The lysines in the KKSKK sequence are required for Na,K-ATPase endocytosis during hypoxia. WT or K4R-α1-A549 cells were exposed to 1.5 %O2 for 60 min. Na,K-ATPase endocytosis was determined as described above. Top, quantitative densitometric scan of three experiments (mean ± SEM), **p<0.01, bottom representative Western blot of the α subunit abundance. B, WT or K4R-GFP-α1-A549 were incubated with MG132 (20 μM) and then exposed to 21 or 1.5% O2 for 60 min. At the end of the incubation cell lysates were isolated and Na,K-ATPase-ubiquitin conjugates were immunoprecipitated with an antibody against GFP. Immunoprecipitates were immunoblotted against ubiquitin. The membranes were then stripped and reblotted against GFP as a loading control. A representative western blot is shown, n=3. C, GFP-α1-A549 cells were exposed to 1.5% O2 for 20 min, and Na,K-ATPase GFP α1 subunit was immunoprecipitated using a polyclonal GFP antibody. An in vitro phosphorylation reaction with the immunoprecipitated Na,K-ATPase, purified PKC, and [γ32P]ATP was conducted. A representative autoradiogram of the α1 subunit phosphorylation is shown with the corresponding immunoblot performed with a monoclonal anti-GFP antibody (n=3).
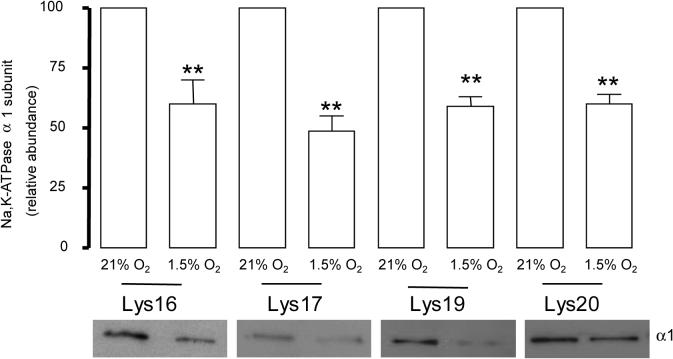
We studied whether the presence of any of these lysines individually or a combination of lysines was necessary for the hypoxia induced Na,K-ATPase endocytosis. We transfected cells with constructs where only one lysine in the sequence KKSKK was preserved (Lys-16 , Lys 17, Lys 19 or Lys 20 α1–Na,K-ATPase). When one of the lysines was present, we did not find alterations in the rate of internalization as compared with the cells expressing the Na,K-ATPase carrying the 4 lysines (Figure 5), indicating that these residues have a redundant function in the internalization sequence.
Figure 5.
Lysines in the KKSKK have a redundant function. WT or Lys 16, Lys 17, Lys 19, Lys 20 α1-A549 cells were exposed to 1.5 % O2 for 60 min. At the end of the incubation, Na,K-ATPase endocytosis was studied as described above. Top, quantitative densitometric scan of three experiments (mean ± SEM), **p<0.01, bottom representative Western blot of the α1-subunit abundance.
Discussion
Ion-motive ATPases and protein synthesis are the major energy-consuming processes of cells [1]. Thus, during hypoxia regulation of Na,K-ATPase activity is important for the cells to adapt to the low O2 levels. We have previously reported that phosphorylation of Na,K-ATPase α1 subunit is necessary for its endocytosis [2, 21]. However, the mechanisms by which Na,K-ATPase is internalized from the plasma membrane have not been fully elucidated Here, we report that phosphorylation of the Na,K-ATPase at Ser 18 is necessary for ubiquitination at the neighboring lysines to occur. Using a proteasomal inhibitor or cells with a temperature-sensitive mutation in the E1 of the ubiquitin conjugation pathway, we have observed that inactivation of the ubiquitin system prevents Na,K-ATPase endocytosis during hypoxia (Figure 1). Inhibitors of proteasomal function have been shown to prevent endocytosis and intracellular targeting of plasma membrane proteins [25]. This disruption of trafficking can occur independently of their ability to inhibit proteosomal degradation [26]. For example, proteasome inhibitors prevented EGF-receptor delivery to multivesicular bodies [23] and the delivery of the growth hormone receptor to the lysosome for degradation even though this receptor is not a proteasome target [26, 27]. Our data suggest that ubiquitination may act as a signal for endocytosis via the ubiquitination of the Na,K-ATPase itself or that one of the components of the endocytic sorting machinery is targeted for the proteasome. Ubiquitination has being suggested to play a role in plasma membrane protein endocytosis but whether the ubiquitination of the cargo protein is necessary or not remains controversial. For example for G-coupled receptor endocytosis ubiquitination of both the receptor and adaptor proteins has been described but while the adaptor protein ubiquitination is necessary for endocytosis, receptor ubiquitination is necessary for its degradation at the lysosome [28]. Here, we propose a role for Na,K-ATPase ubiquitination in endocytosis because of the increase of Na,K-ATPase-ubiquitin conjugates during hypoxia occurs at the plasma membrane (Figure 2), is prevented in the absence of Ser-18 (Figure 3) and specific lysines in the Na,K-ATPase α1 subunit are necessary for the hypoxia induced endocytosis (Figure 4).
Phosphorylation of Na,K-ATPase at Ser-18 is a trigger for endocytosis and mutation of Ser 18 to an alanine prevented this process [2, 21]. Here, we report that mutation of Ser 18 prevents ubiquitination of the α1 subunit during hypoxia, suggesting that phosphorylation is necessary for ubiquitination. It has been proposed for some membrane receptors, that ligand binding induced phosphorylation controls ubiquitination and that phosphorylation generates a docking site for the ubiquitination machinery [29]. Previous reports suggest that for some plasma membrane proteins, the sites of ubiquitination and phosphorylation are adjacent to each other [29]. Thus, based on proximity to the phosphorylation site we mutated the four lysines adjacent to Ser 18 and we observed that in cells bearing a K4R mutation in the Na,K-ATPase α subunit failed to undergo endocytosis and was not ubiquitinated (Figure 3). To determine whether the inhibition of endocytosis and phosphorylation did not occur as a consequence of decreased phosphorylation, we assessed the Na,K-ATPase α-1 subunit phosphorylation state in the Na,K-ATPase α subunit bearing a K4R mutation; however, the ability of PKC to phosphorylate this molecule was not different from the one obtained with the WT- Na,K-ATPase α1 subunit.
Some sequence motifs have been reported to be responsible for binding ubiquitin to cell membrane proteins [20]. Among them it has been suggested that the lysine residues included in a (D/E) XK(S/T) motif are potential targets for the ubiquitination of plasma membrane proteins [30]. The Na,K-ATPase α1 subunit bears a similar motif: DKK18SKK where the target lysines are adjacent to the phosphorylable Ser 18. However, as shown in Figure 5, the presence of only one of these lysines restored the ability of the Na,K-ATPase to internalize. Thus, ubiquitination at any one of these lysines residues appears to be sufficient for Na,K-ATPase endocytosis, which is similar to previous reports [10].
In summary, we provide evidence that Na,K-ATPase endocytosis requires the phosphorylation of Ser 18 which in turn is necessary for ubiquitination at the sequence KKSKK in the cytoplasmatic N-terminus of the Na,K-ATPase α1-subunit, which triggers Na,K-ATPase for endocytosis as a mechanism of cell adaptation to hypoxia.
Acknowledgments
The authors want to thank M. Butti for technical assistance. This work was supported in part by HL-PO1- 71643.
Glossary
The abbreviations used are
- DAG
1,2-dioleoyl-sn-glycerol
- PS
L-α-phosphatidyl-L-serine
- BLM
basolateral membranes
- PBS
phosphate-buffered saline
- E1
ubiquitin-activating enzyme
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Michiels C. Am J Pathol. 2004;164:1875. doi: 10.1016/S0002-9440(10)63747-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dada LA, Chandel NS, Ridge KM, Pedemonte C, Bertorello AM, Sznajder JI. J. Clin. Invest. 2003;111:1057. doi: 10.1172/JCI16826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen Z, Krmar RT, Dada L, Efendiev R, Leibiger IB, Pedemonte CH, Katz AI, Sznajder JI, Bertorello AM. Am. J. Respir. Cell Mol. Biol. 2006;35:127. doi: 10.1165/rcmb.2006-0044OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hershko A, Ciechanover A. Annu Rev Biochem. 1998;67:425. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 5.Hicke L, Dunn R. Ann. Rev. of Cell and Dev. Biol. 2003;19:141. doi: 10.1146/annurev.cellbio.19.110701.154617. [DOI] [PubMed] [Google Scholar]
- 6.Hicke L. Trends Cell Biol. 1999;9:107. doi: 10.1016/s0962-8924(98)01491-3. [DOI] [PubMed] [Google Scholar]
- 7.Strous G, Govers R. J Cell Sci. 1999;112:1417. doi: 10.1242/jcs.112.10.1417. [DOI] [PubMed] [Google Scholar]
- 8.Staub O, Gautschi I, Ishikawa T, Breitschopf K, Ciechanover A, Schild L, Rotin D. EMBO J. 1997;16:6325. doi: 10.1093/emboj/16.21.6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strous GJ, van Kerkhof P, Govers R, Ciechanover A, Schwartz AL. EMBO J. 1996;15:3806. [PMC free article] [PubMed] [Google Scholar]
- 10.Haglund K, Sigismund S, Polo S, Szymkiewicz I, Di Fiore PP, Dikic I. Nat Cell Biol. 2003;5:461. doi: 10.1038/ncb983. [DOI] [PubMed] [Google Scholar]
- 11.Hicke L. Cell. 2001;106:527. doi: 10.1016/s0092-8674(01)00485-8. [DOI] [PubMed] [Google Scholar]
- 12.Katzmann DJ, Babst M, Emr SD. Cell. 2001;106:145. doi: 10.1016/s0092-8674(01)00434-2. [DOI] [PubMed] [Google Scholar]
- 13.de Melker AA, van der Horst G, Calafat J, Jansen H, Borst J. J Cell Sci. 2001;114:2167. doi: 10.1242/jcs.114.11.2167. [DOI] [PubMed] [Google Scholar]
- 14.Lecuona E, Dada LA, Sun H, Butti ML, Zhou G, Chew T-L, Sznajder JI. FASEB J. 2006:fj.06. doi: 10.1096/fj.06-6503fje. [DOI] [PubMed] [Google Scholar]
- 15.Strous GJ, van Kerkhof P, Govers R, Rotwein P, Schwartz AL. J. Biol. Chem. 1997;272:40. doi: 10.1074/jbc.272.1.40. [DOI] [PubMed] [Google Scholar]
- 16.Ridge KM, Dada L, Lecuona E, Bertorello AM, Katz AI, Mochly-Rosen D, Sznajder JI. Mol Biol Cell. 2002;13:1381. doi: 10.1091/mbc.01-07-0323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorvel J-P, Chavrier P, Zerial M, Gruenberg J. Cell. 1991;64:915. doi: 10.1016/0092-8674(91)90316-q. [DOI] [PubMed] [Google Scholar]
- 18.Bertorello AM, Ridge KM, Chibalin AV, Katz AI, Sznajder JI. Am J Physiol. 1999;276:L20. doi: 10.1152/ajplung.1999.276.1.L20. [DOI] [PubMed] [Google Scholar]
- 19.Kulka RG, Raboy B, Schuster R, Parag HA, Diamond G, Ciechanover A, Marcus M. J Biol Chem. 1988;263:15726. [PubMed] [Google Scholar]
- 20.Bonifacino JS, Traub LM. Annu. Rev. Biochem. 2003;72:395. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- 21.Chibalin AV, Ogimoto G, Pedemonte CH, Pressley TA, Katz AI, Feraille E, Berggren PO, Bertorello AM. J Biol Chem. 1999;274:1920. doi: 10.1074/jbc.274.4.1920. [DOI] [PubMed] [Google Scholar]
- 22.Hicke L, Riezman H. Cell. 1996;84:277. doi: 10.1016/s0092-8674(00)80982-4. [DOI] [PubMed] [Google Scholar]
- 23.Longva KE, Blystad FD, Stang E, Larsen AM, Johannessen LE, Madshus IH, Cell Biol J. 2002;156:843. doi: 10.1083/jcb.200106056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newton AC. J. Biol. Chem. 1995;270:28495. doi: 10.1074/jbc.270.48.28495. [DOI] [PubMed] [Google Scholar]
- 25.Melikova MS, Kondratov KA, Kornilova ES. Cell Biol Int. 2006;30:31. doi: 10.1016/j.cellbi.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 26.Takayama Y, May P, Anderson RGW, Herz J. J. Biol. Chem. 2005;280:18504. doi: 10.1074/jbc.M410265200. [DOI] [PubMed] [Google Scholar]
- 27.van Kerkhof P, dos Santos CMA, Sachse M, Klumperman J, Bu G, Strous GJ. Mol. Biol. Cell. 2001;12:2556. doi: 10.1091/mbc.12.8.2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shenoy SK, McDonald PH, Kohout TA, Lefkowitz RJ. Science. 2001;294:1307. doi: 10.1126/science.1063866. [DOI] [PubMed] [Google Scholar]
- 29.Kelm KB, Huyer G, Huang JC, Michaelis S. Traffic. 2004;5:165. doi: 10.1111/j.1600-0854.2004.00168.x. [DOI] [PubMed] [Google Scholar]
- 30.Marchal C, Haguenauer-Tsapis R, Urban-Grimal D. J. Biol. Chem. 2000;275:23608. doi: 10.1074/jbc.M001735200. [DOI] [PubMed] [Google Scholar]