Abstract
Background
The Gateway recombinatorial cloning system allows easy and rapid joining of DNA fragments. Here we report the construction and evaluation of three different Gram-positive vectors that can be used with the Multisite Gateway cloning system to rapidly produce new gene arrangements in plasmid constructs for use in a variety of Gram-positive bacteria.
Results
Comparison of patterns of reporter gene expression with conventionally constructed clones show that the presence of residual recombination (att) sites does not have an effect on patterns of gene expression, although overall levels of gene expression may vary. Rapid construction of these new vectors allowed vector/gene combinations to be optimized following evaluation of plasmid constructs in different bacterial cells and demonstrates the benefits of plasmid construction using Gateway cloning.
Conclusion
The residual att sites present after Gateway cloning did not affect patterns of promoter induction in Gram-positive bacteria and there was no evidence of differences in mRNA stability of transcripts. However overall levels of gene expression may be reduced, possibly due to some post-transcriptional event. The new vectors described here allow faster, more efficient cloning in range of Gram-positive bacteria.
Background
The study of gene expression in bacteria has advanced rapidly over the last two decades. In addition to traditional mutagenesis approaches, the characterization of gene function increasingly requires the investigation of DNA segments containing promoters and their associated regulatory sequences. While many cloning and expression vectors have been developed for use in Gram-negative bacteria, there is a paucity of equivalent materials for use when working in Gram-positive bacteria.
Recombinatorial cloning systems, such as Invitrogen's 'Gateway' provide an alternative to conventional cloning that uses restriction enzymes and ligation. The Gateway system uses directed recombination between modified attachment (att) sites derived from E. coli bacteriophage λ. The λ integration system mediates two recombination reactions to promote either integration or excision of the phage genome from the E. coli chromosome. During integration the phage attP site recombines with the related, but non-identical, bacterial attB site producing hybrid attL and attR sequences located to the left and right of the phage genome, respectively. This recombination is mediated by integrase (Int) and integration host factor (IHF). To achieve excision of the phage from the chromosome attLand attR are recombined to regenerate attB and attP in a reaction mediated by Int in combination with the excisionase (Xis) and IHF [see [1]]. For the Gateway system, modified att recombination sites have been developed with recombination specificity, such that the variant attB1 will recombine with attP1 but not with other attP variants. Introducing these variant att sites at the ends of fragments to be cloned allows them to be recombined with vectors containing cognate attB sites, maintaining the orientation of DNA fragments during the in vitro recombination [2-4]. To facilitate the recombinatorial cloning, the appropriate enzymes are supplied by the manufacturer as BP clonase, that mediates attB/attP recombination events, and LR clonase to mediate recombination between attL/attR sequences.
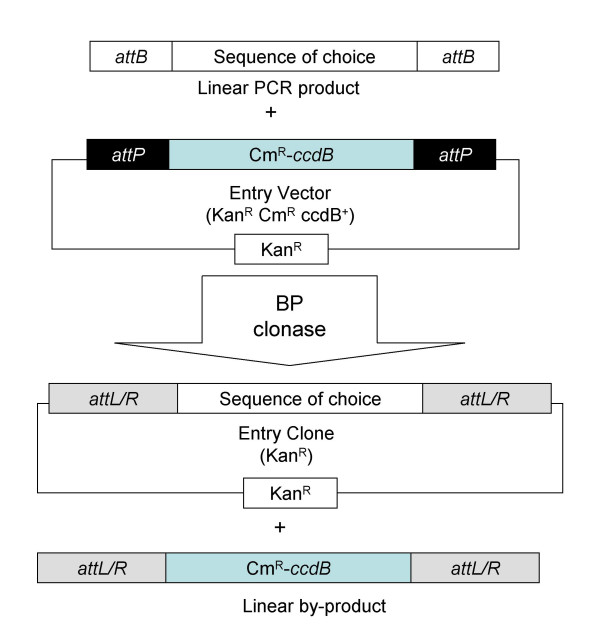
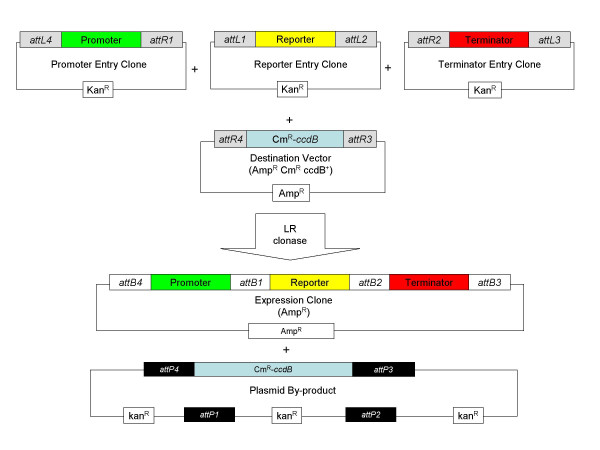
The Multisite Gateway cloning technology is designed to place three DNA fragments adjacent to each other in a specific order and orientation. The procedure is carried out in two stages, the first of which is a BP reaction to transfer a linear DNA fragment flanked by attB sites (generated by PCR) into a plasmid, termed an Entry Vector, containing cognate attP sites. This creates Entry Clones containing the desired DNA fragment flanked by attR or attL sites. These can be selected by loss of the counter-selectable marker [ccdB; [5]] present in the original Entry Vector after transformation into E. coli (see Fig. 1). To create three fragment gene fusions, three such Entry Clones containing different attL/attR variants are created and then utilised in a second recombination step with a Destination Vector containing appropriate attR sites and ccdB using LR clonase (Fig. 2). Selection of correct Destination Clones is again facilitated by counter selection of ccdB that should be lost from the Destination Vector during the recombination event. The major advantage of this system is its ease and efficiency but residual att sites are left flanking DNA fragments (Fig. 3) and there has been some concern that these may affect gene expression from the plasmid constructs created.
Figure 1.
Construction of Entry Clones using the BP reaction. The BP recombination transfers a DNA fragment flanked by attB sites (white boxes) into a plasmid containing cognate attP sites (black boxes), termed an Entry Vector. The resultant Entry Clone contains the desired DNA fragment flanked by attR or attL sites (grey boxes), depending on the orientation of the attB and attP sites.
Figure 2.
Construction of Multisite Expression Clones using LR reaction. Three Entry Clones with the DNA fragments of interest flanked by attL4 and attR1 (Promoter), attL1 and attL2 (Reporter gene), and attR2 and attL3 (transcriptional terminator) recombine with a Destination Vector containing attR4 and attR3. During the LR reaction attL sites react only with their cognate attR sites resulting in a Destination Clone containing all three fragments fused together into the desired expression cassette. Expression Clone constructs are selected on the basis of the antibiotic resistance gene in the Destination vector. The by-product of this reaction is non-replicative and its loss from the cell is further ensured by the presence of the ccdB counter-selectable marker gene.
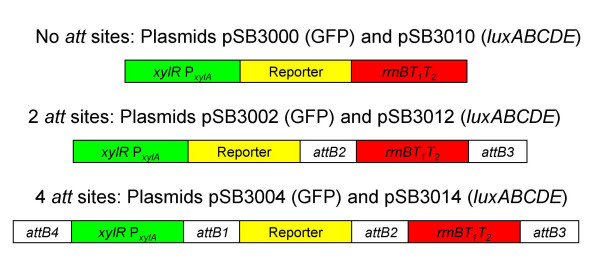
Figure 3.
Structure of Expression Cassettes. Schematic of the different xylose-controllable expression cassettes created in pMK4, showing the plasmid name, corresponding reporter gene present and location of att sites for each construct.
The Gateway system has been primarily developed for Gram-negative bacteria or eukaryotic systems. Recently Bae and Schneewind [6] have described the development of simple Gateway vectors for use in Gram-positive bacteria, where only one fragment at a time is recombined into a Destination vector. We present here the construction of Multisite Gateway Destination Vectors based upon 3 different Gram-positive plasmid replicons that will allow the use of this rapid cloning technique in a wide range of Gram-positive bacterial hosts. These were used to construct reporter gene plasmids with the general structure Promoter:Reporter:Terminator which were then used to introduce both lux and gfp reporter genes into Staphylococcus aureus, Listeria monocytogenes and Bacillus subtilis. To determine whether the residual att sites have any effect on gene expression, reporter gene expression from a Promoter:Reporter:Terminator cassette constructed using either Gateway or conventional cloning methods were compared.
Results
Constructing a Gram-positive destination vector
A prerequisite for the Multisite Gateway system is a Destination vector containing a CmR-ccdB cassette flanked by attR4 and attR3 sites. This was amplified from pDESTR4-R3 (Invitrogen) using primers PmeattR3 and PmeattR4 (Table 1) to generate a 1770 bp PCR product. This amplicon was cut with PmeI and ligated into the Gram-positive shuttle vectors pMK4 (SmaI site), pHB201 (PvuII/PmeI sites) and pUNK1 (SmaI site). These recombinant plasmids were used to transform E. coli DB3.1 (gyrA) that can tolerate the ccdB gene. Resulting plasmids were characterised by restriction mapping and sequence analysis. These new Gram-positive destination vectors were given the designation pDEST-pMK4, pDEST-pHB201 and pDEST-pUNK1, respectively.
Table 1.
Primers for PCR and sequencing
| Gateway primers | Sequence (5-3') | Use |
| Term attB2F | GGGGACAGCTTTCTTGTACAAAGTGGAAATCAGAACGCAGAAGCGGTC | Amplification of terminator |
| Term attB3R | GGGGACAACTTTGTATAATAAGTTGTTTAAACTGGCAGTTTATGGCGGGC | Amplification of terminator |
| Lux E attB2R | GGGGACCACTTTGTACAAGAAAGCTGGGTTCAACTATCAAACGCTTCGG | Amplification of lux reporter |
| GFP attB2R | GGGGACCACTTTGTACAAGAAAGCTGGGTGACACATTTATTTGTATAGTTC | Amplification of gfp reporter |
| GFPopt attB1F | GGGGACAAGTTTGTACAAAAAAGCAGGCTAGGAGGAATAAAAAATGAGTAAAGGCGAAGAAC | Amplification of gfp reporter |
| XylRA attB4F | GGGGACAACTTTGTATAGAAAAGTTGCGTTGACTTAACTAACTTATAG | Amplification of xylR PxylA |
| XylRA attB1R | GGGGACTGCTTTTTTGTACAAACTTGTTGA TTTAAGTGAACAAGTTTATC | Amplification of xylR PxylA |
| XylA attB4F | GGGGACAACTTTGTATAGAAAAGTTGAAACATTGAAATAAACATTTATTTTGTATATGATG | Amplification of PxylA |
| XylA attB1R | GGGGACTGCTTTTTTGTACAAACTTGTTGATTTAAGTGAACAAGTTTATC | Amplification of PxylA |
| LuxA attB1F | GGGGACAAGTTTGTACAAAAAAGCAGGCTAGGAGGACTCTCTATGAAATTTGGAAAC | Amplification of lux reporter |
| SASP attB4F | GGG GAC AAC TTT GTA TAG AAA AGT TGT TTA AAC GCT GGA ACC TTT TGT TCC CAA AAG | Amplification of PsspA |
| SASP attB1R | GGG GAC TGC TTT TTT GTA CAA ACT TGT TTT ATT TAG TAT GGT TGG GTT AAC TGG | Amplification of PsspA |
| attR3 | GGGGCGTTTAAACAAGCTTGTAAAACGACGGCCAGTG | sequencing |
| PMEattR4 | GGGTTTAAACAACTTTGTATAGAAAAGTTGAACGAG | sequencing |
| Ccdb Rev | AGGAAGGGATGGCTGAGGTC | sequencing |
| Cm R4-R3F | CAGGCGGGCAAGAATGTGA | sequencing |
Entry clone construction
A number of Entry Clones using the Entry Vectors from the Gateway construction kit were then prepared to allow us to rapidly create reporter plasmids for use in Gram-positive bacteria. In each case the required sequence was amplified by PCR using the primers listed in Table 1 and introduced into the entry vectors using the BP recombination reaction (see Fig. 2) and each of the resulting Entry Clones was characterised by restriction mapping and sequence analysis. First a 3' Entry Clone was created using plasmid pDONOR-P2R-P3 containing the transcriptional terminator (rrnBT1T2) amplified from pTRC99A as E. coli terminators have been previously demonstrated to function in Gram-positive bacteria [7]. Secondly a series of reporter gene entry clones were created using pDONOR-221. These contained either the gfp or luxABCDE, or both reporter systems (gfp:luxABCDE) [8-10]. Finally a series of 5' entry clones were created using plasmid pDONOR-P4-P1R. The promoter fragments chosen were those that we have previously used to create reporter gene constructs in Gram positive bacteria and included xylose utilisation gene (PxylA [11]) amplified from B. megaterium chromosomal DNA and the small acid soluble protein (SASP) gene promoter (PsspA: [12]) amplified from B. subtilis chromosomal DNA. In addition PxylA was also amplified from B. megaterium along with its cognate regulator protein XylR (XylR PxylA [11]).
Construction of reporter plasmids using Gateway recombination
After generating the Entry Clones containing the promoters, reporter genes and transcriptional terminator, the MultiSite Gateway LR reaction was performed with the new Gram-positive destination vectors to create expression clones with the following structure, attB4-promoter-attB1-reporter gene-attB2-terminator-attB3 (see Fig. 2). Each LR reaction mixture was transformed into E. coli Top10 competent cells to select for loss of the ccdB gene. Transformants were then screened for the expression of the appropriate reporter genes and in each case the structure of clones was confirmed by restriction and sequence analysis. All final expression constructs created using this Gateway system contain 4 attB sites (see Fig. 2).
Construction of expression clones for Staphylococcus aureus
Previously we have successfully used reporter gene constructs based on the shuttle vector pMK4 for studies in Gram-positive bacteria [8-10], therefore pDEST-pMK4 was chosen as the first destination vector to be evaluated. Constructs were created containing xylR PxylA fused to either the gfp or luxABCDE followed by the terminator sequence (pSB3004 and pSB3014, respectively; see Fig. 3 and Table 2) and these were transformed into S. aureus RN4220.
Table 2.
Plasmids used in study
| Plasmid | Features of plasmid | Reference/source |
| pTMP100 | pMK4 containing S. aureus PrpsJ:gfp:luxABCDE | Hill laboratory collection |
| pTRC99A | rrnB T1, rrnB T2 | 27 |
| pDONRP4-P1R | Entry vector to clone attB4 and attB1 flanked PCR products, Kmr, Cmr, ccdB+ | Invitrogen |
| pDONR221 | Entry vector to clone attB1 and attB2 flanked PCR products, Kmr, Cmr, ccdB+ | Invitrogen |
| pDONR P2R-P3 | Entry vector to clone attB2 and attB3 flanked PCR products, Kmr, Cmr, ccdB+ | Invitrogen |
| pDESTR4-R3 | Destination vector, contains attR4/R3 sites. Recombines with entry clones in a Multisite Gateway LR reaction, Apr, Cmr, ccdB+ | Invitrogen |
| pHB201 | B. subtilis ori-pBR322, ori-1060, cat86:lacZα Cmr Emr | 15 |
| pMK4 | Gram-Positive Shuttle vector, ApR, Cmr | 28 |
| pSB2018 | pMK4 containing xylR PxylA:gfp | 8 |
| pSB2026 | pMK4 containing xylR PxylA:luxABCDE | Hill laboratory collection |
| pUNK1 | Gram-positive shuttle vector, Emr | 14 |
| pDEST-pMK4 | pMK4 containing attR4 and attR3 sites for recombination with entry clones in an LR reaction. Apr, Cmr, ccdB+ | This work |
| pDEST-pUNK1 | pUNK1 containing attR4 and attR3 sites for recombination with entry clones in an LR reaction. Emr, Cmr, ccdB+ | This work |
| pDEST-pHB201 | pHB201 containing attR4 and attR3 sites for recombination with entry clones in an LR reaction. Emr, Cmr, ccdB+ | This work |
| pSB3000 | 3 fragment expression cassette in pMK4. xylR PxylA:gfp:rrnBT1T2 (0 attB) | This work |
| pSB3002 | 3 fragment expression cassette in pMK4. xylR PxylA:gfp:rrnBT1T2 (2 attB) | This work |
| pSB3004 | 3 fragment expression cassette in pMK4. xylR PxylA:gfp:rrnBT1T2 (4 attB) | This work |
| pSB3005 | 3 fragment expression cassette in MK4. PxylA:gfp:luxABCDE rrnBT1T2 (4 attB) | This work |
| pSB3007 | 3 fragment expression cassette in pUNK1. PxylA:gfp:luxABCDE:rrnBT1T2 (4 attB) | This work |
| pSB3010 | 3 fragment expression cassette in pMK4. xylR PxylA:luxABCDE:rrnBT1T2 (0 attB) | This work |
| pSB3012 | 3 fragment expression cassette in pMK4. xylR PxylA:luxABCDE:rrnBT1T2 (2 attB) | This work |
| pSB3014 | 3 fragment expression cassette in pMK4. xylR PxylA:luxABCDE:rrnBT1T2 (4 attB) | This work |
| pSB3024 | 3 fragment expression cassette in pHB201. PsspAgfp:luxABCDE:rrnBT1T2 (4 attB) | This work |
To investigate whether the presence of the attB sites between the various elements of the expression cassettes affected patterns of gene expression, a series of parallel constructs in pMK4 were created by conventional cloning. To remove the att sites flanking the promoter in pSB3004, the promoter and gfp reporter gene were replaced with an equivalent fragment from pSB2018 [8] that contains no att sites (see methods section for details of plasmid construction). Similarly to remove the attB sites flanking the promoter in the luxABCDE expression vector pSB3014, the 'bla xylR PxylAluxAB fragment was replaced with an equivalent fragment from pSB2026 [9] which again contains no attB sites. The resulting gfp and lux constructs were designated pSB3002 and pSB3012, respectively. These constructs have two attB sites remaining, namely attB2 and attB3 flanking the terminator (Fig. 3).
To construct a plasmid with an equivalent structure to pSB3004 but without any attB sites, a terminator sequence was inserted into plasmid pSB2018 in a PstI site located immediately downstream of the PxylAgfp sequence. The resulting plasmid was designated pSB3000 and is equivalent to pSB3002 but contains no attB sites flanking the terminator (Fig. 3). To create the equivalent luxABCDE reporter plasmid with no attB sites, the gfp gene was excised from pSB2018 and a luxABCDE fragment inserted in its place. The resulting plasmid was digested with PstI and the rrnBT1T2 fragment cloned downstream of the lux operon. This clone was designated pSB3010 (Fig. 3) and sequencing analysis was used to confirm the presence of the transcriptional terminator, promoter and reporter genes in all new reporter constructs and successful deletion of attB sites.
These experiments created pMK4-based plasmids where no attB sites were present (pSB3000 and pSB3010) or with only two attB sites flanking the terminator sequence (pSB3002 and pSB3012) or where 4 attB sites were present (pSB3004 and pSB3014) (see Table 2 and Fig. 3). All of these plasmids contained the reporter genes under the control of the xylose inducible promoter, PxylA, and were transformed into S. aureus RN4420.
Evaluation of expression clones in Staphylococcus aureus
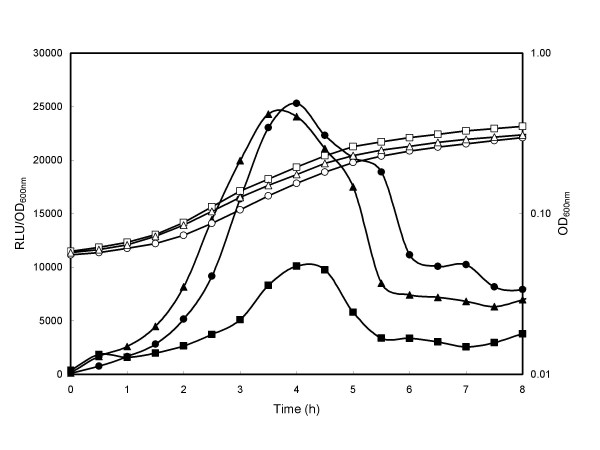
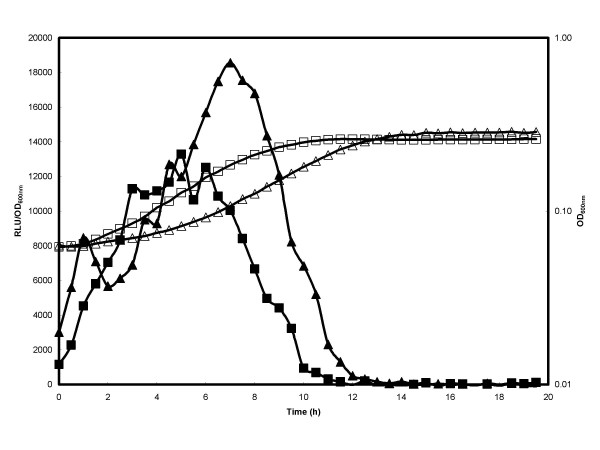
First the growth and expression of the PxylA lux expression plasmids (pSB3010 [no attB sites], pSB3012 [[2]attB sites] and pSB3014 [[4]attB sites]) were compared in rich broth when the promoter systems were fully induced by addition of xylose throughout growth (Fig. 4). No significant differences were seen in the growth of bacteria harboring analogous plasmids and, as previously reported [9], expression of the lux operon from this promoter was seen to reach a maximum during the logarithmic growth phase. Bioluminescence from expression plasmids pSB3010 and pSB3012 was approximately equivalent however light emission from pSB3014, which has two additional attB sites flanking the promoter region, consistently showed a lower light level, with typically half the bioluminescence intensity of the other two plasmid constructs tested. Similar differences in light level from the 4 att constructs were also noted when lux expression from the plasmids in L. monocytogenes was compared (data not shown).
Figure 4.
Effect of att sites on reporter gene expression in S. aureus. S. aureus RN4220 with lux expression plasmids pSB3010 (○, ●; 0 att sites), 2 pSB3012 (△, ▲ 2 att sites) or pSB3014 (□, ■; 4 att sites) were grown in LB medium containing 0.5% (w/v) xylose at 37°C. Growth (OD 600 nm; open symbols) and luminescence (Relative Light Units; RLU closed symbols) were monitored over time. Reporter gene data are presented as RLU/OD600 nm to account for increasing cell number during the experiment.
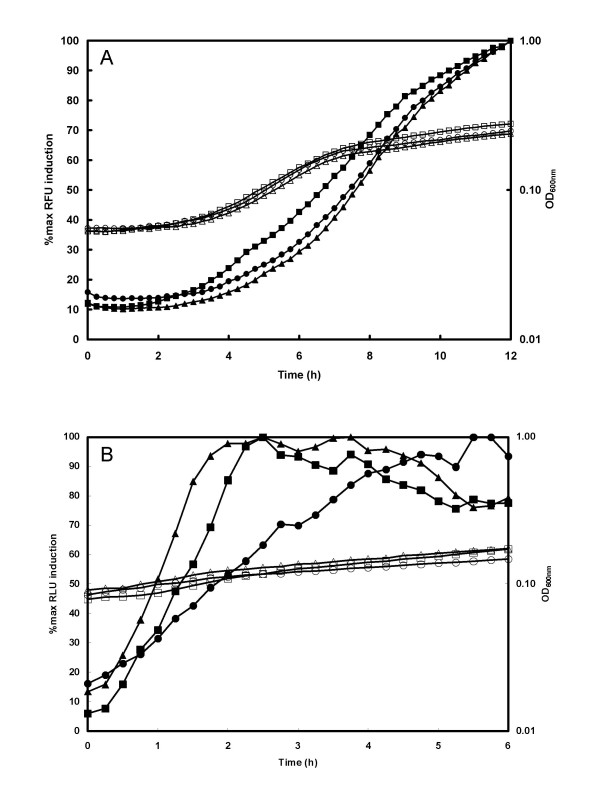
To try to elucidate whether the lower bioluminescence observed when the promoter was flanked by attB sites was due to differences in the rate of transcription or translation, a second series of experiments were carried out in minimal media where the reporter gene expression was only induced by addition of 0.5% (w/v) xylose once bacteria had entered the exponential phase of growth, and the rate of gene induction was then followed. Both lux (pSB3010, pSB3012 and pSB3014) and gfp (pSB3000 [no attB sites], pSB3002 [[2]attB sites] and pSB3004 [[4]attB sites]) expression plasmids were used for these experiments (Fig. 5). The induction of PxylA led to a rapid increase in fluorescence (Fig. 5a) and luminescence (Fig. 5b) for all reporter constructs. Sequences adjacent to ribosome binding sites have previously been shown to affect translational efficiency [13]. In our study no significant differences were found in the maximum rates of induction of reporter activity when analogous plasmids were compared. This indicates that neither the initiation of transcription nor translation were affected by attB sites proximal to the promoter or RBS of the reporter genes.
Figure 5.
Reporter gene induction kinetics. S. aureus RN4220 containing reporter plasmids pSB3004 (□, ■; 4 att sites), pSB3002 (△, ▲; 2 att sites) and pSB3000 (○, ●; no att sites) were grown to mid-log phase in Tris Minimal Succinate medium with 5 μg ml-1 Cm and then diluted 1/20 into fresh medium supplemented with 0.5% xylose. Triplicate samples were placed in a 96 well microtitre plate and incubated at 37°C in a Tecan Genios Pro. Panel A: fluorescence (solid symbols, Relative Fluorescence Units; RFU) and absorbance (open symbols) were measured at 10 min intervals. Plasmids used were pSB3004 (□, ■; 4 att sites), pSB3002 (□, ▲; 2 att sites) and pSB3000 (○, ●; no att sites). Panel B. luminescence (solid symbols, Relative Light Units; RLU)) and absorbance (open symbols) were measured at 10 min intervals. Plasmids used were pSB3014 (□, ■; 4 att sites), pSB3012 (△, ▲; 2 att sites) and pSB3010 (○, ●; no att sites). Data is presented as % maximal signal to allow direct comparison of expression kinetics despite the fact that light levels from each construct were different.
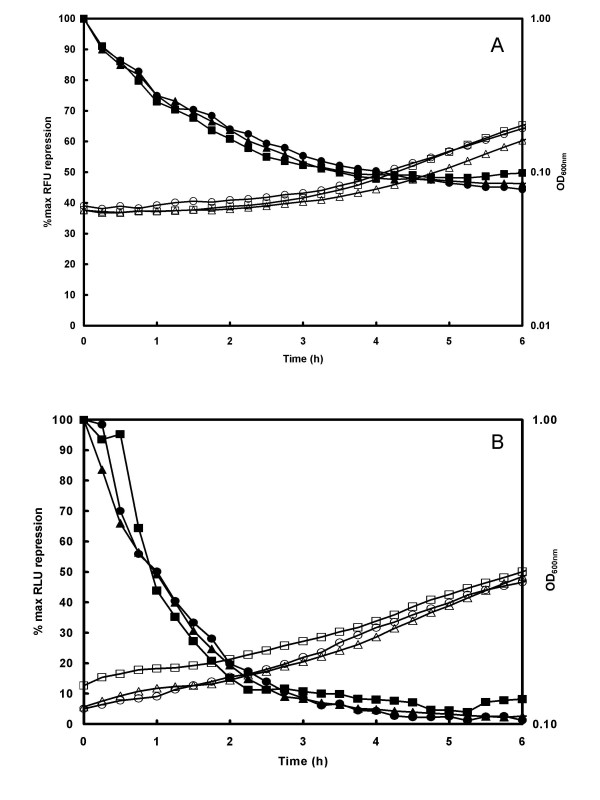
To ascertain if the presence of attB sites had any effect on mRNA stability, repression of reporter gene transcription was monitored following removal of xylose from xylose-induced cultures. As shown in Figures 6a and 6b, removal of xylose from logarithmically growing cells containing the reporter plasmids resulted in an immediate reduction of signal from both gfp and lux expression plasmids. A faster decrease in signal was seen in bacteria harbouring the lux constructs compared to the equivalent gfp constructs, as would be expected due to the longer half-life of the GFP reporter molecule [9]. However the rate of loss of signal was the same for all constructs indicating that the presence of attB sites within the reporter gene structure did not significantly alter the t1/2 of the mRNA.
Figure 6.
Reporter gene repression kinetics. S. aureus RN4220 containing reporter plasmids pSB3004 (□, ■; 4 att sites), pSB3002 (△, ▲; 2 att sites) and pSB3000 (○, ●; no att sites) were grown to mid-log phase in Tris Minimal Succinate medium with 5 μg ml-1 Cm and 0.5% xylose. Bacteria were washed and resuspended in an equal volume of medium containing 5 μg ml-1 Cm and 1% glucose. Aliquots were placed in a 96 well microtitre plate and incubated at 37°C in a Tecan Genios Pro. Panel A: fluorescence (solid symbols, (Relative Fluorescence Units; RFU) and absorbance (open symbols) were measured at 10 min intervals. Plasmids used were pSB3004 (□, ■; 4 att sites), pSB3002 (□, ▲; 2 att sites) and pSB3000 (○, ●; no att sites). Panel B: luminescence (solid symbols, Relative Light Units; RLU) and absorbance (open symbols) were measured at 10 minute intervals. Plasmids used were pSB3014 (□, ■; 4 att sites), pSB3012 (□, ▲; 2 att sites) and pSB3010 (○, ●; no att sites). Data is presented as % maximal signal to allow direct comparison of repression kinetics despite the fact that light levels from each construct were different.
Evaluation of expression clones in Listeria monocytogenes
Plasmid pMK4 is also known to replicate in L. monocytogenes [8] and in this case a dual reporter plasmid containing both gfp and the Gram-positive optimised lux operon was created for evaluation in L. monocytogenes. Plasmid pSB3005 contained the PxylA promoter fused to the gfp:luxABCDE dual reporter operon and the rrnBT1T2 transcriptional terminator in pDEST-pMK4. When L. monocytogenes NCTC 7973 cells containing pSB3005 were grown in the presence of selective antibiotics, cultures showed an extended lag phase but eventually grew at normal rates and achieved a cell density expected for L. monocytogenes cultures (see Fig. 7). When L. monocytogenes(pSB3005) cells were visualised by fluorescence microscopy after overnight growth with antibiotic selection, only approx. 20% of the cells expressed GFP, suggesting either a high rate of selection of non-expressing clones or plasmid instability.
Figure 7.
Comparison of vectors in L. monocytogenes. L. monocytogenes 7973 plasmids based on the shuttle vector pMK4 (△, ▲; pSB3005) or pUNK1 (□ ■; pSB3007) containing the gfp:luxABCDE operon under the control of the PxylApromoter (see Table 2). Cells were grown in MWB at 37°C and luminescence (closed symbols, Relative Light Units; RLU)) and growth measurements (open symbols) were taken at intervals. Reporter gene data are presented as RLU/OD600 nm to account for increasing cell number during the experiment.
While pMK4 has been found to be stable in S. aureus [9,10], some plasmid instability had been observed in L. monocytogenes (Hill, unpublished data) therefore plasmid stability experiments were performed. To rule out any effect on plasmid stability caused by expression of the dual reporter gene operon, L. monocytogenes 7973(pMK4) cells were also examined. After overnight culture (15 generations) without antibiotic the average percentage loss of pMK4 plasmids was 95% demonstrating that the plasmids were not stably maintained in L. monocytogenes without selection. Individual values for plasmid loss with or without the reporter gene were 94% (pMK4) or 97% (pSB3005), respectively, and these values were not significantly different (P = 0.05). Therefore instability could not be attributed to expression of the dual reporter gene operon. When bacteria were grown in the presence of Cm (7 μgml-1) selection, average plasmid loss for both plasmids was still high but decreased to 73%. This was a surprising result since the presence of the antibiotic was expected to completely stabilise the plasmid. Again no significant difference in values for reporter plasmid and parent vector were found.
To address this problem, a new reporter gene construct (pSB3007) was created in the vector pDEST-pUNK1 [14] containing PxylA:gfp:luxABCDE:rrnBT1T2. Both pSB3007 and pUNK1 were transformed into L. monocytogenes 7973 and plasmid stability experiments carried out. Average plasmid loss of pUNK1 plasmids from cells grown in the absence of antibiotic was only 25% (pUNK1) and 12% (pSB3007). For bacteria grown in the presence of Erm (5 μgml-1) average plasmid loss did not improve and was similar for pUNK1 and pSB3007 at approx. 22%. This demonstrated that the pUNK1 plasmids are intrinsically more stable in L. monocytogenes cells than the pMK4-based constructs and shows that expression of the reporter genes does not lead to higher levels of plasmid loss. Little additional plasmid stability was achieved for the pUNK1 plasmids when antibiotic selection was applied, showing that these plasmids are appropriate to use in circumstances where antibiotics cannot be used for plasmid selection.
To investigate whether the expression of the dual reporter operon was stable, microscopic examination of bacteria containing pSB3007 grown in the presence of Erm was carried out. Very few cells were non-fluorescent, confirming that the majority of plasmid-containing cells also expressed the reporter genes. Expression of the lux genes was evaluated during growth of L. monocytogenes(pSB3007) in BHI broth in the presence of Erm. In this case the extended lag phase identified when growing the pMK4-based plasmid did not occur (see Fig. 7). Bioluminescence levels increased as cells entered the exponential phase of growth and began to decline in early stationary phase (Fig. 7), following the pattern of expression previously reported for this promoter in L. monocytogenes [8]. This again confirmed that the presence of the att sites in these constructs does not change the pattern of expression of the reporter gene.
Evaluation of sspA expression clones in Bacillus subtilis
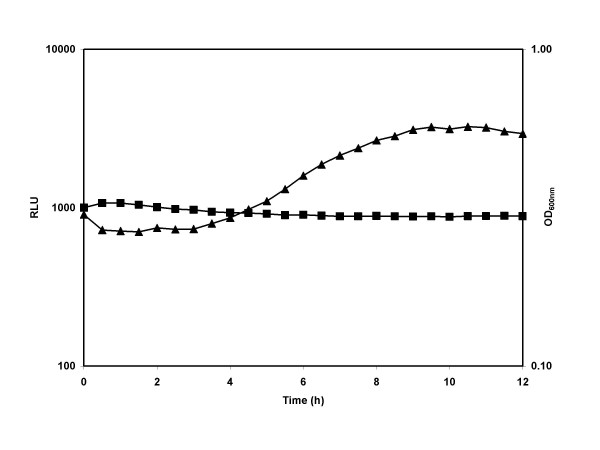
The differences in pMK4 plasmid stability seen in different hosts demonstrated the importance of vector choice when working in different Gram-positive bacteria. Therefore to develop a Gateway Destination vector for Bacilli, a plasmid containing the pTA1060 replicon known to be stable through sporulation in B. subtilis was chosen (pHB201; [12,15]). The Destination vector pDEST-pHB201 was used to create an Expression Clone containing the promoter from the sporulation-associated SASP gene (PsspA)fused to the dual reporter operon gfp:luxABCDE (pSB3024; see Table 2). SASP is synthesised only during sporulation to protect DNA within the spore [16]. The plasmid was transformed into the pyrB B. subtilis strain, BGSC 1A393, that does not grow in minimal media without added uracil. Cells were grown in rich broth and then transferred into a sporulation minimal media (SMM) without uracil supplement to encourage initiation of sporulation due to nutrient limitation [17]. As expected, no growth occurred in the SMM media, but a strong induction of reporter gene expression was seen after 3 h (Fig. 8). Light levels reach a maximum of 3000 RLU after 10 h and then began to decline. This peak of gene expression suggests that a large number of the cells in the population began sporulating at the same time, and corresponds with the expected σG-dependent expression of PsspA in the developing forespore after sporulation initiation in B. subtilis [18].
Figure 8.
Pattern of PsspA promoter induction in Bacillus. B. subtilis(pSB3024) containing the Gram-positive optimised dual gfp:luxABCDE operon under the control of PsspA (see Table 2) was grown overnight in LB supplemented with 5 μg ml-1 Erm at 37°C. Cells were then harvested by centrifugation and washed with PBS before diluting 1/50 into SMM supplemented with 5 μg ml-1 Erm. Cultures were then incubated at 37°C and luminescence (▲, Relative Light Units; RLU)) and absorbance measurements (■) were taken at intervals.
Discussion
The work presented here underlines the flexibility of the MultiSite Gateway Technology to assemble multiple DNA fragments precisely, efficiently, and directionally in a defined order and orientation, without subcloning, thereby negating the need for restriction enzymes and DNA ligases. One concern raised about this cloning system is the presence of the residual attB sites in the final Expression Clone constructs. Here we show that no significant differences are seen in rates of gene induction due to the presence of att sites within the expression cassettes, although in this model system the amount of resultant protein seems to be lower where attB sites are introduced proximal to the promoter and RBS. Comparison of GFP and Lux data indicated that mRNA half-life stability was not affected by differences in mRNA structure between the conventional and Gateway clones in S. aureus.
Another concern often expressed when using reporter genes for in vivo studies is that the expression of the reporter will place a metabolic burden on the bacteria and therefore alter growth behaviour. In this case we have used a dual reporter gene operon that encodes 6 genes in total and yet there was no evidence of even high level expression affecting bacterial growth. However it is clear that problems of vector maintenance may be of more significance than any metabolic burden of the reporter genes. In addition it is clear that antibiotic selection alone is not sufficient to ensure that all cells in a population retain reporter gene constructs, and that the choice of antibiotic marker gene is also an important factor when designing vectors. The high level of plasmid loss seen with pMK4 and its derivatives in the presence of Cm may be due to the fact that the cat gene confers resistance by inactivation of the antibiotic [19]. Hence during culture, levels of chloramphenicol in the media decrease and allow outgrowth of plasmid-free segregants which no longer possess the plasmid-borne cat. This may explain the long lag seen in the growth experiments where antibiotics were added to the growth media (see Fig. 7). In contrast the ermAM gene present on pUNK1 confers resistance by methylation of the rRNA [20] and therefore the antibiotic concentrations will not decline and maintain selection throughout the experiment. The lack of extended lag phase seen with pUNK1-based vectors supports this hypothesis. This makes the development of the Gateway cloning system even more attractive for Gram-positive bacteria, as it is relatively simple to generate new constructs in different vector when such effects are detected.
In this report we have used the Gateway system to rapidly generate Expression Clone plasmids, however the same Multisite DNA fragment fusion system could also be used in site directed mutagenesis strategies by amplifying upstream and downstream regions of a target gene and recombining these with a marker gene that will replace the target gene following in vivo recombination. Evaluation of Gram-positive suicide Destination vectors is currently underway and will facilitate mutational analysis of genes in Gram-positive bacteria.
Conclusion
The residual attB sites resulting from Gateway cloning do not affect gene expression in Gram-positive bacteria any more than residual restriction sites resulting from conventional cloning. The rapidity and precision of recombinatorial cloning makes it the method of choice where precise juxtaposition cloning of DNA sequences is not required, and simplifies the construction of expression plasmids, where multiple ligation steps are usually needed. A range of vector systems with different origins of replication are needed when working with diverse Gram-positive cell types to ensure that data obtained is as robust as possible.
Methods
Bacterial strains and growth conditions
E. coli TOP10 (F-) and E. coli DB3.1 (F- gyrA462) were cultivated in Luria-Bertani (LB; 5 g-1) broth or agar supplemented with appropriate antibiotics. L. monocytogenes NCTC 7973 (ATCC NO. serovar 1/2a) was grown in brain-heart infusion medium (BHI, Oxoid) or in the chemically defined medium Modified Welshimer's Broth (MWB; [21]). Bacillus subtilis BGSC 1A393 pyrB [22] was cultivated in LB broth or in Spizizens Minimal Media (SMM; 16). S. aureus RN4220 was grown in LB medium or in Tris Minimal Succinate medium (TMS; [23]). Antibiotics Ampicillin (Ap) Erythromycin (Erm) and Chloramphenicol (Cm) were used to select for plasmid maintenance when appropriate. Long term stocks of bacterial strains were stored at -80°C in Microbank™ porous beads.
Amplification of DNA
PCR reactions contained 10 μM of each primer, template DNA at 0.2 ng μl-1 and 2.5 u of AccuPrime Taq DNA Polymerase High Fidelity (Invitrogen) according to the Manufacturer's instructions. Bacillus megaterium NCTC 10342 DNA was used as template DNA for amplification of the xylR PxylA fragment, B. subtilis 168 was used as template for amplification of PsspA and plasmid pTMP100 was used as template for amplification of reporter genes; this plasmid contains the dual reporter genes gfp and luxABCDE under the control of the S. aureus PrpsJ; all genes in the lux operon are modified to achieve enhanced translation in Gram-positive bacteria. PCR reactions were incubated at 94°C for 1 min, followed by 30 cycles of 94°C for 30 sec, 52°C for 30 sec, and 68°C for 3 min (or 7 min for 6 kb fragments), with a final extension period at 68°C for 10 min. If non-specific PCR products were formed, anneal temperatures were increased to a maximum of 60°C.
DNA manipulation & vector construction
Plasmids were prepared using Qiagen DNA purification kits. Restriction enzyme analyses were performed using standard procedures [24]. T4 DNA ligase (Promega) was used for ligation of DNA fragments. PCR products and restriction fragments were purified from low melting point agarose gels using a Promega Wizard PCR purification Kit and DNA fragments eluted in water. For Gateway cloning, the Multisite Gateway Three-Fragment Vector Construction kits were used according to manufactures instructions (Invitrogen).
To create pSB3002 (2 att sites), pSB3004 was cut with ScaI/MscI to release a fragment containing 'bla xylR PxylA gfp'. This was replaced with a ScaI/MscI fragment from pSB2018 (Table 2) containing the same promoter/reporter gene sequences created by conventional cloning and contains no att sites [8]. To create pSB3012 (2 att sites), pSB3014 was restricted with ScaI/MluI to excise the 'bla xylR PxylA luxAB fragment and replaced with a ScaI/MluI from pSB2026 [9] containing the same sequences but without attB sites (see Fig. 3). Plasmid pSB3000 (no att sites) was constructed by insertion of the rrnBT1T2 sequence amplified from pTRC99A, using primers incorporating 5' PstI and 3' NsiI sites, into plasmid pSB2018 in the PstI site immediately downstream of PxylAgfp. The amplicon was restricted with PstI and NsiI and ligated with pSB2018 cut with PstI. To create pSB3010 (no attB sites), pSB2018 was restricted with SmaI/PstI to remove gfp and then ligated with a SmaI/PstI luxABCDE fragment excised from pTMP1 (Table 2). The resulting plasmid was digested with PstI and the PstI/NsiI rrnBT1T2 fragment inserted downstream of the lux operon (Fig. 3).
Bacterial transformation
E. coli transformation was performed using DB3.1 (for maintenance of ccdB+ plasmids by virtue of their gyrA462 mutation) or TOP10 (for selection of Entry and Expression clones) frozen competent cells (Invitrogen). Cells were plated on LB agar supplemented with Cm (30 μgml-1), Kanamycin (Km; 30 μgml-1), Ap(50 μgml-1), or Em (150 μgml-1) as appropriate for the vector used.
Transformation of L. monocytogenes and S. aureus with plasmids were carried out by electroporation [25,26]. L. monocytogenes transformants were plated on BHI agar (Cm 7 μgml-1), S. aureus transformants were selected on LB agar (0.5% xylose, Cm 5 μgml-1). The one step natural transformation method was used to transform B. subtilis [16] and plated on LB agar (Cm 7 μg ml-1). Expression of lux and gfp reporter genes in individual colonies was detected using a Nightowl CCD camera system with integrated fluorescence excitation (Berthold Technologies).
Plasmid stability measurements
Bacteria were inoculated into BHI broth, with or without antibiotic selection for the plasmid vector being used. After overnight culture at 37°C with aeration, samples were diluted and viable count determined using duplicate samples grown on plates with and without appropriate antibiotics. Plasmid loss was determined both after one night of culture (15 generations) and after subculture from this initial flask (23 generations). Plasmid loss was calculated by subtracting the number of antibiotic resistance colonies from the total number of colonies and expressing this value as a percentage of the total number of cells. Results presented are the mean of at least two experiments.
Growth and gene expression measurements
L. monocytogenes or B. subtilis cultures were grown overnight in rich broth supplemented with suitable antibiotics at 37°C with shaking (200 rpm). Samples were centrifuged at 16,000 × g for 2 min at room temperature, washed twice with sterile phosphate buffered saline (PBS) before resuspending in an equal volume of PBS. The washed cell suspension was diluted 1/100 (L. monocytogenes) or 1/50 (B. subtilis) into either chemically defined or rich media supplemented with antibiotics as appropriate.
S. aureus overnight cultures were grown aerobically at 37°C in TMS supplemented with Cm (5 μg ml-1). These were diluted 1/50 into fresh medium and grown for 4 h at 37°C with or without 0.5% xylose. To measure induction of PxylA cultures were grown without xylose then 0.5% xylose was added after 4 h. To measure repression of the xylR PxylA promoter system, cultures were grown for 4 h with 0.5% xylose, then cells were harvested and washed (as above) and transferred into media supplemented with Cm 5 μgml-1 and 1% glucose (for catabolite repression via XylR; [11]).
For all reporter gene measurements, replicate samples (200 μl) were placed into the wells of a 96-clear-bottom microtiter plate (Porvair) and incubated at 37°C in a Tecan Genesis Pro microplate reader. Optical density (600 nm), fluorescence (RFU; Excitation wavelength = 485 nm, Emission wavelength = 535 nm) and light levels (RLU) readings were taken at regular periods over the course of the experiment.
Competing interests
The author(s) declares that there are no competing interests.
Authors' contributions
TMP and SNAQ carried out cloning and growth/reporter studies and helped to draft the manuscript, SRG carried out cloning and growth/reporter studies, CEDR and VS participated in the design of the study and helped to draft the manuscript. PJH conceived of the study, and participated in its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
This work was supported by the UK Biotechnology and Biological Sciences Research Council grants 42/E19331 and 330/E19334, and a program grant from the UK Medical Research Council G9219778. SRG was supported by a University of Nottingham New Route Scholarship.
Contributor Information
Tania M Perehinec, Email: tania.perehinec@nottingham.ac.uk.
Saara NA Qazi, Email: saara.qazi@nottingham.ac.uk.
Sanyasi R Gaddipati, Email: stxsrg@nottingham.ac.uk.
Vyvyan Salisbury, Email: vyv.salisbury@uwe.ac.uk.
Catherine ED Rees, Email: cath.rees@nottingham.ac.uk.
Philip J Hill, Email: phil.hill@nottingham.ac.uk.
References
- Bushman W, Thompson JF, Vargas L, Landy A. Control of directionality in lambda site specific recombination. Science. 1985;230:906–911. doi: 10.1126/science.2932798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley JL, Temple GF, Brasch MA. DNA cloning using in vitro site-specific recombination. Genome Res. 2000;10:1788–1795. doi: 10.1101/gr.143000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y, Sone T, Yoshida S, Yahata K, Hotta J, Chesnut JD, Honda T, Imamoto F. Evidence for high specificity and efficiency of multiple recombination signals in mixed DNA cloning by the Multisite Gateway system. J Biotechnol. 2004;107:233–243. doi: 10.1016/j.jbiotec.2003.10.001. [DOI] [PubMed] [Google Scholar]
- Cheo DL, Titus SA, Byrd DR, Hartley JL, Temple GF, Brasch MA. Concerted assembly and cloning of multiple DNA segments using in vitro site-specific recombination: functional analysis of multi-segment expression clones. Genome Res. 2004;10B:2111–2120. doi: 10.1101/gr.2512204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard P, Gabant P, Bahassi EM, Couturier M. Positive selection vectors using the F plasmid ccdB killer gene. Gene. 1994;148:71–74. doi: 10.1016/0378-1119(94)90235-6. [DOI] [PubMed] [Google Scholar]
- Bae T, Schneewind O. Allelic replacement in Staphylococcus aureus with inducible counter-selection. Plasmid. 2006;55:58–63. doi: 10.1016/j.plasmid.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Peschke U, Beuck V, Bujard H, Gentz R, Le Grice S. Efficient utilization of Escherichia coli transcriptional signals in Bacillus subtilis. J Molec Biol. 1985;186:547–555. doi: 10.1016/0022-2836(85)90129-9. [DOI] [PubMed] [Google Scholar]
- Qazi SNA, Rees CED, Mellits KH, Hill PJ. Development of gfp vectors for expression in Listeria monocytogenes and other low G+C Gram positive bacteria. Microbial Ecol. 2001;41:301–309. doi: 10.1007/s002480000091. [DOI] [PubMed] [Google Scholar]
- Qazi SNA, Counil E, Morrissey J, Rees CED, Cockayne A, Winzer K, Chan WC, Williams P, Hill PJ. agr expression precedes escape from the endosome of Staphylococcus aureus. Infec Immun. 2001;69:7074–7082. doi: 10.1128/IAI.69.11.7074-7082.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qazi SNA, Harrison SE, Self T, Williams P, Hill PJ. Realtime monitoring of intracellular Staphylococcus aureus replication. J Bacteriol. 2003;186:1065–1077. doi: 10.1128/JB.186.4.1065-1077.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmiedel D, Kintrup M, Kuster E, Hillen W. Regulation of expression, genetic organisation and substrate specificity of xylose uptake in Bacillus megaterium. Molec Microbiol. 1997;23:1053–1062. doi: 10.1046/j.1365-2958.1997.2881654.x. [DOI] [PubMed] [Google Scholar]
- Hill PJ, Hall L, Vinicombe DA, Soper CJ, Setlow P, Waites WM, Denyer S, Stewart GSAB. Bioluminescence and spores as biological indicators of inimical processes. J Appl Bacteriol. 1994;76:129S–134S. doi: 10.1111/j.1365-2672.1994.tb04364.x. [DOI] [PubMed] [Google Scholar]
- Vellanoweth RL. Translation and its regulation. In: Sonenshein AL, Hoch JA, Losick R, editor. Bacillus subtilis and Other Gram-Positive Bacteria Biochemistry, Physiology and Molecular Genetics. Washington, DC: ASM Press; 1993. pp. 699–711. [Google Scholar]
- Pilgrim S, Stritzker J, Schoen C, Kolb-Maurer A, Geginat G, Loessner MJ, Gentschev I, Goebel W. Bactofection of mammalian cells by Listeria monocytogene s: improvement and mechanism of DNA delivery. Gene Ther. 2003;10:2036–2045. doi: 10.1038/sj.gt.3302105. [DOI] [PubMed] [Google Scholar]
- Bron S, Bolhuis A, Tjalsma H, Holsappel S, Venema G, van Dijl JM. Protein secretion and possible roles for multiple signal peptidases for precursor processing in Bacilli. J Biotechnol. 1998;64:3–13. doi: 10.1016/S0168-1656(98)00099-6. [DOI] [PubMed] [Google Scholar]
- Setlow P. Resistance of bacterial spores. In: Storz G, Hengge-Aronis R, editor. Bacterial stress responses. Washington, D.C. ASM Press; 2000. pp. 217–230. [Google Scholar]
- Harwood CR, Cutting SM. Molecular Biological Methods for Bacillus. Chichester: John Wiley & Sons; 1990. [Google Scholar]
- Errington J. Bacillus subtilis sporulation: regulation of gene expression and control of morphogenesis. Microbiol Rev. 1993;57:1–33. doi: 10.1128/mr.57.1.1-33.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin TJ, Snow GA. Biochemistry and Molecular Biology of Antimicrobial Drug Action. 6. USA, Springer; 2005. [Google Scholar]
- Thakker-Varia S, Ranzini AC, Dubin DT. Ribosomal RNA methylation in Staphylococcus aureus and Escherichia coli: effect of the "MLS" (erythromycin resistance) methylase. Plasmid. 1985;14:152–161. doi: 10.1016/0147-619X(85)90075-7. [DOI] [PubMed] [Google Scholar]
- Premaratne RJ, Lin WJ, Johnson EA. Development of an improved chemically defined minimal medium for Listeria monocytogenes. Appl Environ Microbiol. 1991;57:3046–3048. doi: 10.1128/aem.57.10.3046-3048.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potvin BW, Kelleher RJ, Gooder HW. Pyrimidine biosynthetic pathway of Bacillus subtilis. J Bacteriol. 1975;123:604–605. doi: 10.1128/jb.123.2.604-615.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebulsky MT, Hohnstein D, Hunter MD, Heinrichs DE. Identification and characterization of a membrane permease involved in ironhydroxamate transport in Staphylococcus aureus. J Bacteriol. 2000;182:4394–4400. doi: 10.1128/JB.182.16.4394-4400.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2. USA, Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Augustin J, Gotz F. Transformation of Staphylococcus epidermidis and other staphylococcal specieswith plasmid DNA by electroporation. FEMS Microbiol Lett. 1990;66:203–208. doi: 10.1111/j.1574-6968.1990.tb03997.x. [DOI] [PubMed] [Google Scholar]
- Park SF, Stewart GSAB. High-efficiency transformation of Listeria monocytogenes by electroporation of penicillin treated cells. Gene. 1990;94:129–132. doi: 10.1016/0378-1119(90)90479-B. [DOI] [PubMed] [Google Scholar]
- Brosius J, Dull TJ, Sleeter DD, Noller HF. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Molec Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- Sullivan MA, Yasbin RE, Young FE. New shuttle vectors for Bacillus subtilis and Escherichia coli which allow rapid detection of inserted fragments. Gene. 1984;29:21–26. doi: 10.1016/0378-1119(84)90161-6. [DOI] [PubMed] [Google Scholar]