Abstract
Background
The saprophagous and coprophagous maggots of the drone fly Eristalis tenax (Insecta, Diptera) have evolved the unique ability to survive in aquatic habitats with extreme microbial stress such as drains, sewage pools, and farmyard liquid manure storage pits. Therefore, they represent suitable models for the investigation of trade-offs between the benefits resulting from colonization of habitats lacking predators, parasitoids, or competitors and the investment in immunity against microbial stress. In this study, we screened for genes in E. tenax that are induced upon septic injury. Suppression subtractive hybridization was performed to selectively amplify and identify cDNAs that are differentially expressed in response to injected crude bacterial endotoxin (LPS).
Results
Untreated E. tenax maggots exhibit significant antibacterial activity in the hemolymph which strongly increases upon challenge with LPS. In order to identify effector molecules contributing to this microbial defense we constructed a subtractive cDNA library using RNA samples from untreated and LPS injected maggots. Analysis of 288 cDNAs revealed induced expression of 117 cDNAs corresponding to 30 novel gene clusters in E. tenax. Among these immune-inducible transcripts we found homologues of known genes from other Diptera such as Drosophila and Anopheles that mediate pathogen recognition (e.g. peptidoglycan recognition protein) or immune-related signaling (e.g. relish). As predicted, we determined a high diversity of novel putative antimicrobial peptides including one E. tenax defensin.
Conclusion
We identified 30 novel genes of E. tenax that were induced in response to septic injury including novel putative antimicrobial peptides. Further analysis of these immune-related effector molecules from Eristalis may help to elucidate the interdependency of ecological adaptation and molecular evolution of the innate immunity in Diptera.
Background
Eristalis tenax belongs to the hover flies, family Syrphidae (Diptera), with more than 5,000 described species, which occurred late in evolution and radiated in the Early Tertiary [1]. In contrast to the predominantly flower-feeding habitats of adult syrphids, their maggots are found in a very diverse array of habitats. Those of subfamily Eristalinae are saprophagous, coprophagous and aquatic filterfeeders. Combined analysis of molecular and morphological characters supports both frequent shifts between larval feeding habitats within the Eristalinae and the monophyly of this subfamily [2]. The rat-tailed maggots of E. tenax are characterized by their extended anal breathing tube that functions as a snorkel and enables survival in waters with anaerobic conditions. They realize a unique ecological niche by preferably living in stagnant aquatic environments with high organic and microbial contamination. Because of the preference of E. tenax larvae for dirty waters with anaerobic conditions, they are reliable and prominent indicators in the biological assessment of water quality for extremely high pollution with organic material [3]. They thus represent a suitable model to investigate the trade-offs between the benefits resulting from colonization of habitats lacking predators, parasitoids, or competitors and the investment in immunity against microbial stress.
The innate immunity of insects relies on immediate processes including cellular phagocytosis, encapsulation, hemolymph coagulation, and phenoloxidase activation leading to melanization. On the other hand sustainable defense is achieved by the massive synthesis of antimicrobial peptides like defensins and cecropins [4,5]. Insect immunity has been best analyzed in the fruit fly D. melanogaster [6,7], but results of comparative genome-wide analysis of immunity-related genes in D. melanogaster, Anopheles gambiae, and Aedes aegypti reveal that effector molecules from these dipteran species exhibit large variations [8]. For example, four defensin and four cecropin genes exist in Anopheles, making these more numerous than in Drosophila whereas some antimicrobial peptides that were identified in Drosophila are absent in Anopheles and Aedes [8]. The maggots of E. tenax have been recognized to be particularly resistant to microbial stress, because they are able to survive in aquatic habitats that are usually not colonized by other dipteran larvae or animals [3]. Hence we were interested to identify the immune-related genes of this insect.
To selectively identify immune-induced genes we used the subtractive suppression hybridization (SSH) method which amplifies differentially expressed cDNAs and simultaneously suppresses amplification of common cDNAs. This technique has been proven as a suitable tool for identification of immune-related genes in insects [9-11] and other invertebrates [12,13]. Here, we report analysis of the immune-inducible transcriptome of E. tenax maggots, collected from storage pits of liquid manure. This enabled identification of genes encoding novel proteins that are potentially involved in pathogen-recognition and immune signaling. In addition, we identified numerous potential antimicrobial peptides that most probably contribute to the observed immune defense that is obviously important to control the wide array of pathogens present in the habitat of Eristalis larvae.
Results and discussion
Subtracted cDNA library of immune challenged E. tenax larvae
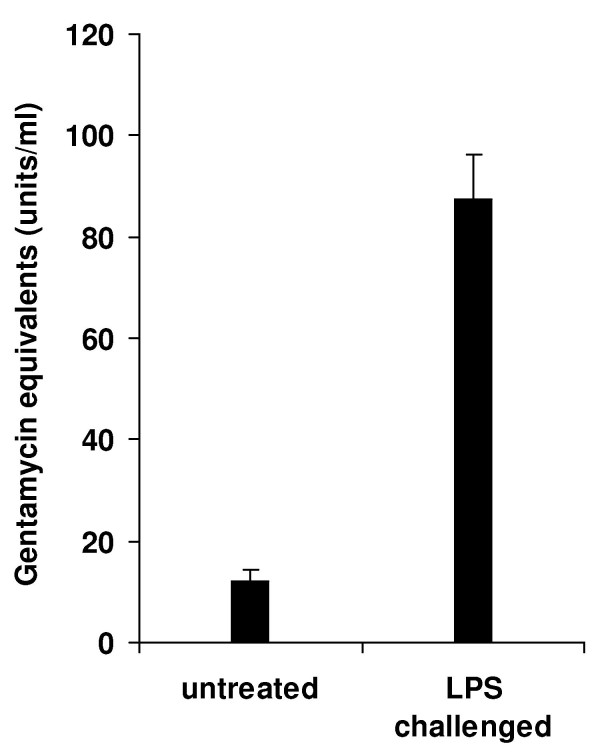
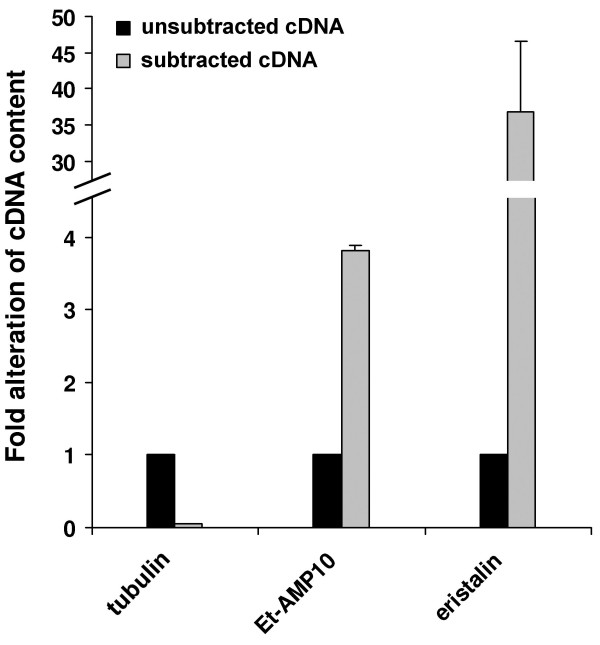
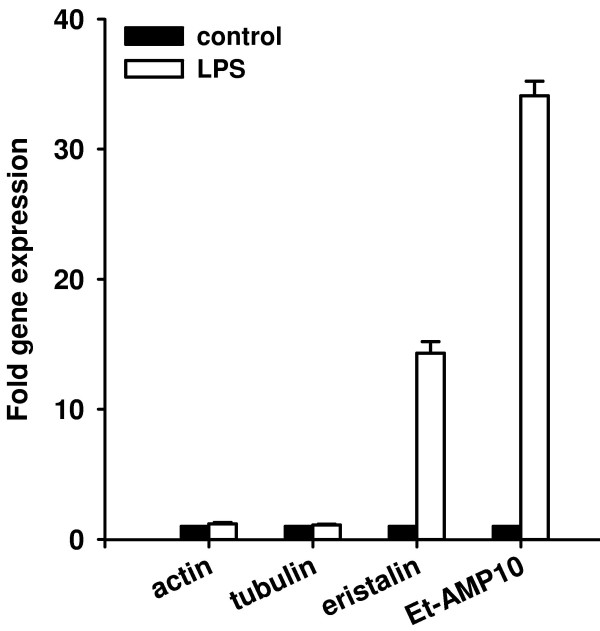
We observed a significant E. coli inhibitory activity of hemolymph samples from untreated E. tenax larvae that further increases upon immune challenge (Fig. 1). In order to construct a subtracted cDNA library enriched in immune-inducible genes we used purified RNA from LPS injected and untreated animals combined with a PCR-based SSH method. To induce strong and broad immune responses we injected a commercially available purified LPS preparation, which is known to contain impurities like nucleic acids, proteins and peptidoglycans, and which according to the supplier is commonly used as elicitor of immune responses in vertebrates and invertebrates. In order to confirm that the subtraction process has been performed efficiently, we analyzed the abundance of transcripts for the house-keeping gene α-tubulin and for two genes, the Et-AMP10 and eristalin. The latter ones were found in the present study to be induced in response to immune challenge in Eristalis. Quantitative real-time PCR analyses revealed that transcripts of α-tubulin were reduced 18 fold, whereas transcripts of Et-AMP10 and eristalin were enriched for 4 and 38 fold, respectively (Fig. 2). This is in agreement with values from the protocols of the manufacturer and indicates successful subtraction of the cDNA library. A total of 288 clones were randomly picked and subjected to colony PCR. Plasmids of 117 colonies that were positively screened in blot hybridization, indicating induced expression in response to LPS challenge, were isolated and sequenced. The sequences obtained (summarized in table 1) were deposited at EMBL European Bioinformatics Institute, and compared to databases of the National Center for Biotechnology Information using the program BLASTX. InterProScan at the EMBL European Bioinformatics Institute was used for an integrated search in PROSITE, Pfam and PRINTS databases in order to predict conserved motifs, signal sequences and transmembrane regions.
Figure 1.
Immune challenge of rat-tailed maggots of E. tenax induces the production of antimicrobial effector molecules. Antibacterial activities represented by gentamycin equivalents of hemolymph from untreated and LPS injected larvae are shown. Results represent mean values of independent determinations ± S.D.
Figure 2.
Confirmation of the enrichment of immune-related transcripts in the subtracted cDNA library by quantitative real-time PCR analysis. The relative amount of cDNAs of α-tubulin, Et-AMP10, and eristalin in the subtracted cDNA library (gray bars) is shown relative to their amount in unsubtracted cDNA library (black bars). The cDNA amount of the house-keeping gene α-tubulin was reduced about 18 fold by the subtraction procedure. In contrast, potentially immunity-related genes Et-AMP10 and eristalin were found to be 4 and 38 fold enriched in the subtracted cDNA library, respectively. Results represent mean values of three independent determinations ± S.D.
Table 1.
cDNAs from the subtracted E. tenax library
| Cluster | EMBL accession No. | Highest BLASTX match | PFAM | E value |
| Protein biosynthesis | ||||
| AM706409 | Ribosomal protein S8 (Culicoides sonorensis) | PF01201 | 7e-09 | |
| AM706410 | Ribosomal protein L6 (D. melanogaster) | PF01159 | 3e-37 | |
| AM706411 | Lysyl-tRNA synthetase (D. melanogaster) | PF00152 | 6e-111 | |
| AM706412 | eIF5c (D. melanogaster) | PF02020 | 8e-84 | |
| Repression of cell proliferation | ||||
| AM706413 | CREG (D. melanogaster) | 3e-46 | ||
| α-tubulin-like protein | ||||
| AM706414 | AlphaTub85E (D. melanogaster) | PF03953 | 5e-18 | |
| dsRNA-binding domain-like protein | ||||
| AM706415 | Replicase polyprotein (Drosophila C virus) | PF00035 | 5e-05 | |
| Pattern recognition protein | ||||
| AM706416 | PGRP-SB1 (D. melanogaster) | PF01510 | 6e-64 | |
| Signal transduction | ||||
| AM706417 | Relish (Glossina morsitans morsitans) | PF00023 | 7e-35 | |
| AM706418 | Protein kinase (D. melanogaster) | 5.2 | ||
| Kazal-type serine protease inhibitor | ||||
| AM706419 | enhancer of split m1 protein (D. simulans) | PF00050 | 4e-07 | |
| Putative antimicrobial peptides | ||||
| Et-AMP1 | AM706420 | Defensin (Phlebotomus duboscqi) | PF01097 | 3e-12 |
| Et-AMP2 | AM706421 | Bactenecin-7 (Bos taurus) | 0.96 | |
| Et-AMP3 | AM706422 | Tachycitin (Tachypleus tridentatus) | PF01607 | 3e-06 |
| Et-AMP4 | AM706423 | Salivary protein SG3 (Anopheles stephensi) | 7e-15 | |
| Et-AMP5 | AM706424 | α-helical cecropin-like | NSM1 | |
| Et-AMP6 | AM706425 | Ser/Thr/Lys-rich putatively disulfide bridged | NSM | |
| Et-AMP7 | AM706426 | Putatively disulfide bridged peptide with a pI 10,5 | NSM | |
| Et-AMP8 | AM706427 | Putatively disulfide bridged peptide with a pI 10,3 | NSM | |
| Et-AMP9 | AM706428 | Putatively disulfide bridged peptide with a pI 9,3 | NSM | |
| Et-AMP10 | AM706429 | Putatively disulfide bridged anionic peptide | NSM | |
| Et-AMP11 | AM706430 | Glycine-rich peptide 1 | NSM | |
| Et-AMP12 | AM706431 | Glycine-rich peptide 2 | NSM | |
| Et-AMP13 | AM706432 | Glycine-rich peptide 3 | NSM | |
| Et-AMP14 | AM706433 | Glycine-rich peptide 4 | NSM | |
| Et-AMP15 | AM706434 | Linear peptide 1 | NSM | |
| Et-AMP16 | AM706435 | Linear peptide 2 | NSM | |
| Et-AMP17 | AM706436 | Linear peptide 3 | NSM | |
| Et-AMP18 | AM706437 | Linear peptide 4 | NSM | |
| Et-AMP19 | AM706438 | Linear peptide 5 | NSM | |
1NSM, no significant match
We describe here the identification of immune-inducible proteins in E. tenax that are potentially involved in recognition of microbes, immune-related signaling, and antimicrobial effector mechanisms.
Sensing of infection
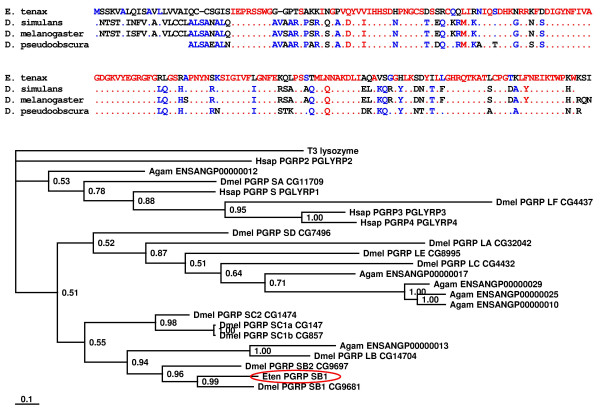
Innate immunity depends on pathogen recognition that is mainly mediated by host proteins called pattern recognition receptors. In insects, two prominent members, the Gram-negative binding proteins (GNBP) and peptidoglycan recognition proteins (PGRP) are involved in the activation of prophenoloxidase cascade and of Toll/IMD pathways resulting in the massive production of immune effector molecules [6]. Here, we identified transcripts of a protein that shares sequence similarities with PGRP-SB1 from several Drosophila species (Fig. 3A). This peptidoglycan recognition protein (PGRP) is one of a battery of pattern recognition proteins in insects that are essential for recognition of invading microbes and induction of immune responses [14]. Drosophila and Anopheles have 13 and 7 PGRP genes that are transcribed into at least 17 and 9 proteins, respectively [14]. Mammals possess a family of 4 PGRPs that function as both recognition proteins and effector molecules exhibiting bactericidal activities [15]. A Bayesian protein tree (Fig. 3B) was generated with sequences from Eristalis, humans, D. melanogaster, and A. gambiae and revealed that Eristalis PGRP groups nearest to Drosophila PGRP-SB1.
Figure 3.
E. tenax PGRP is homologous to PGRP-SB1 from Drosophila. (A) BlastP search with E. tenax PGRP-SB1 sequence resulted in similar sequences from several Drosophila species and the differences are indicated in the alignment. Red color indicates 90% and blue color 50% consensus. The amino-terminal 24 amino acids represent a putative signal sequence. Accession numbers of PGRP-SB1 proteins are: E. tenax, AM706416; D. simulans, Q70PX8; D. melanogaster, Q70PY2; D. pseudoobscura, Q29DT4. (B) A Bayesian protein tree was generated using the sequence of Eristalis PGRP (Eten PGRP SB1, indicated by a red circle) and sequences of human (Hsap PGRPs), D. melanogaster (Dmel PGRPs), and A. gambiae PGRPs (Agam ENSANGPs) and we found that Ersitalis PGRP grouped nearest to Drosophila PGRP-SB1. T3 lysozyme (N-acetylmuramoyl-L-alanine amidase, P20331) of the bacteriophage T3 was used as out-group. The scale bar represents the substitutions per site according to the model of amino acid evolution applied.
Signaling
We identified a protein sharing highest similarity with the relish protein of Glossina morsitans morsitans (Uniprot: AAZ91474). Relish or Nuclear factor (NF)-kappa-B p110 subunit is a key molecule of the Imd pathway and is phosphorylated by LPS-activated I-kappa-B kinase complex before being cleaved [16]. The Relish-p110 subunit is cleaved within seconds after immune challenge into Relish-p49 and Relish-p68 subunits. The latter translocates into the nucleus and activates the transcription of numerous immune-related genes [6]. The NF-kappa-B/I-kappa-B signaling pathways are evolutionarily well conserved and widely distributed among the animal kingdom [17].
Many serine proteinase inhibitors have evolved in vertebrates and invertebrates to regulate vital serine proteinase cascades. These contribute to melanization and host defense responses in insects [18,19] as well as to hemostasis, fibrinolysis, and complement system in vertebrates [20,21]. Here, we identified one proteinase inhibitor that belongs to kazal-type serine proteinase inhibitors. Kazal domains are characterized by a well-preserved amino acid sequence containing three disulfide bridges, and display varying specificity for serine proteinases [22].
One identified cDNA encodes a protein that exhibits highest sequence similarities to the replicase polyprotein of the Drosophila C virus. This virus belongs to the picornavirus family (enveloped single-stranded RNA viruses) that also includes the prominent polio and foot-and-mouth disease viruses [23]. However, it should be mentioned here that the similarities between the sequences depends mainly on a dsRNA-binding domain-like region. This region is also present in a variety of cellular RNA-binding proteins with different structures and diverse functions including e.g. the human interferon-induced protein kinase [24].
Protein biosynthesis and cell proliferation
We identified several transcripts of factors potentially involved in Eristalis protein biosynthesis including ribosomal proteins S8 and L6, lysyl-tRNA synthetase, and the translation initiation factor eIF5c. This is reasonable, since the massive production of antimicrobial peptides upon immune challenge depends on the activation of the cellular translation machinery in cells and tissues of the immune system [9,13,25]. Nevertheless, one important study reported the antibacterial activity of ribosomal protein L1 fragment of Helicobacter pylori [26]. Additionally, Park et al. [27] found that lysyl-tRNA synthetase was secreted from intact human cells as a signaling molecule to trigger proinflammatory response by binding to macrophages and peripheral blood mononuclear cells to enhance their migration and the production of TNF-α. Since lysyl-tRNA synthetase is highly conserved in evolution it is possible that it also serves as a signaling molecule in insects.
One transcript shared high sequence similarities with the evolutionarily conserved cellular repressor of E1A-stimulated genes (CREG). A finely-tuned balance exists between cell proliferation, cell division arrest, and apoptosis in the vertebrate immune response, and this may also be true for insects. CREG is a secreted glycoprotein that inhibits cell proliferation and enhances cell differentiation [28,29]. In humans, CREG was shown to bind to the cation-independent mannose 6-phosphate/insulin-like growth factor II receptor, and this receptor has been shown to be required for CREG-induced growth suppression. In insects, CREG has been shown to be expressed in adult female salivary glands of A. gambiae [30].
We found one transcript that exhibits similarity to the carboxyl terminus of α-tubulin from other organisms. Although α-tubulin is a major constituent of microtubules and a well-known housekeeping gene, many genes and pseudogenes exist that form the α-tubulin superfamily [31]. Induction of α-tubulin-like mRNA after immune challenge has also been observed in leech [25], and another study demonstrated that the dengue virus 2 directly binds to a 48 kDa tubulin or tubulin-like protein of C6/36 mosquito cells [32]. These results suggest that at least some α-tubulin-like proteins may be involved in immune responses. However, in this study α-tubulin itself was depleted by the subtraction process (Fig. 2) suggesting that it is not immune-induced and the identified sequence may be derived from a pseudogene or related isoform.
Effector molecules
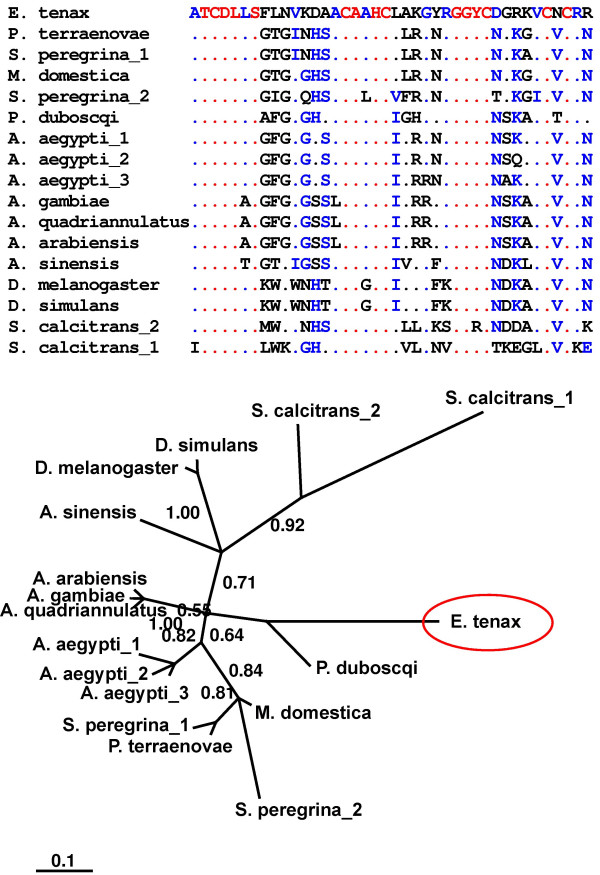
The most striking finding among the genes inferred to be up-regulated in response to septic injury is the identification of numerous putative antimicrobial peptides in E. tenax. Defensins have been found in a variety of animals and plants [33-35] and even in the fungus Pseudoplectania nigrella [36], which is suggestive for an evolutionarily conserved role in innate immunity. In agreement, we identified an E. tenax defensin that we named eristalin. Eristalin contains a predicted signal and pro-sequence and shares sequence similarities to other insect defensins (Fig. 4A). In addition, our phylogenetic analysis using sequences of eristalin and other defensin sequences from dipterans suggests that the defensins of E. tenax, S. peregrina, and S. calcitrans are more deviated in sequence. Maggots of all three of this species live on feces (Fig. 4B) suggesting that independent occupation of such 'extreme' ecological niches is accompanied by divergent evolution of immune-related effector molecules.
Figure 4.
Sequence alignment of eristalin with other dipteran defensins. (A) Eristalin was aligned with other dipteran defensins. For clarity only sequences of mature defensins were used for alignment. Red color indicates 90% and blue color 50% consensus. (B) A Bayesian protein tree was generated and we found that eristalin, defensin_1 and defensin_2 of Stomoxys calcitrans, and defensin_1 of Sarcophaga peregrina show some higher deviation in sequence when compared to other dipteran defensins. Accession numbers of defensins are (from top to bottom): AM706420, P10891, P18313, Q86BU1, P31530, P83404, P91793, P81602, P81603, Q17027, Q38L94, Q38LE1, Q4ZJJ8, P36192, Q86BW0, O16137, O16136. The scale bar represents the substitutions per site according to the model of amino acid evolution applied.
The putative antimicrobial peptide Et-AMP2 shares highest similarities to bactenecin-7 (Fig. 5) a cathelicidin-derived antimicrobial peptide of bovine neutrophil granules that exhibits antibacterial activity predominantly against gram-negative bacteria [37]. Bactenecin-7 belongs to the Pro/Arg-rich antibiotics [38] which also include porcine PR-39 [39], insect drosocin, abaecin, and lebocin [40]; however, no homologues were identified in humans. The antibacterial effects of Pro/Arg-rich peptides are likely due to an impaired function of the respiratory chain and of energy-dependent activities in the inner membrane of susceptible microorganisms [41].
Figure 5.
Sequence alignment of Et-AMP2 with Drosophila drosocin along with bactenecin-7 from sheep and bovine. For clarity only sequences of mature peptides of ET-AMP2 (AM706421), bactenecin-7 from sheep (UniProt: P50415) and bovine (UniProt: P19661), and of drosocin (UniProt: P36193) are shown. The predicted signal sequence of Et-AMP2 was omitted.
ET-AMP3 shows high similarity to the antimicrobial tachycitin of the horseshoe crab [42] and ET-AMP4 to the salivary protein SG3, with as yet unknown function, from Anopheles stephensi [43]. Et-AMP5 is a cationic peptide with putative α-helical content and with similarities to cecropins or to antimicrobial cathelicidin-derived peptides like bovine BMAP-27 [44]. Et-AMP6, 7, 8, and 9 are putatively disulfide-bridged cationic peptides, while Et-AMP10 is a putatively disulfide bridged anionic peptide. None share similarities with any known proteins or peptides but all exhibit Lys/Arg clusters in the proximity of hydrophobic clusters; this is typical for many antimicrobial peptides [4].
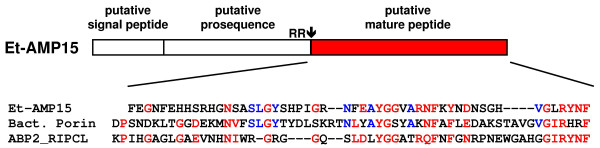
We also identified four isoforms (Et-AMP11 to 14) of glycine-rich peptides with a glycine content of over 20% (Fig. 6). The list of glycine-rich antimicrobial peptides is rapidly growing and includes prominent members like the antifungal holotricin-3 from the chafer Holotrichia diomphalia [45], the antibacterial acanthoscurrin from the spider Acanthoscurria gomesiana [46], and the antibacterial armadillidin from the crustacean Armadillidium vulgare [47]. Interestingly, the observation that several isoforms of a glycine-rich peptide are present in a single organism is in agreement with the identification of infection-induced glycine-rich peptides in C. elegans [48] and of putative glycine-rich attacins in the lepidopteran Antherea mylitta [49]. Similarly, we identified five isoforms (Et-AMP15 to 19) of a linear peptide that are cationic with a glycine content of over 10% and share sequence similarities to the carboxyterminal half of bacterial outer-membrane porins and to a putative antimicrobial peptide from the bug Riptortus clavatus [50] (Fig. 7).
Figure 6.
Sequence alignment of the glycine-rich peptides Et-AMP11, 12, 13, and 14. Sequences of the glycine-rich peptides Et-AMP11, 12, 13, and 14 (AM706430 to AM706433) were aligned. The alignment suggests that the isoforms have most probably been evolved by gene duplication and evolutionary selection to target a variety of pathogens. The amino-terminal 17 amino acids represent a predicted signal sequence. The amino-terminal end of Et-AMP12 is not complete.
Figure 7.
Sequence alignment of the linear peptide Et-AMP15 with a bacterial porin and a putative antimicrobial peptide from Riptortus clavatus. The linear peptide Et-AMP15 (AM706434) is schematically drawn, showing predicted signal peptide, propeptide, and mature peptide. The mature peptide exhibits some sequence similarities with the carboxy-terminal part of bacterial porin from Bordetella parapertussis (Bact. Porin, UniProt: Q7W5A6) and a putative antimicrobial peptide from the bug R. clavatus (ABP2_RIPCL, UniProt: Q27906).
Quantitative real time RT-PCR analysis
In order to precisely determine the immune-induction of genes identified in the present study we used quantitative real-time RT-PCR analysis. The Eristalis defensin eristalin and the putatively disulfide-bridged anionic peptide Et-AMP10 were 14 and 34 fold, respectively, up-regulated upon LPS injection (Fig. 8). The determined induction levels are in a similar range as reported for defensins from other insects [51]. In contrast, the expression of the housekeeping genes actin and tubulin were not influenced by the treatment.
Figure 8.
Quantitative real time RT-PCR analysis of transcriptional levels of selected genes in immune challenged and untreated larvae. The mRNA levels of selected genes in immune challenged animals (black bars) were determined and are shown relative to their expression levels in untreated animals (white bars). The transcriptional rates of eristalin and ET-AMP10 were found to be increased over 14 and 34 fold, respectively, in response to LPS injection. In contrast, the amount of actin and α-tubulin transcripts were not significantly influenced. Results represent mean values of three independent determinations ± S.D.
Conclusion
In the present study we analyzed the immune-inducible transcriptome from rat-tailed maggots of the drone fly E. tenax which have evolved the ability to survive in stagnant, frequently anaerobic aquatic habitats with extreme microbial contamination [3]. Survival in environments with such a strong microbial stress obviously depends on an innate immune system that has been adapted specifically to this ecological niche.
To date, seven distinct antimicrobial peptides have been reported from Drosophila, including the antifungal peptides drosomycin and metchnikowin, and five antibacterial peptides: attacin, cecropin, defensin, diptericin and drosocin [40]. Recently, genome-wide studies with Drosophila, Anopheles, and Aedes reveal that intracellular components of immune signaling pathways are conserved among Diptera [8]. In accordance, we identified potential counterparts in E. tenax that mediate non-self recognition (e.g. PGRP) or immune-related signaling (e.g. relish) in Drosophila and Anopheles. However, genome-wide studies also elucidate differences between both dipterans, particularly in potential extracellular components of the Toll pathway and in effector molecules including antimicrobial peptides [8]. In agreement, we identified numerous putative antimicrobial peptides in E. tenax that have obviously no homologues in D. melanogaster,A. gambiae, or A. aegypti. In total, we found 19 putative antimicrobial peptides some of which show sequence similarities to antimicrobial defensins, bactenecin-7, or tachycitin. 16 peptides are of novel-type including four isoforms of a glycine-rich peptide and five isoforms of a linear peptide. The isoforms may have been evolved by gene duplication and evolutionary selection to target a variety of pathogens, probably in a similar manner to that described for the six α-defensins and four β-defensins from humans [52].
Interestingly, we observed a significant E. coli inhibitory activity of hemolymph samples from untreated E. tenax larvae. This was not normally the case with hemolymph samples from other insect species that we analyzed in our laboratory (e.g. Thermobia domestica [9], Galleria mellonella [10], and Tribolium castaneum). This significant (probably constitutive) expression of antimicrobial peptides might be caused by the uptake of food with a high bacterial load. Recently, it has been demonstrated that Drosophila gut cells are able to sense bacteria-contaminated food which induces epithelial immune responses [6]. Thus, there may be an even richer assembly of immunity-related genes in Eristalis that would not be detected by our SSH approach.
Future work will include chemical or recombinant production of the identified peptides in order to analyze their specific activities against viruses, bacteria, fungi, or protozoa. Currently, we are investigating the potential of Et-AMP10 and eristalin to confer resistance against phytopathogens in transgenic plants similar as recently shown for the antifungal peptide gallerimycin from the greater wax moth Galleria mellonella [53]. In addition, the identified genes may help to elucidate divergent evolution of antimicrobial peptides in Diptera and the interdependence between ecological adaptation and insect immune defenses.
Methods
Insects
Rat-tailed maggots were collected in liquid manure pits at cattle farms near Giessen (Hessen) and the village Siewisch (Brandenburg), Germany. Using a practicable key for determination of hoverfly larvae [54], we identified third instar larvae of the drone fly E. tenax. As typical characters we observed the lack of setae along the lower lateral margins and the last pair of prolegs with most of the large primary crochets facing towards the lateral margins of the body. Identification was confirmed by determination of adults which hatched at the end of October 2005.
Immune challenge of larvae and isolation of hemolymph and mRNA
Last instar E. tenax larvae, each weighing between 200–300 mg, were used for immune-challenge. 10 μl sample volume, corresponding to 100 μg LPS (purified Escherichia coli endotoxin 0111:B4, Cat. No.: L2630, Sigma, Taufkirchen, Germany) per larva were injected dorsolaterally into the hemocoel using 1 ml disposable syringes and 0.4 × 20 mm needles mounted on a microapplicator. For antibacterial activity assays, hemolyph samples were isolated by bleeding injected larvae 24 h post challenge or untreated larvae into plastic tubes. Total RNA was extracted from whole larvae 8 h post injection using the TriReagent isolation reagent (Molecular Research Centre, Cincinnati, Ohio, USA) and a further poly-(A)+ RNA preparation was performed using a mRNA Nucleotrap kit (Macherey Nagel, Germany) according to the instructions of the manufacturers. RNA integrity was confirmed by ethidium bromide gel staining and quantities were determined spectrophotometrically [55].
Antibacterial activity assay
Antibacterial activity of hemolymph samples was measured by an inhibition zone assay using a LPS-defective, streptomycin- and ampicillin-resistant mutant of Escherichia coli K12 strain D31 [56]. In brief, Petri dishes (∅ 100 mm) were filled with 7 ml E. coli suspension, containing 2× YT-nutrient broth (Roth, Karlsruhe, Germany), 1% high-purity agar-agar (Roth), and 2 × 104 viable bacteria in logarithmic growth phase. Holes with a diameter of 4 mm were punched into the agar and filled with 3 μl of cell-free hemolymph. The diameters of the clear zones were measured after 24 h of incubation at 37°C and units/ml were calculated using a calibration curve obtained with dilutions of gentamycin (Sigma).
Construction of a subtracted cDNA library using the SSH method
In order to identify differentially expressed genes during immune response the SSH method was performed using mRNAs from immune challenged and control larvae and the PCR-Select cDNA Subtraction Kit from Clontech (Mountain View, CA, USA), according to the protocol of the manufacturer. Briefly, 1 μg of purified mRNA from immune-challenged and control larvae were reverse transcribed into cDNA using a cDNA synthesis primer, subsequently double stranded cDNA was generated and digested with RsaI. The double stranded cDNA from immunized larvae was ligated in separate aliquots to adaptor 1 or adaptor 2R and were denaturated at 98°C for 90 s and then hybridized at 68°C for 8 h with a 30 fold excess of double stranded cDNA from control larvae. Subsequently, both samples were mixed together again with a 10 fold excess of freshly denaturated double stranded cDNA from control larvae and hybridized in one tube at 68°C for 16 h. The sample was then subjected to two rounds of suppression PCR with PCR-primer 1 and nested primers supplied with the kit. PCR amplifications were performed in a total volume of 25 μl using a PCR cycler (Biometra, Göttingen, Germany) with a heated lid and the Advantage PCR system (Promega, Mannheim, Germany). An initial adapter extension at 72°C for 5 min was followed by a denaturation step at 95°C for 1 min and by 27 cycles of denaturation at 95°C for 15 s, annealing at 66°C for 30 s, and extension at 72°C for 90 s. A final 7-min 72°C step was added to allow complete extension of the products. The secondary PCR was performed with nested primer 1 and 2R on the diluted primary PCR products for 12 cycles under same conditions, except that 68°C was used as annealing temperature. The subtraction efficiency was confirmed by quantitative real-time PCR of the actin gene of subtracted PCR products in comparison to not subtracted PCR products. Resulting PCR products of the secondary subtractive PCR were separated on 1% (w/v) agarose gel electrophoresis with ethidium bromide staining, according to standard procedures [55] and five fractions were obtained by excising five gel pieces of PCR products with different lengths. Subsequently, PCR product fractions were separately purified using the NucleoSpin Extract II kit (Macherey Nagel), ligated into pGEM-T easy vector (Promega) and transformed into TOP10F' cells (Invitrogen, Carlsbad, CA, USA). The library was plated on 2×YT agar plates containing 100 μg/ml ampicillin and incubated at 37°C for 16 h. A preliminary screen of 15 colonies using the FastPlasmid Mini kit (Eppendorf, Hamburg, Germany) followed by EcoRI digestion of isolated plasmids and agarose gel electrophoresis showed that over 90% of clones contained an insert in the vector.
Colony PCR and blot hybridization
Colony PCR was performed on 288 randomly picked colonies with vector specific primers T7-promotor: 5'-TAATACGACTCACTATAGGG-3' and SP6: 5'-ATTTAGGTGACACTATAG-3' (purchased from Thermo electron, Waltham, MA, USA) using a Biometra PCR cycler and the Red Taq PCR system (Sigma, Taufkirchen, Germany). Used PCR conditions were: denaturation at 95°C for 3 min followed by 30 cycles of denaturation at 95°C for 15 s, annealing at 43°C for 15 s, and extension at 72°C for 60 s. A final 7-min 72°C step was added to allow complete extension of the products. 1 μl of resulting PCR products were identically spotted onto two sheets of positively charged nylon membranes (Roche, Lewes, United Kingdom). Membranes were dried and UV cross-linked using a BioRad UV cross-linker (BioRad, München, Germany), according to the instructions of the manufacturer. Digoxigenin labeled probes for hybridization were generated using secondary PCR products of subtracted and non-subtracted cDNAs and the Dig-High Prime Labelling kit (Roche, Lewes, United Kingdom). Hybridization, washing, and detection of digoxigenin labeled DNA was performed in accordance to the user guide instructions of the Dig Easy Hyb Granules, Dig-Wash and Block Buffer Set, Anti-Digoxigenin-AP and NBT/BCIP ready-to-use tablets (Roche).
Sequencing and computer analysis of cDNA sequence data
Plasmid isolation of 117 positively screened colonies was performed with the FastPlasmid Mini kit (Eppendorf) and purified plasmids were custom sequenced by Macrogen Inc. (Seoul, South-Korea). Sequences were used to identify similar sequences of the National Center for Biotechnology Information databases using BLASTX program (BLASTX 2.2.13; http://www.ncbi.nlm.nih.gov/BLAST/). InterProScan http://www.ebi.ac.uk/InterProScan/ was used for an integrated search in PROSITE, Pfam, and PRINTS databases at EMBL-European Bioinformatics Institute and to predict signal sequences and transmembrane regions.
Sequence alignments and phylogenic analyses
Sequence alignments were computed using the blosum62 algorithm [57] at http://bioinfo.genopole-toulouse.prd.fr. For phylogenetic reconstruction, we used the software package MrBayes 3.1.2 [58], which combines Bayesian inference and Markov chain Monte Carlo convergence acceleration techniques known as Metropolis coupling. The best fixed-rate model of amino acid evolution was determined by model jumping among nine possible models. The model with the overall highest posterior probability was WAG [59] for PGRPs and Blosum62 model [60] for defensins. Generations were sampled with the current tree saved at intervals of 100 generations. We used convergence diagnostic (i.e., the standard deviation of split frequencies) to determine whether the run length was sufficient. The average standard deviation of split frequencies at 2 × 106 generations was 0.0076 for PGRPs and at 107 generations 0.0023 for defensins, respectively. This therefore indicated that the two chains that were run converged on similar results. The 50% majority rule tree presented here was constructed from all sampled trees with the first 25% of all trees. Trees were visualized with TREEVIEW 1.6.6 [61]. Posterior probabilities plotted at the nodes can be interpreted as the probability that the tree or clade is correct [62].
Quantitative real time RT-PCR
Quantitative PCR was performed with the real-time PCR system Mx3000P (Stratagene, La Jolla, California, USA) using the FullVelocity SYBR® Green QRT-PCR Master Mix (Stratagene, La Jolla, California, USA), according to the protocols of the manufacturer. In order to confirm the subtraction efficiency of constructed cDNA library 1 ng of unsubtracted and subtracted cDNA, respectively, was used to amplify α-tubulin, Et-AMP10, and eristalin. Used primers were: the universal primers α-tubulin-forward: 5'-GCCAACCAGATGGTCAA-3' and α-tubulin-reverse: 5'-GCTTGGTCTTGATGGTG-3', eristalin-forward: 5-ATGGCTACATGTGATCTGCT-3', eristalin-reverse: 5'-ACGGCAATTGCAGACT-3', Et-AMP10-forward: 5'-ATGGACCCTCTTCTGTGG-3', Et-AMP10-reverse: 5'-TGGGCATCTGACAATA-3'. For gene expression analyses, we used 100 ng total RNA per well and primers described above. In addition, insect universal primers actin-forward: 5'-ATCCTCACCCTGAAGTACCC-3 and actin-reverse: 5-CCACACGCAGCTCATTGTA-3' were used to amplify Eristalis actin. Primers were selected using the primer3 software [63] and were purchased from Thermo electron (Waltham, MA, USA).
Competing interests
The author(s) declare that they have no competing interests.
Authors' contributions
BA designed and carried out the experiments, performed the analyses, and drafted parts of the manuscript. AV collected and immune challenged the animals, participated in the experimental design and coordination, and drafted parts of the manuscript. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
We thank Meike Fischer for excellent technical assistance and Katja Altincicek, Christopher West Wheat, and Rod Snowdon for critical reading of the manuscript. The authors acknowledge financial support from the Justus-Liebig-University of Giessen to BA and from the Deutsche Forschungsgemeinschaft to AV.
Contributor Information
Boran Altincicek, Email: Boran.Altincicek@agrar.uni-giessen.de.
Andreas Vilcinskas, Email: Andreas.Vilcinskas@agrar.uni-giessen.de.
References
- Grimaldi D, Engel M. Evolution of insects. Cambridge University Press, New York; 2005. [Google Scholar]
- Stahls G, Hippa H, Rotheray G, Muona J, Gilbert F. Phylogeny of Syrphidae (Diptera) inferred from combined analysis of molecular and morphological characters. Syst Entomol. 2003;28:433–450. doi: 10.1046/j.1365-3113.2003.00225.x. [DOI] [Google Scholar]
- Chapman D. Water Quality Assessments – A Guide to Use of Biota, Sediments and Water in Environmental Monitoring. Second. University Press, Cambridge; 1996. [Google Scholar]
- Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. doi: 10.1038/415389a. [DOI] [PubMed] [Google Scholar]
- Brahmachary M, Krishnan SP, Koh JL, Khan AM, Seah SH, Tan TW, Brusic V, Bajic VB. ANTIMIC: a database of antimicrobial sequences. Nucleic Acids Res. 2004:D586–589. doi: 10.1093/nar/gkh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre B, Hoffmann JA. The host defense of Drosophila melanogaster. Annu Rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- Cherry S, Silverman N. Host-pathogen interactions in drosophila: new tricks from an old friend. Nat Immunol. 2006;7:911–917. doi: 10.1038/ni1388. [DOI] [PubMed] [Google Scholar]
- Waterhouse RM, Kriventseva EV, Meister S, Xi Z, Alvarez KS, Bartholomay LC, Barillas-Mury C, Bian G, et al. Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science. 2007;316:1738–1743. doi: 10.1126/science.1139862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altincicek B, Vilcinskas A. Identification of immune-related genes from an apterygote insect, the firebrat Thermobia domestica. Insect Biochem Mol Biol. 2007;37:726–731. doi: 10.1016/j.ibmb.2007.03.012. [DOI] [PubMed] [Google Scholar]
- Seitz V, Clermont A, Wedde M, Hummel M, Vilcinskas A, Schlatterer K, Podsiadlowski L. Identification of immunorelevant genes from greater wax moth (Galleria mellonella) by a subtractive hybridization approach. Dev Comp Immunol. 2003;27:207–215. doi: 10.1016/S0145-305X(02)00097-6. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Johnson TJ, Myers AA, Kanost MR. Identification by subtractive suppression hybridization of bacteria-induced genes expressed in Manduca sexta fat body. Insect Biochem Mol Biol. 2003;33:541–559. doi: 10.1016/S0965-1748(03)00028-6. [DOI] [PubMed] [Google Scholar]
- Altincicek B, Vilcinskas A. Analysis of the immune-related transcriptome of a lophotrochozoan model, the marine annelid Platynereis dumerilii. Front Zool. 2007;4:18. doi: 10.1186/1742-9994-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Liu H, Han Y, Fan L, Zhang Q, Liu J, Yu X, Zhang L, Chen S, Dong M, Wang L, Xu A. Profile of acute immune response in Chinese amphioxus upon Staphylococcus aureus and Vibrio parahaemolyticus infection. Dev Comp Immunol. 2007;31:1013–1023. doi: 10.1016/j.dci.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Dziarski R. Peptidoglycan recognition proteins (PGRPs) Mol Immunol. 2004;40:877–886. doi: 10.1016/j.molimm.2003.10.011. [DOI] [PubMed] [Google Scholar]
- Lu XF, Wang MH, Qi J, Wang HT, Li XN, Gupta D, Dziarski R. Peptidoglycan recognition proteins are a new class of human bactericidal proteins. J Biol Chem. 2006;281:5895–5907. doi: 10.1074/jbc.M511631200. [DOI] [PubMed] [Google Scholar]
- Silverman N, Zhou R, Stoven S, Pandey N, Hultmark D, Maniatis T. A Drosophila I kappa B kinase complex required for Relish cleavage and antibacterial immunity. Genes Dev. 2000;14:2461–2471. doi: 10.1101/gad.817800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XW, Tan NS, Ho B, Ding JL. Evidence for the ancient origin of the NF-kappa B/I kappa B cascade: Its archaic role in pathogen infection and immunity. Proc Natl Acad Sci USA. 2006;103:4204–4209. doi: 10.1073/pnas.0507044103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theopold U, Schmidt O, Soderhall K, Dushay MS. Coagulation in arthropods: defence, wound closure and healing. Trends in Immunol. 2004;25:289–294. doi: 10.1016/j.it.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Kanost MR. Serine proteinase inhibitors in arthropod immunity. Dev Comp Immunol. 1999;23:291–301. doi: 10.1016/S0145-305X(99)00012-9. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Yeager M. Molecular evolution of the vertebrate immune system. Bioessays. 1997;19:777–786. doi: 10.1002/bies.950190907. [DOI] [PubMed] [Google Scholar]
- Pike RN, Buckle AM, le Bonniec BF, Church FC. Control of the coagulation system by serpins – Getting by with a little help from glycosaminoglycans. FEBS Journal. 2005;272:4842–4851. doi: 10.1111/j.1742-4658.2005.04880.x. [DOI] [PubMed] [Google Scholar]
- Laskowski M, Kato I. Protein inhibitors of proteinases. Annu Rev Biochem. 1980;49:593–626. doi: 10.1146/annurev.bi.49.070180.003113. [DOI] [PubMed] [Google Scholar]
- Cherry S, Perrimon N. Entry is a rate-limiting step forviral infection in a Drosophila melanogaster model of pathogenesis. Nature Immunol. 2004;5:81–87. doi: 10.1038/ni1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd CG, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- Lefebvre C, Cocquerelle C, Vandenbulcke F, Hott D, Huot L, Lemoine Y, Salzet M. Transcriptomic analysis in the leech Theromyzion tessulatum: involvement of cystatin B in innate immunity. Biochem J. 2004;380:617–625. doi: 10.1042/BJ20040478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putsep K, Branden CI, Boman HG, Normark S. Antibacterialpeptide from H-pylori. Nature. 1999;398:671–672. doi: 10.1038/19439. [DOI] [PubMed] [Google Scholar]
- Park SG, Kim HJ, Min YH, Choi EC, Shin YK, Park BJ, Lee SW, Kim S. Human lysyl-tRNA synthetase is secreted to trigger proinflammatory response. Proc Natl Acad Sci USA. 2005;102:6356–6361. doi: 10.1073/pnas.0500226102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veal E, Eisenstein M, Tseng ZH, Gill G. A cellular repressor of E1A-stimulated genes that inhibits activation by E2F. Mol Cell Biol. 1998;18:5032–5041. doi: 10.1128/mcb.18.9.5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacher M, Di Bacco A, Lunin VV, Ye Z, Wagner J, Gill G, Cygler M. The crystal structure of CREG, a secreted glycoprotein involved in cellular growth and differentiation. Proc Natl Acad Sci USA. 2005;102:18326–18331. doi: 10.1073/pnas.0505071102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arca B, Lombardo F, Valenzuela JG, Francischetti IMB, Marinotti O, Coluzzi M, Ribeiro JAC. An updated catalogue of salivary gland transcripts in the adult female mosquito, Anopheles gambiae. J Exp Biol. 2005;208:3971–3986. doi: 10.1242/jeb.01849. [DOI] [PubMed] [Google Scholar]
- Sullivan KF. Structure and utilization of tubulin isotypes. Annu Rev Cell Biol. 1988;4:687–716. doi: 10.1146/annurev.cb.04.110188.003351. [DOI] [PubMed] [Google Scholar]
- Chee HY, AbuBakar S. Identification of a 48 kDa tubulin or tubulin-like C6/36 mosquito cells protein that binds dengue virus 2 using mass spectrometry. Biochem Biophys Res Commun. 2004;320:11–17. doi: 10.1016/j.bbrc.2004.05.124. [DOI] [PubMed] [Google Scholar]
- Cornet B, Bonmatin JM, Hetru C, Hoffmann JA, Ptak M, Vovelle F. Refined three-dimensional solution structure of insect defensin A. Structure. 1995;3:435–448. doi: 10.1016/S0969-2126(01)00177-0. [DOI] [PubMed] [Google Scholar]
- Hughes AL. Evolutionary diversification of the mammalian defensins. Cell Mol Life Sci. 1999;56:94–103. doi: 10.1007/s000180050010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boman HG. Antibacterial peptides: basic facts and emerging concepts. J of Intern Med. 2003;254:197–215. doi: 10.1046/j.1365-2796.2003.01228.x. [DOI] [PubMed] [Google Scholar]
- Mygind PH, Fischer RL, Schnorr KM, Hansen MT, Sonksen CP, Ludvigsen S, Raventos D, Buskov S, Christensen B, De Maria L, et al. Plectasin is a peptide antibiotic with therapeutic potential from a saprophytic fungus. Nature. 2005;437:975–980. doi: 10.1038/nature04051. [DOI] [PubMed] [Google Scholar]
- Benincasa M, Scocchi M, Podda E, Skerlavaj B, Dolzani L, Gennaro R. Antimicrobial activity of Bac7 fragments against drug-resistant clinical isolates. Peptides. 2004;25:2055–2061. doi: 10.1016/j.peptides.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Frank RW, Gennaro R, Schneider K, Przybylski M, Romeo D. Amino acid sequences of two proline-rich bactenecins. Antimicrobial peptides of bovine neutrophils. J Biol Chem. 1990;265:18871–18874. [PubMed] [Google Scholar]
- Cabiaux V, Agerberth B, Johansson J, Homble F, Goormaghtigh E, Ruysschaert JM. Secondary structure and membrane interaction of PR-39, a Pro+Arg-rich antibacterial peptide. Eur J Biochem. 1994;224:1019–1027. doi: 10.1111/j.1432-1033.1994.01019.x. [DOI] [PubMed] [Google Scholar]
- Bulet P, Stocklin R. Insect antimicrobial peptides: Structures, properties and gene regulation. Protein Pept Lett. 2005;12:3–11. doi: 10.2174/0929866053406011. [DOI] [PubMed] [Google Scholar]
- Skerlavaj B, Romeo D, Gennaro R. Rapid membrane permeabilization and inhibition of vital functions of gram-negative bacteria by bactenecins. Infect Immun. 1990;58:3724–3730. doi: 10.1128/iai.58.11.3724-3730.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabata S, Nagayama R, Hirata M, Shigenaga T, Agarwala KL, Saito T, Cho JK, Nakajima H, Takagi T, Iwanaga S. Tachycitin, a small granular component in horseshoe crab hemocytes, is an antimicrobial protein with chitin-binding activity. J Biochem. 1996;120:1253–1260. doi: 10.1093/oxfordjournals.jbchem.a021549. [DOI] [PubMed] [Google Scholar]
- Valenzuela JG, Francischetti IMB, Pham VM, Garfield MK, Ribeiro JMC. Exploring the salivary gland transcriptome and proteome of the Anopheles stephensi mosquito. Insect Biochem Mol Biol. 2003;33:717–732. doi: 10.1016/S0965-1748(03)00067-5. [DOI] [PubMed] [Google Scholar]
- Skerlavaj B, Gennaro R, Bagella L, Merluzzi L, Risso A, Zanetti M. Biological characterization of two novel cathelicidin-derived peptides and identification of structural requirements for their antimicrobial and cell lytic activities. J Biol Chem. 1996;271:28375–28381. doi: 10.1074/jbc.271.45.28375. [DOI] [PubMed] [Google Scholar]
- Lee SY, Moon HJ, Kurata S, Natori S, Lee BL. Purification and cDNA cloning of an antifungal protein from the hemolymph of Holotrichia diomphalia larvae. Biol Pharm Bull. 1995;18:1049–1052. doi: 10.1248/bpb.18.1049. [DOI] [PubMed] [Google Scholar]
- Lorenzini DM, da Silva PI, Fogaca AC, Bulet P, Daffre S. Acanthoscurrin: a novel glycine-rich antimicrobial peptide constitutively expressed in the hemocytes of the spider Acanthoscurria gomesiana. Dev Comp Immunol. 2003;27:781–791. doi: 10.1016/S0145-305X(03)00058-2. [DOI] [PubMed] [Google Scholar]
- Herbiniere J, Braquart-Varnier C, Greve P, Strub JM, Frere J, Van Dorsselaer A, Martin G. Armadillidin: a novel glycine-richantibacterial peptide directed against gram-positive bacteria in the woodlouse Armadillidium vulgare (Terrestrial Isopod, Crustacean) Dev Comp Immunol. 2005;29:489–499. doi: 10.1016/j.dci.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Couillault C, Pujol N, Reboul J, Sabatier L, Guichou JF, Kohara Y, Ewbank JJ. TLR-independent control of innate immunity in Caenorhabditis elegans by the TIR domain adaptor protein TIR-1, an ortholog of human SARM. Nature Immunol. 2004;5:488–494. doi: 10.1038/ni1060. [DOI] [PubMed] [Google Scholar]
- Gandhe AS, Arunkumar KP, John SH, Nagaraju J. Analysis ofbacteria-challenged wild silkmoth, Antheraea mylitta (lepidoptera) transcriptome reveals potential immune genes. BMC Genomics. 2006;7:184. doi: 10.1186/1471-2164-7-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Ueno S, Kamiya K, Kobayashi J, Matsuoka H, Ando K, Chinzei Y. Cloning of mRNA sequences for two antibacterial peptides in a hemipteran insect, Riptortus clavatus. Zool Sci. 1996;13:111–117. doi: 10.2108/zsj.13.111. [DOI] [PubMed] [Google Scholar]
- Altincicek B, Linder M, Linder D, Preissner KT, Vilcinskas A. Microbial metalloproteinases mediate sensing of invading pathogens and activate innate immune responses in the lepidopteran model host Galleria mellonella. Infect Immun. 2007;75:175–183. doi: 10.1128/IAI.01385-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crovella S, Antcheva N, Zelezetsky I, Boniotto M, Pacor S, Falzacappa MVV, Tossi A. Primate beta-defensins – Structure, function and evolution. Current Protein & Peptide Science. 2005;6:7–21. doi: 10.2174/1389203053027593. [DOI] [PubMed] [Google Scholar]
- Langen G, Imani J, Altincicek B, Kieseritzky G, Kogel KH, Vilcinskas A. Transgenic expression of gallerimycin, a novel antifungal insect defensin from the greater wax moth Galleria mellonella, confers resistance to pathogenic fungi in tobacco. Biol Chem. 2006;387:549–557. doi: 10.1515/BC.2006.071. [DOI] [PubMed] [Google Scholar]
- Rotheray G. Colour guide to hoverfly larvae Dipterist Digest No 9. Derek Whiteley, Sheffield, England; 1993. [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. John Wiley and Sons, New York; 1987. [Google Scholar]
- Boman HG, Nilsson-Faye I, Paul K, Rasmusen TJ. Insect immunity I. Characteristics of an inducible cell-free antibacterial reactionin hemolymph of Samia cynthia. Infect Immun. 1974;10:136–145. doi: 10.1128/iai.10.1.136-145.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page RDM. TREEVIEW: An application to display phylogenetic trees on personal computers. Computer Applications in the Biosciences. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck J, Rannala B. Frequentist properties of Bayesian posterior probabilities of phylogenetic trees under simple and complex substitution models. Syst Biol. 2004;53:904–913. doi: 10.1080/10635150490522629. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Humana Press, Totowa, NJ; 2000. Bioinformatics Methods and Protocols: Methods in Molecular Biology. [DOI] [PubMed] [Google Scholar]