Abstract
A combination of genetic variations, epimutations and environmental factors may be involved in the etiology of complex neurodevelopmental disorders like schizophrenia. To study such disorders, we use apomorphine-unsusceptible (APO-UNSUS) Wistar rats and their phenotypic counterpart apomorphine-susceptible (APO-SUS) rats that display a complex phenotype remarkably similar to that of schizophrenic patients. As the molecular basis of the APO-SUS/UNSUS rat model, we recently identified a genomic rearrangement of the Aph-1b gene. Here, we discovered between the two rat lines differences other than the Aph-1b gene defect, including a remarkable cluster of genetic variations, two variants corresponding to topoisomerase II-based recombination hot spots and an epigenetic (DNA methylation) difference in cerebellum and (hypo)thalamic but not hippocampal genomic DNA. Furthermore, genetic variations were found to correlate with the degree of apomorphine susceptibility in unselected Wistar rats. Together, the results show that a number of genetic and epigenetic differences exist between the APO-SUS and -UNSUS rat genomes, raising the possibility that in addition to the Aph-1b gene defect the newly identified variations may also contribute to the complex APO-SUS phenotype.
Keywords: AP-PCR, Genetic and epigenetic variations, Neurodevelopmental disorders, APO-SUS rat model, Aph-1b
Introduction
Schizophrenia is a neurodevelopmental disorder affecting nearly 1% of the world’s population (Jablensky et al. 1987), and is characterized by positive and negative symptoms (Kay and Opler 1987). The aetiology of schizophrenia and other related disorders, such as schizoaffective and bipolar disorder, is still unclear. Twin, family and adoption studies have suggested that complex interactions at the genetic and environmental level underlie the aetiology of schizophrenia (Gottesman 1991). It is thought that gene variations by themselves do not result in schizophrenia, but they can establish a predisposition status that, when combined with environmental stressors, may lead to schizophrenia pathogenesis. Numerous environmental factors, such as viral infections (Mednick et al. 1988), insufficient folate and methionine levels (Regland 2005), or repeated psychological stress (Goldstein 1987), can influence brain development of prenatal or early postnatal individuals with a genetic predisposition for neuropsychiatric disorders. Due to the heterogeneity in genetic and environmental interactions, most of the genes and pathways for schizophrenia and for other complex disorders are still unknown.
To get insight into the gene (or genes) that may be involved in schizophrenia pathogenesis, a rat model was developed with schizophrenia-like features. This model was based on the behavioural response of Wistar rats to the dopamine agonist apomorphine (Cools et al. 1990). The apomorphine-susceptible (APO-SUS) rat line displayed many features of psychopathology, with similar disturbances at the behavioural, physiological, endocrinological and pharmacological level as seen in schizophrenics (Ellenbroek and Cools 2002). For example, APO-SUS rats have a reduced prepulse inhibition and latent inhibition (Ellenbroek et al. 1995), display a higher plasma release of adrenocorticotropin (ACTH) and corticosteroids in response to novelty (Rots et al. 1995), are more sensitive to dopamimetic drugs (Ellenbroek et al. 2000), and have a higher susceptibility to inflammatory and infectious diseases when compared to apomorphine-unsusceptible (APO-UNSUS) rats (Kavelaars et al. 1997). We therefore wondered about the molecular-genetic basis underlying the APO-SUS/-UNSUS rat model and recently identified a genetic difference between the two rat lines (Coolen et al. 2005). Whereas APO-UNSUS rats harbour three gene copies of the γ-secretase component Aph-1b, APO-SUS rats have only one or two copies. This gene-dosage imbalance was due to an unequal crossing over event (nonallelic homologous recombination) between two direct repeats (a segmental duplication) within the Aph-1b locus. In addition, we observed a direct link between the Aph-1b genotypes and a number of phenotypic APO-SUS and -UNSUS characteristics (Coolen et al. 2005). Approximately 10 years after developing the APO-SUS and -UNSUS lines a second, independent breeding procedure was started that resulted in rats with features similar to those displayed by the original APO-SUS and -UNSUS rat lines (Ellenbroek and Cools 2002). Interestingly, the replicated rat lines also resulted in APO-UNSUS rats with three Aph-1b gene copy numbers and APO-SUS rats with only one or two gene copies (Coolen et al. 2005).
In the present study, we wondered whether genetic variations other than the Aph-1b gene-dosage imbalance may be present between the APO-SUS and -UNSUS rats, and whether epigenetic factors may be involved as well. Epigenetics has been defined as heritable changes in gene expression that do not occur by changes in the DNA sequence, but by modifications in DNA methylation and chromatin remodeling (Wolffe and Matzke 1999), or, in its widest sense, as any change in an organism that is not due to genetic factors (Van de Vijver et al. 2002). Increasing evidence suggests that epigenetic modifications play a role in disease susceptibility (reviewed by Jirtle and Skinner 2007). We used the arbitrarily primed-polymerase chain reaction (AP-PCR) fingerprinting technique (Welsh and McClelland 1990) to analyse the genomes and epigenomes (DNA methylation) of the APO-SUS and -UNSUS rats. Comparison of the AP-PCR fingerprints generated from the genomic DNAs of the two rat lines revealed genetic as well as epigenetic alterations and we conclude that, besides in the Aph-1b locus, a number of other variations are present in the APO-SUS and -UNSUS genomes and epigenomes.
Materials and methods
Experimental animals
The generation of the APO-SUS and -UNSUS rat lines with a high or low susceptibility for apomorphine, respectively, has been described previously (Cools et al. 1990). The present experiments were performed with male APO-SUS and -UNSUS rats belonging to the 32nd (original lines) and 18th (replicate lines) generation. At post-natal day 60 (PND60), APO-SUS and -UNSUS rats were sacrificed and the hippocampus, cerebellum and the combined thalamus/hypothalamus (further denoted as (hypo)thalamus) were isolated. To establish their apomorphine susceptibility, unselected male Wistar rats of the Nijmegen outbred population (PND60) were injected with apomorphine (1.5 mg/kg s.c.) and gnawing scores were measured in a gnawing box for 45 min, as described previously (Cools et al. 1990). Immediately following the measurements, the rats were sacrificed and the same tissues (hippocampus, cerebellum and (hypo)thalamus) were removed. All rats were bred and reared in the Central Animal Facility of the Radboud University Nijmegen under approved animal protocols and in accordance with institutional guidelines.
Arbitrarily primed-PCR
Genomic DNAs were isolated from hippocampus, cerebellum and (hypo)thalamus using standard procedures involving the use of proteinase K and phenol extraction. Two micrograms of genomic DNA were digested with 20 units of RsaI, 20 units RsaI in combination with the methylation-sensitive enzyme HpaII, or 20 units RsaI and MspI (MBI Fermentas) in a total volume of 40 μl at 37°C for 16 h. HpaII does not cut DNA if the internal cytosine (CCGG) is methylated, whereas MspI is insensitive to DNA methylation. Using such combinations of methylation-sensitive and -insensitive enzymes allows genome-wide screening for differences at the genetic level (single-nucleotide polymorphisms––SNPs, duplications, insertions, deletions and recombinations) as well as the epigenetic (DNA methylation) level. Restriction enzymes were heat inactivated by incubating the reactions at 65°C for 20 min. Digested DNA (100 ng) was amplified using AP-PCR (Welsh and McClelland 1990) with a single primer. PCRs were performed in a total volume of 25 μl containing 10 mM Tris-HCl (pH 8.3), 2.5 mM MgCl2, 50 mM KCl, 0.001% gelatin, 0.25% Nonidet P-40, 0.25% Tween-20, 200 μM each of the four deoxynucleotide triphosphates, ∼1 μCi of [α-32P]dCTP (3000 Ci/mmol, Amersham Corp.), 25 pmol of primer (AP-1: 5′-AACCCTCACCCTAACCCCGG-3′, AP-7: 5′-AACCCTCACCCTAAGGCGCG-3′, AP-777: 5′-CACTCCTCTACAAGGTGCCG-3′ or Topo: 5′-GCCTCCTTGCAGGTCTTT-3′), and 0.8 units of Taq polymerase (MBI Fermentas). Reactions were carried out in a thermal cycler (Perkin-Elmer) with five cycles of low stringency (94°C for 30 s, 40°C for 60 s, 72°C for 1.5 min), followed by 30 cycles of high stringency (94°C for 15 s, 55°C for 15 s, 72°C for 1 min). Two microliters of the PCR products were analysed on high-resolution 5% polyacrylamide gels under denaturing conditions (7 M urea) for 4–4.5 h at 70 W. Gels were dried and radiolabelled DNA was visualized by autoradiography at −70°C (CEA AB, Sweden).
Cloning and sequencing of AP-PCR fragments
AP-PCR fragments generated from APO-SUS and -UNSUS rat genomic DNAs were excised from the dried gels and incubated in 50 μl MilliQ at 80°C for 10 min. The eluted DNA (two microliters) was reamplified with the same primer as used for the AP-PCR to generate sufficient amounts of template for subsequent cloning. The reactions were carried out for 40 cycles of 94°C for 1 min, 55°C for 30 s, 72°C for 1 min, under the same conditions as described in the AP-PCR protocol (except that [α-32P]dCTP was not included). The PCR products were purified, cloned into the pGEM-T easy vector (Promega) and sequenced with a T7 or Sp6 primer according to the manufacturer’s instructions using the ABI310 machine (Applied Biosystems).
Sequencing and genotyping of chromosomal region 9q22
A 1948-bp fragment that harbours the nucleotide sequence corresponding to product 3 was obtained by PCR on genomic DNA derived from (hypo)thalamic tissue of an APO-UNSUS rat using forward primer 5′-GGGAAGCAACGCATCCTG-3′ and reverse primer 5′-CATATCAAAGCACCAAGTCCACAG-3′. The DNA was subsequently purified and directly sequenced using the ABI310 machine (Applied Biosystems). Genotyping of chromosomal region 9q22 was performed with PCR using primers specific for either the APO-SUS or APO-UNSUS genomic sequence. Briefly, PCRs were performed in a total volume of 20 μl containing 50 ng genomic DNA, 10 mM Tris-HCl (pH 8.3), 2.5 mM MgCl2, 50 mM KCl, 0.001% gelatin, 0.25% Nonidet P-40, 0.25% Tween-20, 200 μM each of the four deoxynucleotide triphosphates, 0.6 μM of each primer (FW: 5′-AACACTTGGACTCATTCTCACTGG-[G (SUS) or T (UNSUS)]-3′ and RV: 5′-CCTGGATGGAATGTTGACAC-[C (SUS) or T (UNSUS)]-3′), and 0.8 units of Taq polymerase (MBI Fermentas). Reactions were carried out at 94°C for 60 s, 58°C for 60 s and 72°C for 60 s for 35 cycles. Products were analysed on a 1% agarose gel.
Quantification and statistics
Quantification of AP-PCR products was performed using the Labworks 4.0 program (UVP BioImaging Systems, Cambridge, UK) and statistical evaluation was performed by means of an unpaired Student’s t-test.
Results
AP-PCR DNA fingerprint patterns of the APO-SUS and APO-UNSUS rat genomes and epigenomes
In order to identify differences between the genomes and epigenomes of APO-SUS and -UNSUS rats, we performed a comparative analysis of fingerprints of AP-PCR products generated from genomic DNAs of the two rat lines. Initially, genomic DNAs isolated from APO-SUS and -UNSUS (hypo)thalamic tissue and digested with RsaI in combination with the methylation-sensitive restriction enzyme HpaII (CCGG) was analysed using arbitrary primers AP-1, AP-7 or AP-777. These primers were selected from a total set of ten primers because they gave fingerprints with reproducible and discrete products (data not shown). Typical AP-PCR fingerprints obtained with the three selected arbitrary primers are shown in Fig. 1. With each arbitrary primer ∼30 chromosomal fragments were reproducibly amplified. DNAs digested with RsaI and the methylation-insensitive enzyme MspI served as controls to determine whether the observed differences were due to a differential methylation of the CCGG sequence or a genetic polymorphism in this sequence. AP-PCR analysis with primer AP-1 revealed 16 products corresponding to fragments without an HpaII site (“genetic fragments”) and 23 products corresponding to fragments containing an HpaII site (“epigenetic fragments”). Analysis with AP-PCR primer AP-7 showed 17 genetic and 11 epigenetic fragments, and with AP-777 primer 22 genetic and 6 epigenetic fragments.
Fig. 1.
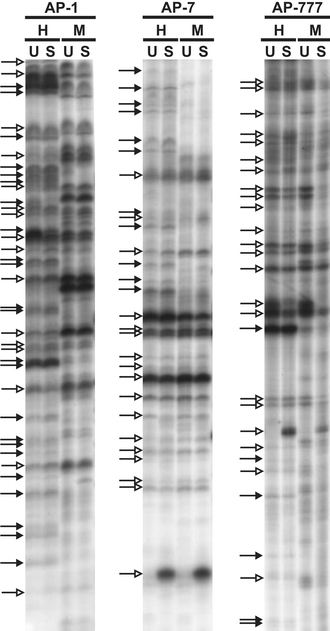
AP-PCR analysis of genomic DNAs from APO-UNSUS (U) and APO-SUS (S) (hypo)thalamus. AP-PCR was performed with primers AP-1, AP-7 and AP-777 using genomic DNAs digested with RsaI and HpaII (H) or RsaI and MspI (M) as templates. Epigenetic products (methylation-sensitive and thus absent in the MspI lanes) are indicated by closed arrows and genetic products (methylation-insensitive and thus present in the MspI lanes) by open arrows
Genetic variations between the APO-SUS and APO-UNSUS rat genomes
Comparison of the genetic fingerprints generated with primers AP-1, -7 and -777 revealed three reproducible variations between the genomic DNAs from the original (F32) APO-SUS and -UNSUS rats, designated products 1, 2 and 3 (Fig. 2a). Product 1 was less prominent in the APO-SUS than in the APO-UNSUS rats, product 2 was found only in APO-SUS, while the level of product 3 was higher in the APO-SUS than -UNSUS rats. Interestingly, the three products were also present in the replicate (F18) lines and at the same levels, indicating that the replication of the APO-SUS and -UNSUS lines had resulted in a similar genotypic distribution. Next, digestions using RsaI in combination with MspI were used to examine whether the observed differences were due to a genetic or an epigenetic alteration. Following digestion with RsaI and MspI, products 1, 2 and 3 were still found, indicating that the presence of the three AP-PCR products was due to genetic differences (Fig. 2b).
Fig. 2.

Genetic variations in genomic DNAs from APO-SUS (S) and APO-UNSUS (U) (hypo)thalamus. Rats were from the original (F32) or the replicate (F18) rat lines. (A) Products generated by AP-PCR using primers AP-1 (product 1), AP-7 (product 2) or AP-777 (product 3) on genomic DNAs digested with the methylation-insensitive enzyme RsaI and the methylation-sensitive enzyme HpaII. ND = not determined. (B) Products 1, 2 and 3 generated by AP-PCR on genomic DNAs digested with RsaI and HpaII (H) or with the two methylation-insensitive enzymes RsaI and MspI (M). MspI products served as controls to determine whether products 1, 2 and 3 were due to differential methylation or to a genetic polymorphism. The fact that products 1, 2 and 3 were still present following MspI digestion indicates that they represent products without an HpaII site (“genetic fragments”). (C) Products generated by AP-PCR using a topoisomerase II binding site consensus sequence (5′-GCCTCCTTGCAGGTCTTT-3′) on genomic DNAs digested with the methylation-insensitive enzymes EcoRI (product 4) or MboI (product 5). Arrows indicate increased amounts of the AP-PCR products
We previously discovered that the Aph-1b gene-dosage imbalance between the APO-SUS and -UNSUS rats is the result of a DNA recombination event between the two Aph-1b genes. Furthermore, we identified the region in which the recombination occurred, namely in a region of 1106 nucleotides that is identical between the two genes and encompasses exon 5 (Coolen et al. 2005). In the present study, we decided to examine in detail the site of recombination and found a topoisomerase II binding site (5′-ACCCACCTGCTGGTGTCC-3′) in the DNA region harbouring the recombination site. Topoisomerase II binding sites (with the vertebrate consensus sequence 5′-RNYNNCNNGYNGKTNYNY-3′) (Spitzner and Muller 1988) are known to be hotspots where DNA recombination events occur easily (Craig and Nash 1983). We therefore wondered whether other topoisomerase II binding sites could have led to additional differences between the APO-SUS and -UNSUS rat genomes. Interestingly, using a primer based on the topoisomerase II binding site consensus for PCR analysis of genomic DNAs digested with EcoRI or MboI revealed two differences between the APO-SUS (n = 3) and -UNSUS (n = 2) rat genomes of the original (F32) lines, designated products 4 and 5 (Fig. 2c). Product 4 was present in APO-SUS but not in APO-UNSUS rat genomic DNA. In the replicate APO-SUS and -UNSUS lines (F18), the genomes of two APO-UNSUS rats did also not contain product 4, whereas it was present in two of the four APO-SUS rats tested. Product 5 was present in three of the four APO-UNSUS rats examined (in both the original and the replicate lines), whereas it was not observed in the seven APO-SUS rats tested (Fig. 2c).
Epigenetic variations between the APO-SUS and APO-UNSUS rat genomes
We then wondered whether, besides the five genetic variations, also epigenetic variations would be present between the APO-SUS and -UNSUS rat lines. AP-PCR analysis using primer AP-1 on RsaI- and HpaII-digested genomic DNAs from APO-SUS and -UNSUS (hypo)thalamus revealed one epigenetic variation, designated the E1-product (Fig. 3a). The difference was observed in both the original APO-SUS and -UNSUS rats as well as the replicated lines. An ∼1.4-fold reduced amount of the E1-product was observed in APO-SUS when compared with APO-UNSUS genomic DNAs (n = 12, P < 0.05). Since DNA methylation may be tissue specific, we decided to examine the E1-product in two other brain tissues. The level of the E1-product was ∼2-fold reduced in genomic DNAs from the cerebellum of APO-SUS compared to APO-UNSUS rats (n = 4, P < 0.01), while no difference in the E1-levels was found in the hippocampus of the APO-SUS and -UNSUS rats (n = 4).
Fig. 3.

Epigenetic variation in genomic DNAs from APO-SUS (S) and APO-UNSUS (U) (hypo)thalamus. Rats were from the original (F32) or the replicate (F18) rat lines. (A) Product E1 generated by AP-PCR using primer AP-1 on genomic DNAs digested with RsaI and HpaII. Arrow indicates a representative example of an AP-PCR product that did not show variable amounts using primer AP-1 (randomly chosen out of 38 products) and was used for normalization. (B) The AP-PCR E1-product generated from genomic DNAs from cerebellum, hippocampus and (hypo)thalamus. All rats were from different nests. The products indicated by the arrow were used for normalisation. Amounts for the epigenetic E1-product were significantly different between the APO-SUS and APO-UNSUS rats in the cerebellum (**P < 0.01; n = 4, plus s.e.m.) and (hypo)thalamus (*P < 0.05; n = 12, plus s.e.m.), but not in the hippocampus (n = 4, plus s.e.m.)
Chromosomal localizations of the genetic and epigenetic variations between the APO-SUS and -UNSUS rat genomes
To identify the locations of the genetic and epigenetic variations within the APO-SUS and -UNSUS rat genomes, AP-PCR fragments 1, 2, 3 and E1 were excised from the gel and each fragment was reamplified with the primer used for the original AP-PCR reaction. DNA sequence analysis of the amplified PCR fragments and database searches with the obtained nucleotide sequences revealed the chromosomal localisations of the four fragments; product 1 was located on chromosome 19q11, ∼3.3 kb upstream of the first exon of the GAIP-interacting protein, C terminus (GIPC) gene; product 2 was part of a repeat sequence located on chromosome 2q34; product 3 was located in the first intron of the myosin 1b gene on chromosome 9q22; the epigenetic variation E1 was located on chromosome 6q31, downstream of the Jun dimerization protein 2 (NP_446346.1) and upstream of the ATF-like basic leucine zipper transcriptional factor B-ATF (SF-HT-activated gene 2; XP_216745.2). Since product 3 was localized within a gene, we decided to analyse this chromosomal region in more detail. Sequence analysis of the DNA region corresponding to AP-PCR fragment 3 and its surrounding region revealed a remarkably high number of genetic variations: 10 of the 1948 base pairs analysed were different between the APO-SUS and -UNSUS genomes (Fig. 4). Comparison of the nucleotide sequences of the APO-SUS and -UNSUS DNA regions with the corresponding database sequence (geneID: 117057) revealed a 100% identity between the database and the APO-UNSUS sequences, indicating that the APO-SUS genome has diverged from the database sequence. Next, more APO-SUS and -UNSUS rats were genotyped for this region. All APO-UNSUS rats tested (n = 5) indeed showed the database sequence, whereas the APO-SUS rats tested (n = 5) all contained the relatively high number of variations in this chromosomal region.
Fig. 4.

Nucleotide sequence of chromosomal region 9q22, corresponding to product 3, in APO-SUS and APO-UNSUS rats. APO-SUS/-UNSUS genomic variations are indicated between brackets; the first nucleotide represents the APO-UNSUS sequence, the second nucleotide the APO-SUS sequence. The nucleotide sequences corresponding to the annealing sites of arbitrary primer AP-777 are underlined
The newly identified genetic and epigenetic variations, and apomorphine susceptibility in Wistar rats
We wondered whether the molecular-genetic difference between the APO-SUS and -UNSUS rats (the Aph-1b gene-dosage imbalance) would also be present in the Nijmegen outbred population of Wistar rats, or if the imbalance was generated during the breeding of the rat lines. PCR analysis of the Aph-1b locus in the 50 Wistar rats examined revealed in all cases the presence of three copies of the Aph-1b gene, suggesting that the reduction of Aph-1b copies in the APO-SUS rats had been induced during the breeding of the rats.
We next wondered whether the newly identified genetic and epigenetic variations in the APO-SUS and -UNSUS genomes were also induced during the breeding of the two rat lines, or if these variants are already present in Wistar rats. For this purpose, we performed AP-PCR analysis of (hypo)thalamic genomic DNAs from Wistar rats using primers AP-1, -7 and -777. The fingerprints revealed the presence of the APO-SUS as well as the -UNSUS variants of the genetic products 1, 2 and 3 (n = 8) and the epigenetic E1-product (n = 4) in Wistar rats (Fig. 5), indicating that the newly identified variations were not induced during breeding of the APO-SUS and -UNSUS lines, but were already present in the original Wistar population.
Fig. 5.
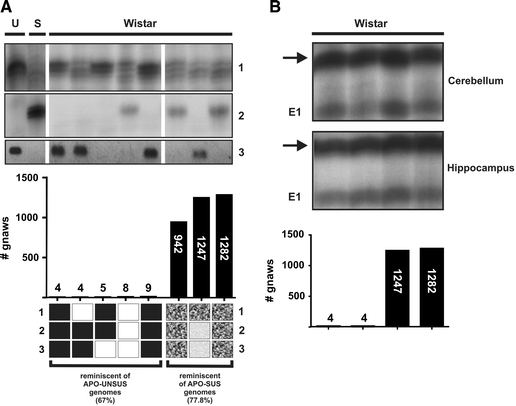
AP-PCR analysis (products 1, 2 and E1) and specific PCR (product 3) analysis of genomic DNAs from Wistar rats with low or high apomorphine susceptibility. (A) The presence or absence of products 1, 2 and 3 (genetic differences) was analysed in genomic DNAs from the (hypo)thalamus of Wistar rats with low (<10 gnaws in 45 min) or high (>500 gnaws in 45 min) apomorphine susceptibility. U: APO-UNSUS, S: APO-SUS. Lower panel: the amounts of the three products present in the Wistar rats were compared with the amounts found in the APO-SUS and -UNSUS rats. (■) the genotype of the genetic products 1, 2 or 3 in the Wistar rats with low apomorphine susceptibility is similar to the products 1, 2 and 3 genotype of the APO-UNSUS rats; (□) the genotype in the Wistar rats with low apomorphine susceptibility deviates from the APO-UNSUS genotype;
( ) the genotype in the Wistar rats with high apomorphine susceptibility is comparable with the genotype in the APO-SUS rat;
(
) the genotype in the Wistar rats with high apomorphine susceptibility is comparable with the genotype in the APO-SUS rat;
( ) the genotype in the Wistar rats with high apomorphine susceptibility deviates from the genotype in the APO-SUS rats. (B) AP-PCR analysis of the epigenetic E1-product (E1) on genomic DNAs from cerebellum and hippocampus of four Wistar rats. The product indicated by the arrow is an example of a product that did not show variable amounts and was used for normalisation of the E1-product. The amounts of the E1-products generated from the cerebellum or hippocampus were not different between the two Wistar rats with low gnawing scores (<10 gnaws in 45 min) and the two Wistar rats with high gnawing scores (>500 gnaws in 45 min)
) the genotype in the Wistar rats with high apomorphine susceptibility deviates from the genotype in the APO-SUS rats. (B) AP-PCR analysis of the epigenetic E1-product (E1) on genomic DNAs from cerebellum and hippocampus of four Wistar rats. The product indicated by the arrow is an example of a product that did not show variable amounts and was used for normalisation of the E1-product. The amounts of the E1-products generated from the cerebellum or hippocampus were not different between the two Wistar rats with low gnawing scores (<10 gnaws in 45 min) and the two Wistar rats with high gnawing scores (>500 gnaws in 45 min)
To study whether the presence or absence of the genetic AP-PCR products observed in the APO-SUS and -UNSUS rats (products 1, 2 and 3) was linked to the apomorphine susceptibility of Wistar rats, we examined a group of five rats with low susceptibility to apomorphine (<10 gnaws per 45 min) and a highly susceptible group of three rats (>500 gnaws per 45 min). Product 1, which was less abundant in APO-SUS than -UNSUS rats, was not present in the three Wistar rats with high gnawing scores, but was also not found in two of the five Wistar rats with low gnawing scores. Of the eight rats examined, the APO-SUS-specific product 2 was found in two Wistar rats with a high and only one rat with a low degree of apomorphine susceptibility. The DNA region corresponding to product 3 was found twice as the APO-UNSUS sequence in the group consisting of the low-apomorphine-susceptible Wistar rats and once in the group of the high-apomorphine-susceptible rats (Fig. 5a). These results indicate that none of the three products was directly linked to the apomorphine susceptibility of the Wistar rats. However, apomorphine susceptibility may not be the result of only a single genetic variation, but rather of multiple genetic and epigenetic alterations. Interestingly, 67% of the three genetic products present in the Wistar rats with low apomorphine susceptibility were also found in the APO-UNSUS genome, and 78% of the three products in Wistar rats with high apomorphine susceptibility were APO-SUS-specific variants (Fig. 5a), indicating that a combination of the three genetic products may well be linked to apomorphine susceptibility. To study the link between the epigenetic E1-product and apomorphine susceptibility, two Wistar rats with low gnawing scores and two with high gnawing scores were examined for the amount of the E1-product in the cerebellum. Similar amounts of the E1-product as detected in APO-UNSUS rats were found in one Wistar rat with a low and in one Wistar rat with a high gnawing score. The amount of the E1-product detected in APO-SUS rats was found in one Wistar rat with low and in one Wistar rat with high apomorphine susceptibility, indicating that no direct link exists between the E1-product and apomorphine susceptibility. As observed in the APO-SUS and -UNSUS rats, no variation was detected in the level of the E1-product in hippocampal genomic DNAs from the four Wistar rats tested (Fig. 5b).
Discussion
In this study, we investigated the genetic and epigenetic background of the phenotypically well-characterized APO-SUS and -UNSUS rats. Unravelling the molecular basis of this rat model may help in our understanding of complex human neurodevelopmental disorders, since many of the characteristics of the APO-SUS rat line are also observed in schizophrenic patients (Ellenbroek and Cools 2002). We recently identified a gene-dosage imbalance in the Aph-1b locus of the APO-SUS and -UNSUS rats, leading to a reduced expression of the Aph-1b gene in APO-SUS rats and a segregation with a number of behavioural parameters (Coolen et al. 2005). Here, we used AP-PCR analysis as an approach to explore the presence of any other alterations in the two genomes and epigenomes. We indeed identified additional genetic and epigenetic variations, indicating that the Aph-1b locus might not be solely responsible for the observed phenotypes of the two rat lines and suggesting a multi-genetic and -epigenetic origin of the differences observed between the APO-SUS and -UNSUS rats. Importantly, we found similar AP-PCR patterns in both the original and replicate APO-SUS and -UNSUS lines. It is therefore highly unlikely that the observed variations between the APO-SUS and -UNSUS rat genomes were simply due to coincidence.
Detailed analysis of one of the newly identified genetic alterations (in the DNA region corresponding to product 3) revealed in APO-SUS rats a cluster of variations in the myosin 1b gene. Hence, this cluster appears to be a hotspot for genetic instability. Product 1 was located upstream of the GIPC gene encoding a protein interacting with membrane-associated and transmembrane proteins, including the dopamine receptors D2 and D3 (Jeanneteau et al. 2004). The genetic variation in this locus may therefore contribute to the differences in apomorphine susceptibility between the APO-SUS and -UNSUS rat lines. We also identified genetic alterations using a primer corresponding to a topoisomerase II binding site consensus. The rationale for this study was based on our present finding that a topoisomerase II binding site was located at the recombination site in the Aph-1b locus. The two newly identified, topoisomerase II-based genetic variations might point to a more general role for topoisomerase II binding sites in psychopathological mechanisms. We hypothesize that at these sites environmental factors, such as stress during early development, may cause an increase in the incidence of recombination and other mutagenic events, leading to brain dysfunction and affected behavior. Besides the genetic differences, one tissue-specific epigenetic variation was found between the APO-SUS and -UNSUS epigenomes. At present it is not clear what, if any, functional consequence should be attributed to the decrease in the methylation status of this CpG in the (hypo)thalamus and cerebellum, but not in the hippocampus, of APO-SUS relative to APO-UNSUS rats.
Earlier microarray analysis of mRNA expression in the hippocampus of the APO-SUS and -UNSUS rats (∼7000 full-length sequences and ∼1000 EST clusters) revealed that only Aph-1b was differentially expressed (Coolen et al. 2005). The newly identified variations open the possibility that for an explanation of the background of the rat model one has to consider more genes that operate together in a multi-genetic and -epigenetic setting with several susceptibility loci. Thus, based on our present findings, more than one locus may be responsible for the complex phenotype of APO-SUS rats. Recently, it has been established that the contribution of genetic modifiers is also of importance for the outcome of a phenotype, since they can modulate the severity of the affected phenotype and the phenotypic characteristics without having a clear effect on the normal situation (Nadeau 2001; Nadeau and Topol 2006). Hence, as part of the genetic and epigenetic background of the APO-SUS and -UNSUS rat lines, our newly identified variants can be genetic and epigenetic modifiers influencing the phenotypic expression of the model. Insight into the genetic and epigenetic background may provide diagnostic tools, and clues for mechanisms and pathways to explain complex disorders.
Besides the multiple variations in the APO-SUS and -UNSUS rat genomes and epigenomes, Aph-1b will presumably be a major player in the development of the complex APO-SUS phenotype, since it has a broad cellular effect via tissue-specific cleavage of many different substrates (Coolen et al. 2006b) and is functional already during early development (Coolen et al. 2006a). Remarkably, however, the number of Aph-1b gene copies was not linked to apomorphine susceptibility in Wistar rats. Whereas their apomorphine susceptibility varied, all unselected Wistar rats tested harboured three copies of the Aph-1b gene. Therefore, genetic and epigenetic factors other than Aph-1b will likely contribute to the susceptibility for apomorphine. We now indeed found a correlation between the newly identified genetic variations and the apomorphine susceptibility in the Wistar population, confirming our hypothesis that apomorphine susceptibility is caused by a number of genetic and epigenetic factors. A combination of the newly identified variations may have thus initially contributed to the degree of apomorphine susceptibility in the original Wistar rat population. Subsequent apomorphine injections to determine the susceptibility for the drug during the breeding of the APO-SUS and -UNSUS lines may have acted as an environmental stressor, triggering the Aph-1b recombination event at the topoisomerase II binding site only in rats with a high susceptibility for apomorphine. The induced gene-dosage imbalance of the Aph-1b gene, probably in combination with other genetic or epigenetic factors, could then have led to the complex phenotype observed in the APO-SUS rats.
In conclusion, the present findings suggest that psychopathological disturbances may be the result of multiple genetic as well as epigenetic factors. We infer that our newly identified variations are susceptibility loci for schizophrenia-like features in the rat and may give new insights into the genetic and epigenetic background of complex neurodevelopmental disorders.
Acknowledgements
We thank M. Verheij and L. Lubbers for animal breeding and U. Nguyen for technical assistance. This work was partially supported by grant T5-209 from the Dutch Top Institute Pharma (Leiden, The Netherlands).
Footnotes
Edited by Stephen Maxson.
References
- Coolen MW, van Loo KM, Ellenbroek BA, Cools AR, Martens GJ (2006a) Ontogenic reduction of Aph-1b mRNA and gamma-secretase activity in rats with a complex neurodevelopmental phenotype. Mol Psychiatry 11:787–793 [DOI] [PubMed]
- Coolen MW, van Loo KM, van Bakel NN, Ellenbroek BA, Cools AR, Martens GJ (2006b) Reduced Aph-1b expression causes tissue- and substrate-specific changes in gamma-secretase activity in rats with a complex phenotype. Faseb J 20:175–177 [DOI] [PubMed]
- Coolen MW, Van Loo KM, Van Bakel NN, Pulford DJ, Serneels L, De Strooper B, Ellenbroek BA, Cools AR, Martens GJ (2005) Gene dosage effect on gamma-secretase component Aph-1b in a rat model for neurodevelopmental disorders. Neuron 45:497–503 [DOI] [PubMed]
- Cools AR, Brachten R, Heeren D, Willemen A, Ellenbroek B (1990) Search after neurobiological profile of individual-specific features of Wistar rats. Brain Res Bull 24:49–69 [DOI] [PubMed]
- Craig NL, Nash HA (1983) The mechanism of phage lambda site-specific recombination: site-specific breakage of DNA by Int topoisomerase. Cell 35:795–803 [DOI] [PubMed]
- Ellenbroek BA, Cools AR (2002) Apomorphine susceptibility and animal models for psychopathology: genes and environment. Behav Genet 32:349–361 [DOI] [PubMed]
- Ellenbroek BA, Geyer MA, Cools AR (1995) The behavior of APO-SUS rats in animal models with construct validity for schizophrenia. J Neurosci 15:7604–7611 [DOI] [PMC free article] [PubMed]
- Ellenbroek BA, Sluyter F, Cools AR (2000) The role of genetic and early environmental factors in determining apomorphine susceptibility. Psychopharmacology (Berl) 148:124–131 [DOI] [PubMed]
- Goldstein MJ (1987) Psychosocial issues. Schizophr Bull 13:157–171 [DOI] [PubMed]
- Gottesman I (1991) Schizophrenia genesis: the origins of madness. W.H. Freeman and Company, New York
- Jablensky A, Sartorius N, Korten A, Ernberg G, Anker M, Cooper JE, Day R (1987) Incidence worldwide of schizophrenia. Br J Psychiatry 151:408–409 [DOI] [PubMed]
- Jeanneteau F, Diaz J, Sokoloff P, Griffon N (2004) Interactions of GIPC with dopamine D2, D3 but not D4 receptors define a novel mode of regulation of G protein-coupled receptors. Mol Biol Cell 15:696–705 [DOI] [PMC free article] [PubMed]
- Jirtle RL, Skinner MK (2007) Environmental epigenomics and disease susceptibility. Nat Rev Genet 8:253–262 [DOI] [PMC free article] [PubMed]
- Kavelaars A, Heijnen CJ, Ellenbroek B, van Loveren H, Cools A (1997) Apomorphine-susceptible and apomorphine-unsusceptible Wistar rats differ in their susceptibility to inflammatory and infectious diseases: a study on rats with group-specific differences in structure and reactivity of hypothalamic-pituitary-adrenal axis. J Neurosci 17:2580–2584 [DOI] [PMC free article] [PubMed]
- Kay SR, Opler LA (1987) The positive-negative dimension in schizophrenia: its validity and significance. Psychiatr Dev 5:79–103 [PubMed]
- Mednick SA, Machon RA, Huttunen MO, Bonett D (1988) Adult schizophrenia following prenatal exposure to an influenza epidemic. Arch Gen Psychiatry 45:189–192 [DOI] [PubMed]
- Nadeau JH (2001) Modifier genes in mice and humans. Nat Rev Genet 2:165–174 [DOI] [PubMed]
- Nadeau JH, Topol EJ (2006) The genetics of health. Nat Genet 38:1095–1098 [DOI] [PubMed]
- Regland B (2005) Schizophrenia and single-carbon metabolism. Prog Neuropsychopharmacol Biol Psychiatry 29:1124–1132 [DOI] [PubMed]
- Rots NY, Cools AR, de Jong J, De Kloet ER (1995) Corticosteroid feedback resistance in rats genetically selected for increased dopamine responsiveness. J Neuroendocrinol 7:153–161 [DOI] [PubMed]
- Spitzner JR, Muller MT (1988) A consensus sequence for cleavage by vertebrate DNA topoisomerase II. Nucleic Acids Res 16:5533–5556 [DOI] [PMC free article] [PubMed]
- Van de Vijver G, Van Speybroeck L, De Waele D (2002) Epigenetics: a challenge for genetics, evolution, and development? Ann N Y Acad Sci 981:1–6 [DOI] [PubMed]
- Welsh J, McClelland M (1990) Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res 18:7213–7218 [DOI] [PMC free article] [PubMed]
- Wolffe AP, Matzke MA (1999) Epigenetics: regulation through repression. Science 286:481–486 [DOI] [PubMed]


