Abstract
The development of standard terminologies such as RadLex is becoming important in radiology applications, such as structured reporting, teaching file authoring, report indexing, and text mining. The development and maintenance of these terminologies are challenging, however, because there are few specialized tools to help developers to browse, visualize, and edit large taxonomies. Protégé (http://protege.stanford.edu) is an open-source tool that allows developers to create and to manage terminologies and ontologies. It is more than a terminology-editing tool, as it also provides a platform for developers to use the terminologies in end-user applications. There are more than 70,000 registered users of Protégé who are using the system to manage terminologies and ontologies in many different domains. The RadLex project has recently adopted Protégé for managing its radiology terminology. Protégé provides several features particularly useful to managing radiology terminologies: an intuitive graphical user interface for navigating large taxonomies, visualization components for viewing complex term relationships, and a programming interface so developers can create terminology-driven radiology applications. In addition, Protégé has an extensible plug-in architecture, and its large user community has contributed a rich library of components and extensions that provide much additional useful functionalities. In this report, we describe Protégé’s features and its particular advantages in the radiology domain in the creation, maintenance, and use of radiology terminology.
Key words: Ontologies, terminologies, vocabulaires, RadLex, software tools
INTRODUCTION
The language of biomedicine is rich and diverse, and many domains recognizing the need to standardize the language they use have been developing terminologies and ontologies. Terminologies and ontologies are related. Terminologies (also called “vocabularies” and “lexicons”) are collections of labels or terms (or “entities”) particular to a subject field or domain of human activity, developed for the purpose of documenting and promoting correct usage. Ontologies are similar to terminologies, but generally contain rich knowledge about their terms (through attributes and relations) with the goal of describing the entities that exist in a domain. Many terminologies and ontologies have been appearing throughout biomedicine, including genomics,1 molecular biology,2 anatomy,3 clinical research,4 and clinical care.5
The radiology community has also recently recognized the need for developing a standardized terminology. Hospitals contain a diversity of computerized information systems, and they often refer to the same procedures, findings, and diagnoses using different terminologies. The lack of standards in terminology conventions creates a barrier for comparing studies across enterprises and among health organizations, and it also hampers research because it is difficult to retrieve indexed case material in a consistent manner.
RadLex is a project to create a comprehensive medical imaging terminology for capturing, indexing, and retrieving a variety of radiology information resources.6 RadLex is being developed by acquiring the terms relevant to the radiology subdisciplines (abdominal, cardiovascular, musculoskeletal, pediatric, thoracic, and neuroradiology). The ultimate goal is to distribute RadLex widely in electronic form so that it can be incorporated into applications such as structured reporting, teaching file authoring, and indexing research data.
RadLex was initially developed using word processors and spreadsheets. However, it soon became evident that these tools were too limited for accessing and managing RadLex. First, as RadLex is a large taxonomy, it is difficult to visualize and navigate RadLex using conventional tools such as word processors and spreadsheets. Second, RadLex contains many attributes associated with each term, and it is difficult to maintain this information in correct structured form as RadLex grows. Third, although text documents and spreadsheets were a simple initial format for collecting the RadLex terms, they are not suitable for distributing RadLex on the Web or for making it accessible to applications. A tool tailored to the needs of terminology development and dissemination in radiology is needed.
Protégé (http://protege.stanford.edu)7 is an open-source tool for editing and managing terminologies and ontologies. It is the most widely used domain-independent, freely available, platform-independent technology for developing and managing terminologies, ontologies, and knowledge bases in a broad range of application domains. The community of registered Protégé users exceeds 70,000. In addition, the large and active Protégé user community is highly engaged in Protégé code development, regularly contributing enhancements to the software (http://protege.stanford.edu/community/wiki.html), as well as participating in online discussion groups devoted to modeling questions, technical-support issues, and requests for new features (http://protege.stanford.edu/community/archives.html).
Protégé has been used as the primary development environment for many ontologies in the life sciences. These projects include the Foundational Model of Anatomy,3 Cerner’s Clinical Bioinformatics ontology,8 the DICE TS,5 and the MGED Ontology.2 Recently, the RadLex project decided to adopt Protégé, and RadLex is currently distributed in Protégé XML format (http://radlex.org/radlex/docs/downloads.html).
In this report, we introduce Protégé and highlight its value to managing terminology in the radiology domain. Besides providing a suite of functionality that helps curators edit and manage terminologies such as RadLex, Protégé provides an extensible architecture to which custom functionality specific to radiology needs can be added, and it also provides a platform for deploying radiology applications that exploit terminologies such as RadLex.
TECHNICAL DESCRIPTION
Protégé Knowledge Model
Protégé is a suite of tools for ontology development and use. Before describing the tool itself, it is important to understand how terminologies and ontologies are built and stored in computers. Briefly, terminologies comprise lists of terms (also called “entities”), and they may also specify attributes (also called “slots”) for those terms (for example, RadLex contains a term called supraaortic valve area, which has an attribute Version Number). Ontologies are similar to terminologies, but contain rich relationships amongst terms, enabling them to represent knowledge in a domain (for example, a relationship SegmentOf linking supraaortic valve area to thoracic arota represents the fact that the supraaortic valve area is a segment of the thoracic aorta).
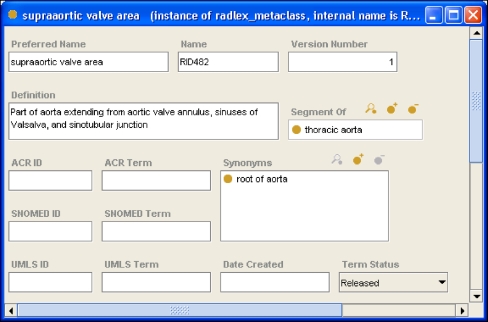
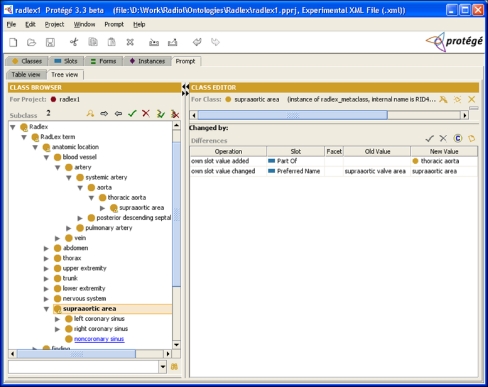
In the Protégé knowledge model, terminologies and ontologies are represented using “frames” (classes, slots, and facets).9 An ontology in Protégé consists of frames and axioms. Classes are the entities (“terms”) used in the domain of discourse. Classes are the sometimes-called “concepts” in terminologies. For example, if we wish to have a term in our RadLex vocabulary representing the supraaortic valve area, we would create the class supraaortic valve area in the Protégé ontology (Fig. 1).
Fig 1.
A Protégé “frame.” This screen capture shows a frame in Protégé representing the class in RadLex (a RadLex term). The class contains several slots (attributes the contained values), such as a preferred name of “supraaortic valve area,” an internal RadLex identifier name (“RID482”), a version number, definition, and other information such as the arterial segment of which it is a part, synonyms, and other information about the term.
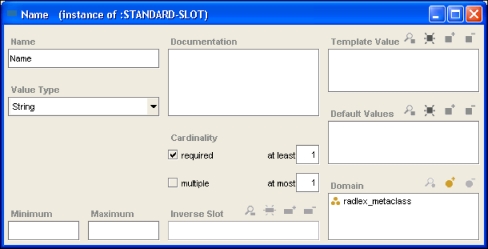
Slots describe properties or attributes of classes. For example, if we wanted all RadLex terms in our ontology to have a string unique identifier called “Name,” which had a nonempty value, then we would create a Name slot and give it the necessary facets (Fig. 2). Facets describe characteristics of slots (such as cardinality, required values, etc). Facets allow us to express the fact that the name of a RadLex term class is required (Fig. 2). Using a set of slots, we can fully describe each RadLex term. For example, we can express the fact that the supraaortic valve area is a segment of the thoracic aorta, that “root of aorta” is a synonym, and that it has a definition and a RadLex identifier of “RID482” (Fig. 1).
Fig 2.
A Protégé “slot.” This screen capture shows a frame in Protégé representing the slot called “Name,” which is used to convey the unique identifier name for a RadLex term. This name slot contains facets–components that constrain its values such as cardinality, value type, minimum, and maximum values. In this case, the facet values are saying that Name is a single-valued string, and it must have a value for every term in RadLex. This slot is associated with all classes in RadLex; for example, for the supraaortic valve area class (Fig. 1), this slot has a value of “RID482.” Another slot called Preferred Name is used to store the actual name of the class, “supraaortic valve area” (Fig. 1).
Axioms specify additional constraints, but will not be described further here as they are not used in RadLex at this time. An instance is a frame built from at least one class (“instantiation”) that carries particular values for the slots. A “knowledge base” includes the ontology (classes, slots, facets, and axioms) as well as instances of particular classes with specific values for slots. The distinction between classes and instances is not an absolute one; however, because terminologies generally do not contain instances, we can simplify and limit our discussion to classes.
Classes, slots, and facets are the basic building blocks of terminologies. A tutorial on using these elements and Protégé for creating terminologies and ontologies is available10 and will not be repeated here. Instead, we will focus on the features of Protégé that are particularly relevant to accessing, editing, and sharing ontologies and using them in applications.
Architecture of Protégé
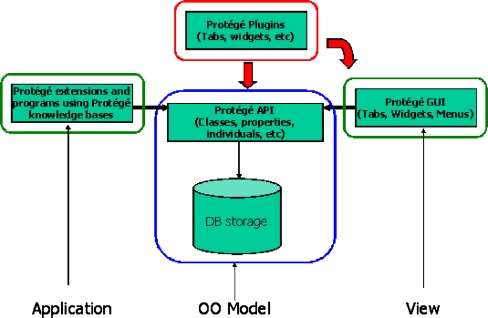
Protégé is implemented in Java and runs on a broad range of software platforms, including Windows, MacOS, Linux, and Unix. Protégé is built using a tiered architecture (Fig. 3), providing an ontology storage layer, a knowledge model layer, a graphical user interface (GUI), and an application programming interface (API). Users running the desktop application interact with ontologies using the GUI, while application programs access ontologies in Protégé via the API. The Protégé GUI uses the API to access the Protégé knowledge model of the ontology. Thus, the Protégé GUI, plug-ins, and user applications all access the ontology through the same API, making the Protégé architecture modular and highly flexible (Fig. 3).
Fig 3.
Protégé architecture. Protégé is built using a tiered architecture. The Protégé knowledge model is an object-oriented model (“OO Model”) of ontologies stored in a persistence layer (DB storage) and accessed via the Protégé API. The API is used both by the protégé GUI application (View layer) with which users access the ontologies, as well as by application programs (Application layer). New functionality is added to Protégé by creating plug-ins, which access ontologies through the Protégé API and with which the user can interact by plugging into the Protégé GUI.
In addition, Protégé has a plug-in architecture, permitting developers to extend Protégé’s core functionality in many ways without needing to modify the Protégé source code. There are more than 90 Protégé plug-ins providing advanced capabilities such as import/export, validation, and visualization of large ontologies (http://protege.cim3.net/cgi-bin/wiki.pl?ProtegePluginsLibraryByTopic), some of which we discuss below.
Import and export of ontology content is a critical feature of any tool, because there are many formats for storing ontologies and terminologies. There are several plug-ins for Protégé, enabling it to import and export ontologies in many different formats, including XML, RDF, OWL, and the native Protégé format.
If the ontology is stored in a file, the entire ontology is read into memory. Protégé also provides a relational database backend, which is useful when working with ontologies too large to reside in memory. Finally, through Protégé’s plug-in mechanism, developers can create their own custom import/export plug-ins to work with custom or specialized formats.
Protégé Application and Graphical User Interface
Protégé can be run as a stand-alone application or through a Protégé client in communication with a remote server (the latter is particularly useful for collaborative ontology development). When the Protégé desktop application is launched, the user can create a new ontology, open an existing ontology, or import an ontology from a variety of formats. Protégé creates a project file that records display-related information and the location of the ontology source file; the project file simplifies the process of reopening ontologies, and it maintains user GUI customizations.
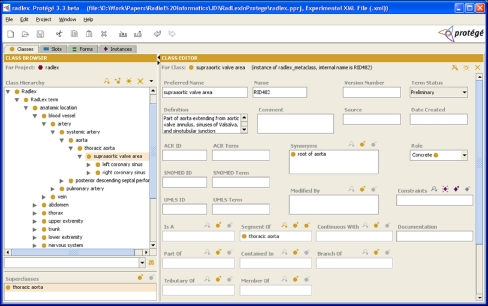
Once an ontology is opened, the user works with it in the Protégé GUI (Fig. 4). The Protégé GUI is organized into separate panels (accessed by tabs along the top of the GUI) that provide different views into the contents of the ontology. The first tab is the Classes tab, which is the most common view used to browse ontologies and terminologies. On this tab, the ontology is shown as an expandable tree on the left of the GUI, and the attributes of a class selected by the user is shown on the right (Fig. 4). In the tree view, terms that are indented and below a term are called “child” terms, whereas the term above is the “parent.” The interpretation of a child term is that it is subsumed by the parent by an “is-a” relationship (unless the tree of classes is displayed according to a different slot). For example, Protégé shows that the RadLex term aorta is a child of systemic artery, which is interpreted as “aorta is-a systemic artery” (Fig. 4).
Fig 4.
RadLex shown in the main Protégé user interface, which provides the ability to build, populate, and view ontologies and terminologies. This screen capture shows the Classes tab, in which the user creates, edits, and browses the terms in RadLex. The taxonomy of Radlex is shown as an expandable tree on the left. The right panel displays the details of a RadLex term selected from the tree on the left. For each class selected, Protégé displays the attributes of that class (right) which users can edit. For example, the left coronary sinus has been selected (left) and it is a branch of the supraaortic valve area. Other tabs in the Protégé GUI include: the Slots tab, for creating and editing slots; the Forms tab, for customizing layout of knowledge-entry forms (such as the one shown on the right); the Instances tab for creating instances of classes and entering particular slot values for those instances; and any other special-purpose “plug-in” tab that the user may want to use.
The tree view of RadLex can facilitate the process of curating the terminology to find problems to be corrected, because the various hierarchies can be easily expanded and browsed to detect inconsistencies. In this tree view, for example, we see that the supraaortic value area is a child of thoracic aorta (Fig. 4; “supraaortic value area is-a thoracic aorta”). As it is not correct to say that “supraaortic valve area is-a thoracic aorta,” the RadLex ontology should be changed in a future release (what was likely intended was to say “supraaortic value area part-of thoracic aorta,” which could be reflected by changing the Part-of value).
The classes tab permits full editing of the ontology. In the tree view of the ontology on this tab, users can drag and drop classes to reorganize the hierarchy, create and rename classes using intuitive GUI paradigms. The values for attributes of classes can also be directly edited. For example, we could change the name of the class, left coronary sinus, by simply selecting this class and changing the value of the “Preferred Name” slot in the GUI (Fig. 4, right panel).
The other tabs accessible from the Protégé GUI are the slots, forms, and instances tab, which are less often used by those curating terminologies. The slots tab enables users to view and edit all slots in an ontology; the instances tab displays all instances associated with the classes; and the forms view permits users to customize the layout of the elements on display forms.
Ontology Difference
Ontologies change over time. They are developed iteratively, with incremental changes contributed by people working collaboratively on them. RadLex will soon be releasing a new version, adding new terms as well as changes to the original terminology based on suggestions from users of the first version. There is a need to maintain and compare versions of ontologies, similar to the need for document version management in large projects. Document versioning systems enable users to compare versions, examine changes, and accept or reject changes. However, a versioning system for ontologies must compare and present structural changes rather than changes in the text of a document.
PROMPT is a plug-in to Protégé providing a suite of tools for aligning and comparing two ontologies. PROMPT contains PROMPTDIFF, a versioning environment for ontologies, allowing developers to track changes in ontologies over time.11,12 PROMPTDIFF is a version-comparison algorithm that produces a structural difference between two versions of an ontology. The results are presented to the users enabling them to view classes that were added, deleted, and moved. The users can then act on the changes, accepting or rejecting them.
Suppose we edited a copy of RadLex to update it based on community feedback. Consider that we move the class supraaortic valve area and place it under the anatomic location class, and rename it to supraaortic area. In addition, suppose we create a new class noncoraonary sinus under supraaortic area, and that we update the Part-of relationship on supraaortic area to say it is Part-of thoracic aorta. When we run PROMPTDIFF, comparing our edited version of RadLex against the original version, the PROMPT interface displays the following differences (Fig. 5):
supraaortic valve area was renamed to supraaortic area
supraaortic area was moved from being under thoracic aorta to being under anatomic location.
noncoraonary sinus is a new class, under supraaortic area
thoracic aorta was added as a value for the Part-of relationship on the supraaortic area class.
Fig 5.
Displaying changes in PromptDiff. The output from PROMPT is shown in after comparing RadLex with a version of the terminology that was edited to move a class, change its name, and add a slot value. Left panel: Changes to the class hierarchy are shown. Different styles represent different types of changes: added class is underlined and in blue; moved classes are flagged out in their old positions and appear in bold in their new ones. Right panel: Individual changes to the slot values for a selected class are shown (in this case, for the supraaortic area class).
In addition to viewing versions of ontologies, PROMPT also includes a tool to align two different ontologies to locate classes that are the same or similar to each other. Such functionality could be useful in RadLex for finding related terms in other vocabularies such as SNOMED and the ACR index.
Ontology Visualization Components
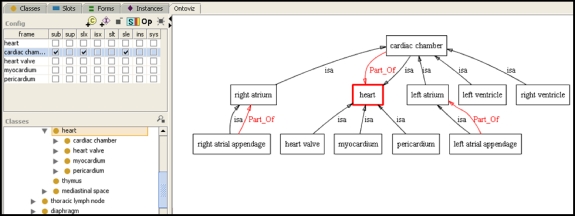
The Protégé GUI provides a tree view of ontologies (Fig. 4). Although an expandable tree is suitable for many ontologies and terminologies, it can be difficult to visualize those that have multiple types of relationships among the terms. A tree shows the relationships among terms using one relationship (usually the is-a relationship). RadLex contains several types of relationships, which are best shown using a graph (Fig. 6). There are several types of ontology visualization components that have been developed for Protégé, two of which we highlight here in the context of RadLex.
Fig 6.
Visualizing RadLex using the OntoViz plug-in for Protégé. This visualization plug-in for Protégé displays an ontology as a graph (right panel). Ontology classes are shown as boxes, and the relationships among them are shown as arcs. The label on the arc is the name of the relationship. The user can select the classes to be displayed in the graph (lower left panel) as well as the types of information to display, such as subclasses, superclasses, and relationships (upper left panel). This visualization paradigm is particularly helpful when an ontology contains more than one type relationship, since the tree view (left) only shows the ontology classes according to one relationship (is-a).
OntoViz is a plug-in to Protégé that creates graph visualizations of ontologies (http://protege.cim3.net/cgi-bin/wiki.pl?OntoViz). OntoViz produces its graphs using the Graphviz software package.13 The types of visualizations are highly configurable and include ability to (1) visualize part of an ontology by selecting specific classes and instances, (2) display slots and slot edges, (3) customize the graph by specifying colors for nodes and edges, and (4) visualize the neighborhood of classes for particular classes or instances by applying various operators (e.g., subclasses, superclasses).
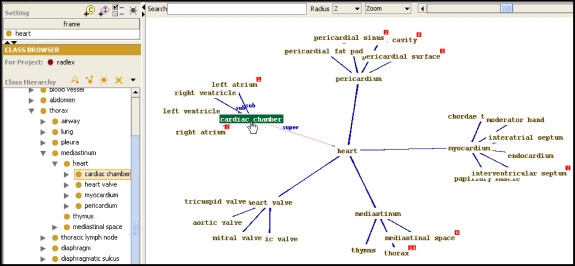
TGVizTab is a plug-in for Progégé for visualizing ontologies as a TouchGraph, a software implementation for producing an interactive graph that can be navigated by clicking on the nodes and edges in the graph. The TouchGraph library has been modified and integrated into TGVizTab for visualizing ontologies.14 TGVizTab is well suited for navigating RadLex, because of the size of the terminology: the TouchGraph display provides a compact and graphical way to visualize large numbers of classes (Fig. 7). Unlike OntoViz, the user can dynamically navigate the ontology and interrogate each of its components in real time. This visualization paradigm makes the terminology display simple and intuitive.
Fig 7.
Visualizing RadLex using the TGVizTab plug-in for Protégé. The TGViz plug-in to Protégé displays an ontology as a TouchGraph (right panel), enabling the user to navigate the ontology as a nested, zoomable graph. The graph is initially created by selecting a root class from the tree view (upper left panel); the root class is shown in the center of the graph (right panel). Classes are shown as the names of the class, and the relationships between classes are shown as arcs, with the type of relationship appearing when the mouse passes over the arcs (upper left part of graph in right panel). Users navigate the ontology by double-clicking on a class name in the graph, which zooms into that class, producing a new TouchGraph rooted at that class.
Collaborative Ontology Development
A key challenge for maintaining a large terminology such as RadLex is to collect and process the feedback from the large community of users. The current model for feedback on terminologies and ontologies is for users to contact the developers or to post comments to online discussion forums. The problem with this approach is that the user feedback is disconnected from the ontology under discussion; it is difficult for the ontology developer to keep track of these discussions, to prioritize the feedback, and to decide which parts of the ontology need updated first.
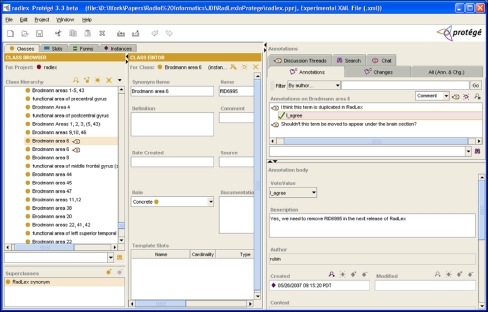
Collaborative Protégé15 is a recent extension of the Protégé system that supports collaborative ontology development by enabling the community to annotate ontologies. A set of annotation components can be accessed by people viewing terminologies such as RadLex, enabling them to associate their comments directly with the parts of the ontology to which they apply. Thus, for example, if a user noticed that there is a duplicate term in RadLex, that person could create an annotation on the class in question in which the nature of the problem is described (Fig. 8). At a later time, the RadLex curator can view the parts of the ontology for which users have made annotations, either acting on them or making annotations on the annotations themselves. All annotations can be viewed by the community as well, enabling a dialog on ontologies that is tied to the specific components under discussion. This mechanism can streamline the process of acquiring and reviewing community feedback on ontologies.
Fig 8.
Collaborative Protégé. Protégé supports collaborative ontology editing by permitting users to annotate ontology components as well as ontology changes. It also supports searching and filtering of user annotations based on different criteria. In this screenshot, a user has identified a duplicate term in RadLex and communicates that to the curators by creating an annotation on the term in question (“Brodmann area 6”). The term to be annotated is selected (left panel), and annotations are created (right panel) to describe the user’s feedback. The RadLex curator can search, filter, and browse annotations created by the community, and create annotations serving as to-do items for incorporating changes and suggestions in future releases of RadLex. This screenshot shows that the curator agrees with the user feedback and creates an annotation reflecting the need for the duplicate term to be removed in the next release of RadLex.
The Collaborative Protégé components provide search and filtering of user annotations based on different criteria. There are also two types of voting mechanisms that can be used for voting of change proposals. All annotations made by one user are seen immediately by other users, enabling a community-based discussion about ontologies to be directly linked to the applicable portions of the ontologies.
Programming Interfaces
Despite the variety of plug-ins available for Protégé, many users will have custom needs related to how they work with ontologies, which will require direct programmatic access to the ontology content. For example, a RadLex curator may wish to find the classes that have missing values for slots (currently, many SNOMED and UMLS identifiers have not yet been collected for RadLex terms). To find these classes in RadLex, a program would be needed to traverse all classes in the RadLex ontology, looking for those that have missing values for the slots in question.
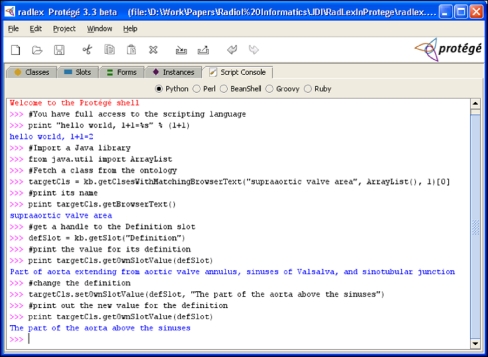
Protégé provides two programming interfaces that developers can use to access its ontology content. The Protégé Script Tab is a plug-in to Protégé that provides a scripting environment in several interpreted languages such as python, pearl, and ruby (http://protege.cim3.net/cgi-bin/wiki.pl?ProtegeScriptTab). The Script Tab provides the simplest and quickest programmatic access to ontologies in Protégé; it accessed via an additional tab in the Protégé GUI into which users can enter commands (Fig. 9). The commands will be applied to the ontology currently loaded into Protégé. Thus, in our example above, we could perform the query to find classes having missing values for slots by entering a few lines of code into the Script Tab and viewing the results. An example of the output of a few simple commands entered into the Script Tab is shown in Figure 9.
Fig 9.
Programmatically interacting with ontologies using the Protégé Script Tab. This screen shot shows some example interactive commands to the Protege API and their results.
In addition to the Script Tab, Protégé provides an API. System developers can use the Protégé API, a Java interface to access and programmatically manipulate Protégé ontologies—this is the same API that the Protégé GUI uses to access ontologies (Fig. 3). Developers can access the Protege Java API from their Java programs by calling the appropriate methods to access the ontology in Protégé and to perform the desired operations. Documentation for guiding developers in using the Protégé API for application and plug-in development is available (http://protege.stanford.edu/doc/dev.html).
APPLICATIONS
While Protégé is a useful tool for working with ontologies and terminologies, the ultimate reason to be creating these artifacts is to use them in applications. For example, if one wanted to use RadLex in a structured reporting application to enforce use of controlled terminology, it would be necessary for that application to access terms in the ontology. Protégé can be integrated with many applications, connecting external programs to ontologies via the Protégé API (as described above). In this section, we highlight an example application relevant to RadLex that was created in this manner—WebProtege.
A large terminology such as RadLex needs to be both widely available and locally editable. A common method for distributing a terminology is via the Web; however, once a user downloads the terminology, it will be become out of date as soon as a new version is posted. An alternative method for disseminating RadLex is to provide a Web-based program for viewing the terminology. In fact, RadLex is currently made available through a dedicated Web application (http://www.radlex.org/radlex/). However, because the version of RadLex that is deployed in the Web application is separate from the working draft, it is challenging to keep the development and production versions of RadLex tightly synchronized.
An approach whereby a single version of RadLex could be edited by curators and disseminated on the Web could be advantageous to make a current working draft available. We undertook to implement this strategy by adopting WebProtege, an ontology-enabled application built on the Protégé platform. The goal was to distribute Radlex in a Web application while providing the means to keep it synchronized with the version in development.
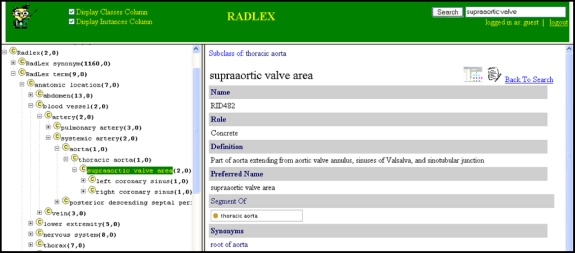
WebProtege is an application that allows users to share, browse, and edit ontologies using a Web browser (http://protege.cim3.net/cgi-bin/wiki.pl?WebProtege). The WebProtege application provides a graphical paradigm similar to the Protégé GUI; users can browse ontologies in a Web browser in a similar manner to viewing them in Protégé (Fig. 10). In addition, users can create and post comments about specific ontology components.
Fig 10.
RadLex ontology accessed over the Internet using WebProtege. This application provides Web access to ontologies; a single ontology file can be edited by curators (Fig. 1) and immediately deployed on the Web without needing to update any software applications. The ontology is shown on the left, and details about selected terms on the right, similar to that provided by the Protégé editing tool (Fig. 4).
The Protégé architecture allows multiple applications as well as Protégé itself to access the same ontology (see Fig. 3). Thus, the maintenance and coordination of the terminology is greatly facilitated, because there need be only one copy of the terminology residing in a central location that all applications access through the Protégé API. A RadLex developer can use the Protégé GUI to edit the terminology, while applications that need RadLex use the Protégé API, accessing the same version that the developer has edited, and users browsing RadLex through WebProtege see the same version of the terminology as well.
In addition to the WebProtege application that we have described here, other applications that use ontologies can be built using similar principles. The common denominator is that in order for an application to use an ontology, the appropriate methods in the Protégé API are called to get references to classes (terms) and their attributes (slots).
DISCUSSION
The need for terminologies and ontologies in radiology is becoming increasingly apparent. Variation in language used in reporting the results of imaging studies is a recognized challenge,16 which can be mitigated by adopting standard terminologies. Radiology educators need a comprehensive lexicon for indexing online educational materials.6 Radiology researchers need their data to be indexed with terminologies so that search and data mining can be more effective. Ontologies can be used to create new intelligent Picture Archiving and Communication System applications.17
In general, there are two types of users of terminologies: terminology developers and terminology consumers. The developers of RadLex are in the former group, and those wishing to use RadLex or to create applications enabled by RadLex are in the latter group. Terminology developers need a tool that meets the functional requirements of terminology management:
Visualization of terminologies: Terminologies can be large. Users need to be able to browse terminologies compactly, collapsing entire trees to hide the complexity from users. Different paradigms for visualization (tree view, graph view, etc) are desirable because different ways of viewing large terminologies could be preferable under different circumstance (tree views give a good overview of taxonomies; graphs are suitable for displaying multiple relationships among terms).
Terminology versioning: It is useful to show how terminologies changed between versions. Traditional “document diff” methods that show text differences are not useful for terminologies because of their complex structure. Specialized tools are needed to show how terminologies changed between versions (term moving to a different place, being split into two or more new terms, etc).
User feedback management: User feedback drives the evolution of terminologies. It is difficult to keep track of all the requested changes to a terminology, especially as the volume of feedback expands. Methods are needed to track the type of comments being made, the terms to which they apply, and whether and how feedback was handled. It is desirable for the feedback to be publicly accessible so that the details of the evolution of the terminology are transparent to the community.
Terminology consumers have requirements related to terminology dissemination and application development:
Terminology dissemination: Applications need to be able to access the current version of the terminology as soon as new versions are produced. It can be cumbersome for developers of applications that use a terminology such as Radlex to keep their version of the terminology current as RadLex evolves; it would be preferable to avoid the need to modify of update applications when new versions of the terminology are released.
Application development: Ultimately, a terminology is only useful if it can be easily incorporated into applications. An application should be able to reference terms in the terminology using simple methods, and application developers should not need to understand the arcane details of terminology storage formats.
In this report, we have described Protégé, an open-source tool that can help users of terminologies and ontologies to develop them and use them in applications. Protégé provides tools to support the functional requirements for terminology developers and consumers as described above. The Protégé tools range from creating the terminology (GUI, visualization, and versioning support) to deployment and application development (user feedback management, API, and application support). We have illustrated these functions using RadLex as an example terminology. In fact, the RadLex project has adopted Protégé for managing the terminology, and Protégé has been useful in meeting the terminology curation needs of the RadLex project to date.
Although Protégé provides functionality that meets many needs of the user community, there are still some unmet challenges. First, terminologies evolve over time, and terms that are used in applications may be retired in future versions of the terminologies. When a term is deleted from a newer version of the terminology, applications that use an older version of the terminology need to be provided an appropriate replacement term.
A second challenge is that the compact visualizations provided by Protégé may not be sufficient in cases of very large terminologies. Although there are some visualization paradigms for large ontologies, it could be helpful to filter the terminology or produce a compact “view” of the ontology specific to context of the user. A final challenge is that users new to terminologies and ontologies may experience a learning curve in becoming acquainted with the concepts and learning how to create terminology-enabled applications. The Protégé web site contains tutorials and introductory material that can help the community get up to speed with this tool and its use in applications.
Acknowledgements
This work was supported by the National Center for Biomedical Ontology, under roadmap-initiative grant U54 HG004028 from the National Institutes of Health. This work was also supported by the Protégé resource, under grant LM007885 from the U.S. National Library of Medicine. We acknowledge the highly talented Protégé research and development team, Tim Redmond, Samson Tu, Tania Tudorache, and Jennifer Vendetti, and Ray Fergerson for developing the Protégé system. We also thank the Protégé user community for their outstanding contributions to the Protégé platform.
References
- 1.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Whetzel PL, et al. The MGED Ontology: a resource for semantics-based description of microarray experiments. Bioinformatics. 2006;22:866–873. doi: 10.1093/bioinformatics/btl005. [DOI] [PubMed] [Google Scholar]
- 3.Rosse C, Mejino JL., Jr A reference ontology for biomedical informatics: The Foundational Model of Anatomy. J Biomed Inform. 2003;36:478–500. doi: 10.1016/j.jbi.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 4.Sioutos N, Coronado S, Haber MW, Hartel FW, Shaiu WL, Wright LW. NCI Thesaurus: a semantic model integrating cancer-related clinical and molecular information. J Biomed Inform. 2007;40:30–43. doi: 10.1016/j.jbi.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Keizer NF, Abu-Hanna A, Cornet R, Zwetsloot-Schonk JH, Stoutenbeek CP. Analysis and design of an ontology for intensive care diagnoses. Methods Inf Med. 1999;38:102–112. [PubMed] [Google Scholar]
- 6.Langlotz CP. RadLex: a new method for indexing online educational materials. Radiographics. 2006;26:1595–1597. doi: 10.1148/rg.266065168. [DOI] [PubMed] [Google Scholar]
- 7.Gennari JH, et al. The evolution of Protege: an environment for knowledge-based systems development. Int J Hum Comput Stud. 2003;58:89–123. doi: 10.1016/S1071-5819(02)00127-1. [DOI] [Google Scholar]
- 8.Hoffman M, Arnoldi C, Chuang I: The clinical bioinformatics ontology: a curated semantic network utilizing RefSeq information. Pac Symp Biocomput:139–150, 2005 [DOI] [PubMed]
- 9.Noy NF, Fergerson RW, Musen MA. The knowledge model of Protege-2000: Combining interoperability and flexibility. Lect Notes Artif Int. 2000;1937:17–32. [Google Scholar]
- 10.Noy NF, McGuinness DL: Ontology Development 101: A Guide to Creating Your First Ontology. Stanford Knowledge Systems Laboratory Technical Report KSL-01-05 and Stanford Medical Informatics Technical Report SMI-2001-0880, 2001
- 11.Noy NF, Kunnatur S, Klein M, Musen MA: Tracking changes during ontology evolution. Semantic Web—Iswc 2004, Proceedings 3298:259–273, 2004
- 12.Noy NF, Musen MA. The PROMPT suite: interactive tools for ontology merging and mapping. Int J Hum Comput Stud. 2003;59:983–1024. doi: 10.1016/j.ijhcs.2003.08.002. [DOI] [Google Scholar]
- 13.Ellson J, Gansner E, Koutsofios L, North SC, Woodhull G. Graphviz—Open source graph drawing tools. Graph Draw. 2002;2265:483–484. doi: 10.1007/3-540-45848-4_57. [DOI] [Google Scholar]
- 14.Alani H: TGVizTab: An Ontology Visualisation Extension for Protégé. Proc. Proceedings of Workshop on Semantic Authoring, Annotation & Knowledge Markup (SAAKM’02), the 15th European Conference on Artificial Intelligence, (ECAI’02): Lyon City
- 15.Tudorache T, Noy NF: Collaborative Protégé. Proc. Workshop on Social and Collaborative Construction of Structured Knowledge: Banff City
- 16.Sobel JL, et al. Information content and clarity of radiologists’ reports for chest radiography. Acad Radiol. 1996;3:709–717. doi: 10.1016/S1076-6332(96)80407-7. [DOI] [PubMed] [Google Scholar]
- 17.Kahn CE, Jr, Channin DS, Rubin DL. An ontology for PACS integration. J Digit Imaging. 2006;19:316–327. doi: 10.1007/s10278-006-0627-3. [DOI] [PMC free article] [PubMed] [Google Scholar]