Abstract
We have created a content-based image retrieval framework for computed tomography images of pulmonary nodules. When presented with a nodule image, the system retrieves images of similar nodules from a collection prepared by the Lung Image Database Consortium (LIDC). The system (1) extracts images of individual nodules from the LIDC collection based on LIDC expert annotations, (2) stores the extracted data in a flat XML database, (3) calculates a set of quantitative descriptors for each nodule that provide a high-level characterization of its texture, and (4) uses various measures to determine the similarity of two nodules and perform queries on a selected query nodule. Using our framework, we compared three feature extraction methods: Haralick co-occurrence, Gabor filters, and Markov random fields. Gabor and Markov descriptors perform better at retrieving similar nodules than do Haralick co-occurrence techniques, with best retrieval precisions in excess of 88%. Because the software we have developed and the reference images are both open source and publicly available they may be incorporated into both commercial and academic imaging workstations and extended by others in their research.
Key words: Content-based image retrieval, open source, lung, computer-aided diagnosis (CAD), extensible markup language (XML), image database, software design, computed tomography, texture feature, nodule
BACKGROUND
In the continuing battle against lung cancer, computed tomography (CT) scanning has been found to increase the detection rate of pulmonary nodules.1 Much work has been done to develop computer assisted diagnosis and detection (CAD) systems for pulmonary nodules in CT. We hypothesize that we can also reduce the uncertainty of the radiologist in identifying suspicious pulmonary nodules by providing a visual comparison of a given nodule to a collection of similar nodules of known pathology. To eventually test this hypothesis we first need to develop a content-based image retrieval (CBIR) system for pulmonary nodules in CT. The human observer (radiologist) manually (or semi-automatically or automatically) segments a nodule from a clinical case. The system computes a set of quantitative descriptors for that nodule (our current work focuses on texture-based descriptors) and compares those descriptors to the descriptors of known nodules. The underlying assertion is that if a known malignant nodule has certain computable features, then unknown nodules with similar computable features would be malignant.
Simply put, our system provides a way of performing a “look-up” on a query image to return similar images from a collection. Much research is being done to see which methods of comparing and retrieving similar images are best. For a detailed description of CBIR systems for the medical field, we suggest the review by Muller et al.2 Our work compares three different sets of texture feature descriptors to determine which one has the best precision in retrieving similar nodules.
There are generally two types of medical CBIR systems: (1) those that retrieve entire anatomic structures, and (2) those that retrieve abnormalities or pathologies within an anatomical structure. The latter problem is more complex than the former, but more useful for CAD. Thus we have focused our efforts on images of pulmonary nodules, rather than images of the entire lung.
The first known large-scale comparison of texture features was done by Ohanian and Dubest in 1992.3 They tested 16 Haralick co-occurrence features, 4 Markov random field features, 16 Gabor filter features, and 4 fractal geometry features on 3200 32 × 32 sub-images and found that co-occurrence performed the best. However, whereas Ohanian and Dubest evaluated the feature types in respect to their ability to classify an image’s texture correctly, we sought to evaluate the features by their performance in an image retrieval system. There are several other CBIR projects currently underway in the medical field in general and particularly with lung CT images. One of these, called ASSERT, is being developed at Purdue University and uses a variety of different image features, including co-occurrence statistics, shape descriptors, Fourier transforms, and global gray level statistics. The system also includes physician-provided ratings of features such as homogeneity, calcification, and artery size.4,5
There are, however, problems associated with content-based retrieval of medical images, such as the difficulty of automatic segmentation, the large variability of feature selection, and the lack of standardized toolkits and evaluation methods.6–8 There have been several efforts over recent years to solve some of these problems. For instance, the Lung Image Database Consortium (LIDC) collection was specifically developed to support evaluation and comparison of chest CAD systems.9 It can be used similarly to develop, evaluate and compare CBIR systems.
There are also a growing number of open source frameworks for medical imaging applications, such as the Visualization Tookit (VTK),10 the Insight Toolkit (ITK)11 for segmentation and registration, and the Image-Guided Surgery Toolkit (IGstk).12 All of these projects are community-driven and freely available on their websites. In addition, the National Cancer Institute is funding the development of an eXtensible Imaging Platform (XIP) through its Cancer Bioinformatics Grid (caBIG) program.13
We believe that the nature of pulmonary nodules (characterized by very small images and significant physician disagreement) justifies the creation of a specialized system for nodule retrieval. Our goal was to build an open source, independent, extensible, CBIR system for pulmonary nodules in CT images and to contribute this system to the growing open source medical imaging community.
MATERIAL AND METHODS
This work is exempt from human subjects research regulation. It makes use of a publicly available, completely de-identified data set (LIDC). We used a portion of the LIDC data consisting of 90 CT studies of the chest, each containing between 100 and 400 Digital Imaging and Communication (DICOM) images. An XML data file containing the expert annotations from the LIDC consortium accompanies each data set.
The LIDC expert annotations include a freehand outline of nodules on each CT slice in which the nodules are visible, along with subjective ratings on a 5- or 6-point scale of the following pathological features: calcification, internal structure, subtlety, lobulation, margin, sphericity, malignancy, texture, and spiculation. Our image extraction routine uses the outlines to mask the original DICOM image and produce individual nodule images exactly as segmented by the LIDC expert viewers.
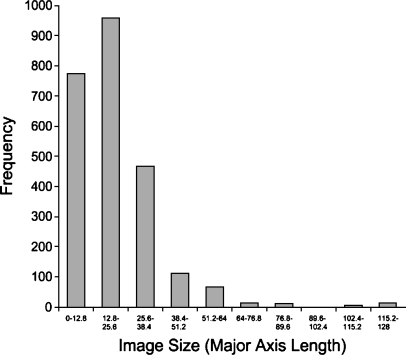
Figure 1 is a histogram of nodule sizes as measured by the standard Response Evaluation Criteria In Solid Tumors (RECIST) criteria of major axis length. Elsewhere, we have used a 2-D area measurement (total pixels) for the nodule size because texture is a surface property, and therefore the number of total pixels is more relevant to texture analysis. We discarded all nodule images smaller than 5 × 5 pixels (around 3 × 3 mm) because images this small would not have yielded meaningful texture data.14 The final database contained 2424 images of 141 unique nodules. The median image size was 15 × 15 pixels, and the median actual size was approximately 10 × 10 mm. The smallest nodules were roughly 3 × 3 mm, whereas the largest were more than 70 × 70 mm. Eighty-eight percent of the images were 20 × 20 mm or smaller.
Fig 1.
Image size histogram.
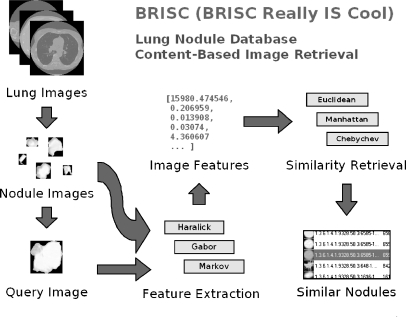
Figure 2 provides an overview of the various stages of the CBIR process:
Extract individual nodule images from the LIDC DICOM images based on data from physician annotations.
Store the extracted data in a database.
Calculate a set of quantitative descriptors that provide a high-level characterization of each image’s texture.
Use various measures to determine how similar one image is to another.
Fig 2.
System overview.
We have previously described the low-level image features used to capture the nodule’s image texture.15
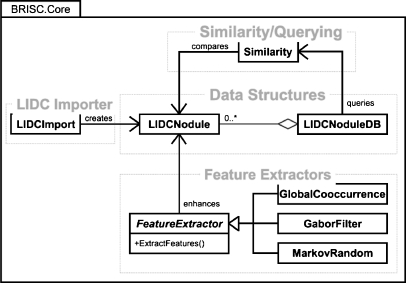
To develop our system, we used Microsoft C# and the .NET 2.0 Framework. The .NET Framework is a software library that provides a large range of pre-built solutions, including collections, file access, and graphical user interfaces. This allowed for rapid development and deployment over the limited period of time allotted for this project. Our design of the core library contains four major components (see Fig. 3 for the class diagram), corresponding to the four stages of the CBIR process described above.
Fig 3.
Class diagram.
LIDC Importer
The LIDCImport module extracts data from the LIDC XML files and saves this data to the formats used by our library. It also initiates the calculation of features for all images in the dataset.
Data Structures
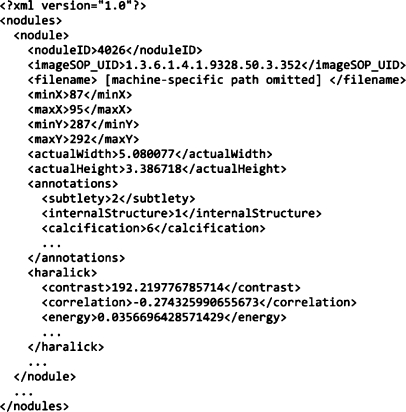
There are two main data structures: LIDCNodule and LIDCNoduleDB. The first represents a single nodule image. It contains data elements that store information about the nodule and its attributes. Because there are usually many images of the same nodule, we have included a field for storing a nodule identification number so that all images of a particular nodule can be retrieved by querying for this number. The LIDCNodule class also stores links to the raw image data on disk and knows how to read/write its data to an XML file. Currently, all feature data are stored in this class. The second data structure encapsulates a collection of LIDCNodule objects. It provides the core functionality of a CBIR system by allowing for the normalization and querying of the image dataset. It also handles the reading and writing of XML files. Figure 4 contains sample excerpts from our XML files.
Fig 4.
Sample XML.
Feature Extractors
There are currently three feature extraction classes: GlobalCooccurrence, GaborFilter, and MarkovRandom. All of these implement the FeatureExtractor interface, which requires a common method called ExtractFeatures. This method takes an LIDCNodule as a parameter and should access DICOM image data for the image, calculate its features, and then save that feature data back into the LIDCNodule object. This cluster facilitates an implementation of the Strategy software design pattern16 for interchangeable image features.
Similarity/Querying
The Similarity class contains functions that implement various similarity measures and are used by the LIDCNoduleDB class to compare nodule features during query operations. The various methods used to compare image features have been previously described.15 Once the database has been created and features have been extracted, the system is able to respond to a query image by producing a list of images from the database that have been determined to be closest to the query image.
There is also a separate package of classes that comprise the user interface portion of our project, but these are entirely application-specific and will not be discussed. We used the openDICOM.net library for all DICOM file handling.17
Precision
Our initial analysis of the LIDC expert annotations showed significant discrepancies between the observers’ annotations, so we decided to base our calculation of the retrieval precision on the assumption that the first results returned by the system for a particular nodule should be other instances of that same nodule, perhaps on a different CT slice or marked and rated by a different radiologist. Thus, in the absence of subjective physician agreement for all nodules, ground truth was determined by objective, a priori knowledge about the nodules. In this way, we have defined precision as:
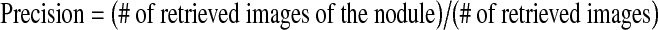 |
We used our system to run a query on each of the 2,424 images of the 141 nodules in the database and examined the mean precision under a variety of conditions:
The queries were run on all nodules in the database with various numbers of items to be retrieved: 1, 2, 3, 5, and 10.
The nodule database was divided into four roughly equal groups based on the size (measured as 2D area) of the nodule images, and precision calculations were run on each group separately with one item retrieved.
The query dataset was restricted to only those nodules on which a certain number of the LIDC viewers gave the nodule the same quantitative “texture” rating (this was given on a five-point scale), and precision calculations were run with one item retrieved.
RESULTS
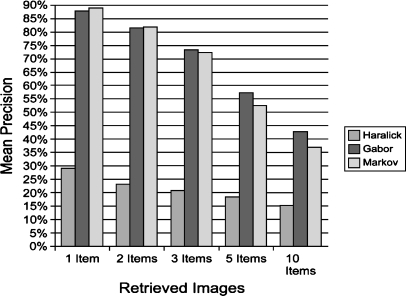
Figure 5 shows that as we vary the number of items retrieved, Gabor and Markov perform nearly identically, with the best mean precision of about 88% when one item is retrieved. The graph also shows that Markov performs similarly to Gabor when fewer than five items are retrieved. However, for five and ten images retrieved, Gabor shows a marked improvement over Markov.
Fig 5.
Mean precision vs. retrieved images.
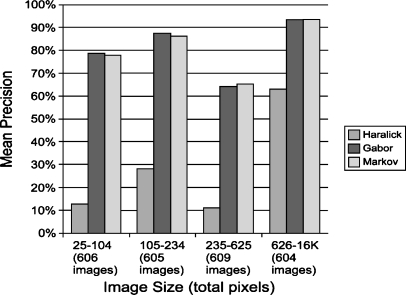
Figure 6 shows the relationship between nodule size and mean retrieval precision. The graph shows that the precision tends to increase for larger images, except for an unexplained decrease in precision in the third group (235–625 total pixels).
Fig 6.
Mean precision vs. image size.
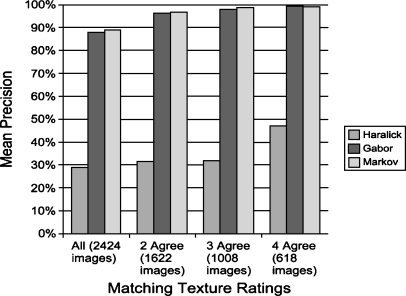
Figure 7 shows the relationship between LIDC expert agreement and the mean precision of image retrieval. When at least two radiologists agreed, the mean precision increased from 88 to 96% for both Gabor and Markov texture models. Once three or four radiologists agreed, the precision increased to nearly 100%.
Fig 7.
Mean precision vs. radiologist agreement.
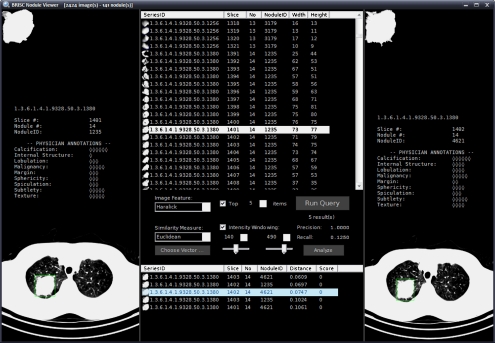
Figure 8 shows a screen capture of the nodule database browser and query interface.
Fig 8.
Nodule viewer.
DISCUSSION
Co-occurrence methods perform noticeably worse than both Gabor and Markov methods with a mean precision of only 29% when retrieving one item. One possible explanation is that the co-occurrence method encodes the texture information at the global (image) level whereas both Gabor and Markov are calculated at the local (pixel) level, which allows for a more robust comparison.
Similarly, Markov and Gabor methods also perform nearly identically and co-occurrence again performs worse when looking at the relationship between lesion size and precision. Generally, these methods appear to perform better on larger images.
Lastly, our results show that as the number of experts in agreement increases so does the precision of the retrieval. This supports a hypothesis that as experts agree on the nature of a lesion the computable descriptors of the lesion become more homogeneous. We have preliminary results that suggest, however, that the features “computed” by the humans and by our software to make similarity decisions are not the same. This is an active area of research.
With respect to open source software, we have developed a system that provides a strong base for research using the LIDC data set. With minor modifications it could also be useful to researchers using other data sets as well. The system was designed to be extensible and easy to use. Because all the feature extraction classes are guaranteed to have an ExtractFeatures method and because standard enumeration methods are implemented for the database class, it is easy to write code that calculates novel features for all nodules in the database. By keeping the logic for the different phases of the image retrieval process in separate modules, we were able to develop the various modules separately and then integrate them for the final project without major difficulties. The modular design also allowed us to automate the retrieval precision calculations.
Recent advances in CT allows for robust extraction and re-assembly of 3D volumes. These advances hold great promise for improving content-based CBIR for lung nodules, because they would increase the sample size of pixels (or in this case “voxels”) for each nodule and reduce errors introduced by inconsistent patient orientation. Our system could be easily extended to include volumetric data analysis, as long as new algorithms are developed to extract and compare features in three dimensions.
CONCLUSIONS
The BRISC Really is Cool (BRISC) project provides a simple base for future work in pulmonary nodule detection and diagnosis. The current design allows for the importing, browsing, and retrieval of lung nodule images from the LIDC database. Local Gabor and Markov methods of texture characterization perform better than global Haralick co-occurrence methods. The precision of image retrieval can be very high, and so this technique has the potential to be useful as an adjunct to radiologist decision making in the context of pulmonary nodules in CT images.
The entire project is available online at http://brisc.sourceforge.net. This work was supported by the National Science Foundation under Grant No.0453456.
Acknowledgements
We thank Dr. Samuel G. Armato III from the University of Chicago, local principal investigator for the Lung Image Data Consortium, for providing an explanation of their database. We also thank Mailan Pham and Ruchaneewan Susomboon for providing their code for the co-occurrence and MRF texture implementation.
Contributor Information
Michael O. Lam, Phone: +1-540-3836971, Email: michael.o.lam@gmail.com
Tim Disney, Email: tim.disney@gmail.com.
Daniela S. Raicu, Phone: +1-312-3625512, FAX: +1-312-3626116, Email: dstan@cti.depaul.edu
Jacob Furst, Phone: +1-312-3625158, FAX: +1-312-3626116, Email: jfurst@cti.depaul.edu.
David S. Channin, Phone: +1-312-9264233, FAX: +1-312-9264220, Email: dsc@northwestern.edu
References
- 1.Henschke CI, McCauley DI, Yankelevitz DF, Naidich DP, McGuinness G, Miettinen OS, Libby DM, Pasmantier MW, Koizumi J, Altorki NK, Smith JP. Early lung cancer action project: overall design and findings from baseline screening. The Lancet. 1999;354:99–105. doi: 10.1016/S0140-6736(99)06093-6. [DOI] [PubMed] [Google Scholar]
- 2.Muller H, Michoux N, Bandon D, Geissbuhler A. A review of content-based image retrieval systems in medical applications - clinical benefits and future directions. International Journal of Medical Informatics. 2004;73(1):1–23. doi: 10.1016/j.ijmedinf.2003.11.024. [DOI] [PubMed] [Google Scholar]
- 3.Ohanian PP, Dubest RC. Performance evaluation for four classes of textural features. Pattern Recogn. 1992;25(8):819. doi: 10.1016/0031-3203(92)90036-I. [DOI] [Google Scholar]
- 4.Shyu C-R, Brodley C, Kak A, Kosaka A, Aisen AM, Broderick LS. Assert:A physician-in-the-loop content-based retrieval system for HRCT image databases. Comput Vis Image Underst. 1999;75(1–2):111–132. doi: 10.1006/cviu.1999.0768. [DOI] [Google Scholar]
- 5.Aisen AM, Broderick LS, Winer-Muram H, Brodley CE, Kak AC, Pavlopoulou C, Dy J, Shyu C-R, Marchiori A. Automated storage and retrieval of thin-section ct images to assist diagnosis: system description and preliminary assessment. Radiology. 2003;228(1):265–270. doi: 10.1148/radiol.2281020126. [DOI] [PubMed] [Google Scholar]
- 6.Smeulders AW, Worring M, Santini S, Gupta A, Jain R. Content-based image retrieval at the end of the early years. IEEE Trans Pattern Anal Mach Intell. 2000;22(12):1349–1380. doi: 10.1109/34.895972. [DOI] [Google Scholar]
- 7.Antani S, Long LR, Thoma GR: Content-based image retrieval for large biomedical image archives. In Proceedings of 11th World Congress on Medical Informatics (MEDINFO) 2004 (September) [PubMed]
- 8.Muller H, Michoux N, Bandon D, Geissbuhler A. A review of content-based image retrieval systems in medical applications—clinical benefits and future directions. International Journal of Medical Informatics. 2004;73(1):1–23. doi: 10.1016/j.ijmedinf.2003.11.024. [DOI] [PubMed] [Google Scholar]
- 9.LIDC Lung Nodule Image Database. National Cancer Imaging Archive https://imaging.nci.nih.gov/ncia/). Accessed 24 May 2007
- 10.The Visualization Toolkit http://www.vtk.org/). Accessed 10 July 2007
- 11.Pieper S, Lorenson B, Schroeder W, Kikinis R: The na-mic kit: Itk, vtk, pipelines,grids and 3d slicer as an open platform for the medical image computing community. In Proceedings of the Third IEEE International Symposium on Biomedical Imaging (ISBI ’06), 2006
- 12.Cleary K. IGSTK: The book. Gaithersburg, MD: Signature Book; 2007. [Google Scholar]
- 13.Prior F: XIP (eXtensible Imaging Platform)-NCI’s open source workstation. RSNA annual meeting and scientific assembly, Chicago, 2006 (November)
- 14.Kim D-Y, Kim J-H, Noh S-M, Park J-W. Pulmonary nodule detection using chest ct images. Acta Radiol. 2003;44:252–257. doi: 10.1034/j.1600-0455.2003.00061.x. [DOI] [PubMed] [Google Scholar]
- 15.Lam M, Disney T, Pham M, Raicu D, Furst J, Susomboon R: Content-based image retrieval for pulmonary computed tomography nodule images. In Proceedings of SPIE 6516, 2007 (March)
- 16.Gamma E, Helm R, Johnson R, Vlissides J. Design patterns:Elements of reusable object-oriented software. Reading, MA: Addison-Wesley; 1995. [Google Scholar]
- 17.openDICOM.NET. Sourceforge http://opendicom.sourceforge.net/). Accessed 24 May 2007.