Abstract
Context
Dilated cardiomyopathy (DCM), a genetically heterogeneous disorder, causes heart failure and rhythm disturbances. The majority of identified DCM genes encode structural proteins of the contractile apparatus and cytoskeleton. Recently, genetic defects in calcium and potassium regulation have been discovered in patients with DCM, implicating an alternative disease mechanism. The full spectrum of genetic defects in DCM, however, has not been established.
Objectives
To identify a novel gene for DCM at a previously mapped locus, define the spectrum of mutations in this gene within a DCM cohort, and determine the frequency of DCM among relatives inheriting a mutation in this gene.
Design, Setting, and Participants
Refined mapping of a DCM locus on chromosome 3p in a multigenerational family and mutation scanning in 156 unrelated pro-bands with DCM, prospectively identified at the Mayo Clinic between 1987 and 2004. Relatives underwent screening echocardiography and electrocardiography and DNA sample procurement.
Main Outcome Measure
Correlation of identified mutations with cardiac phenotype.
Results
Refined locus mapping revealed SCN5A, encoding the cardiac sodium channel, as a candidate gene. Mutation scans identified a missense mutation (D1275N) that cosegregated with an age-dependent, variably expressed phenotype of DCM, atrial fibrillation, impaired automaticity, and conduction delay. In the DCM cohort, additional missense (T220I, R814W, D1595H) and truncation (2550-2551insTG) SCN5A mutations, segregating with cardiac disease or arising de novo, were discovered in unrelated probands. Among individuals with an SCN5A mutation 27% had early features of DCM (mean age at diagnosis, 20.3 years), 38% had DCM (mean age at diagnosis, 47.9 years), and 43% had atrial fibrillation (mean age at diagnosis, 27.8 years).
Conclusions
Heritable SCN5A defects are associated with susceptibility to early-onset DCM and atrial fibrillation. Similar or even identical mutations may lead to heart failure, arrhythmia, or both.
Dilated cardiomyopathy (DCM), an idiopathic form of heart failure and the primary indication for cardiac transplantation,1 has an incidence of 6.0 per 100000 person-years and prevalence of 36.5 per 100000 population.2 Dilated cardiomyopathy is familial in more than 20% of cases,3 implicating genetic defects in single proteins in the disease pathogenesis. Mutations that impair excitation-contraction coupling, which translates electrical excitation of cell membranes into mechanical force production via intracellular calcium, have been demonstrated in DCM and in other heritable cardiac disorders.
Although mutations in genes encoding cardiac ion channels are a molecular substrate for isolated ventricular4–8 and atrial9,10 arrhythmias, the majority of mutations identified in heritable DCM disrupt structural proteins of the cytoskeleton and contractile apparatus.11–14 Mutations in the gene for the nuclear membrane protein lamin A/C, by contrast, have pleiotropic non-cardiac and cardiac manifestations including DCM, atrial fibrillation, and conduction system disease.15,16 The mechanisms by which lamin A/C mutations cause mechanical and/or electrical heart disease are unknown. Notwithstanding, a mechanistic link between heart failure and arrhythmias is further underscored by recent reports of genetic defects in cardiac ion flux in DCM. Recently, altered intra-cellular calcium cycling due to phospholamban mutations was established as a mechanism for familial DCM17; also, genetic defects in the regulatory subunit of the KATP channel that impair membrane potential adjustment in response to metabolic state were identified in patients with DCM.18
In 1986, we reported on a large kindred with autosomal dominant DCM that resembles the phenotype in families later found to have lamin A/C mutations.19 A decade later, we mapped the disease locus to the short arm of chromosome 3 by genetic linkage analysis.20 Thus, another protein, yet to be identified, was implicatedin the pathogenesis of heart failure, arrhythmia, and impaired electrical impulse initiation and propagation. We report herein mutations in SCN5A as a risk factor for heart failure and atrial fibrillation, expanding the spectrum of phenotypic anomalies associated with sodium channel defects.
METHODS
Patients
Probands with idiopathic DCM were identified following referral to a cardiologist. Diagnostic criteria for idiopathic DCM were lack of an identifiable cause for myocardial dysfunction, left ventricular dimensions greater than the 95th percentile indexed for body surface area, and left ventricular shortening fraction less than 28% and/or ejection fraction lessthan 50%, as determined by echocardiography.3 Rhythm and conduction abnormalities were defined using established criteria.20 Probands and their relatives who had enrolled in the study provided written informed consent under protocols approved by the LDS Hospital and/or Mayo Clinic institutional review board.
Medical records and electrocardiograms and echocardiography reports were reviewed. Although some testing of certain family members had been previously performed for clinical indications, most asymptomatic relatives underwent testing for the research study. The clinical description of the multi-generational family used in this study, designated as DC-19 (Figure 1), has been reported.19,20 Phenotypic characterization was updated with additional clinical information and was expanded to include additional family members. Genomic DNA was isolated from peripheral white blood cells. To avoid bias, phenotypic data were interpreted without knowledge of mutation status. Mutations in the known DCM genes encoding cardiac actin,12 α-tropomyosin,21 and metavinculin22 were previously excluded in our cohort of patients with DCM.
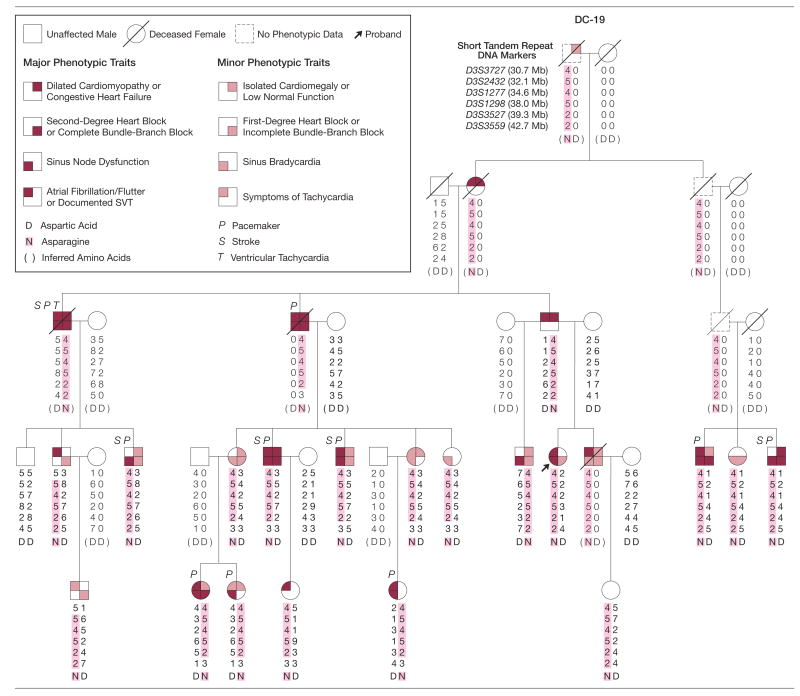
Figure 1.
Pedigree Struture and Clinica Features of Index Family With SCN5A Mutation
Phenotypic traits are variably expressed, designated by shaded quadrants within pedigree symbols. Genotypes for closely spaced DNA markers are shown as numbers, representing different lengths of short tandem repeat marker alleles that distinguish the paternally and maternally inherited chromosomal region. Markers are located within the previously reported disease gene locus on chromosome 3p22-p25. The distance of each marker from the p-arm telomere is indicated by Mb (megabase). The 4 central markers (5-4-5-2) within the shaded chromosome segment define a haplotype, a group of alleles inherited as a unit, common to all family members with cardiac disease. Recombination events indicate that the disease-causing gene resides between markers D3S3727 and D3S3559. SCN5A, located at 38.6-Mb, was investigated as a candidate gene. A point mutation in SCN5A that alters a single amino acid, D1275N, was identified in all affected family members. Mutation-carrier status is indicated by letter symbols for the amino acid at position 1275: aspartic acid (D) in the normal protein and asparagine (N) in the mutant protein. Amino acids shown in parentheses were inferred, together with their corresponding haplotypes. SVT indicates supraventricular tachycardia.
Genotyping and Refined Mapping
Short tandem repeat (STR) DNA markers were identified within the previously identified DCM locus on chromosome 3p in DC-19 (Figure 1). Gel-based genotyping was performed and haplotypes (series of closely spaced markers) were constructed as previously described.20 When possible, genotypes were inferred for individuals from whom DNA samples were not available. Refined mapping was based solely on recombination events in affected family members. (In our previous study SCN5A was erroneously excluded as a positional candidate gene based on a recombination event in an adult classified at the time as unaffected but who has since developed heart disease.20)
Mutation Scanning
Primer pairs for exon-specific PCR amplification of genomic DNA were designed using OLIGO v6.51 Primer Analysis Software (National Biosciences, Plymouth, Minn) and WAVEMAKER version 4.0.32 software (Transgenomic, Omaha, Neb). Primer pairs (n=31) were designed for all 28 translated exons of SCN5A, including an alternatively spliced exon. Heterozygous sequence variants in PCR-amplified DNA fragments were identified by denaturing high-performance liquid chromatography (D-HPLC) heteroduplex analysis (WAVE DNA Fragment Analysis System, Transgenomic), carried out according to the manufacturer’s instructions. Chromatographic elution profiles of amplified fragments were compared against the wild-type homoduplex pattern; samples yielding anomalous traces (Figure 2A) were selected for further analysis by sequencing. Control DNA samples were from a population-based sample (n = 500) with normal electrocardiogram and echocardiogram results.
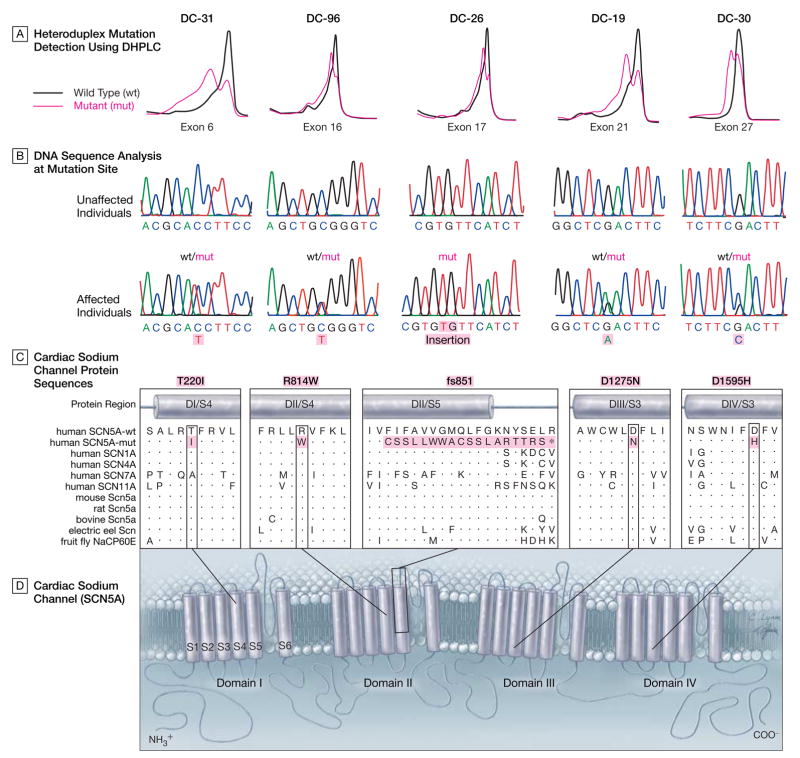
Figure 2.
Identification of SCN5A Mutation and Location of Cardiac Sodium Channel Defects
A, Heteroduplex mutation scans of exons comprising the entire coding region of SCN5A were performed by denaturing high-performance liquid chromatography (DHPLC). Heterozygous variation in DNA sequence was detected in exons 6, 16, 17, 21, and 27 for the 5 family probands in Figure 1 and Figure 3. In contrast to normal exons generating single peaks on chromatographic profiles, exons harboring mutations had anomalous profiles characterized by 2 peaks. B, To determine if detected variations were benign or pathogenic, genomic DNA sequencing was performed. In 4 of the exons (exons 6, 16, 21, and 27), mutations were discovered in 1 copy of the gene, resulting in amino acid substitutions. In the remaining exon (exon 17), insertion of 2 bases disrupts the coding sequence (only the mutant gene is shown). C, Regions in the cardiac sodium channel protein altered by mutations were aligned with protein sequences of other human and nonhuman sodium channels. The 4 missense mutations alter highly conserved amino acids as indicated by vertical boxes wherein identical amino acids are designated by dots. The frameshift mutation creates a series of 18 anomalous amino acids, shown by the horizontal box in the fs851 panel, and an early stop codon, designated by an asterisk. D, Two-dimensional schematic of SCN5A, demonstrating the predicted transmembrane topology. SCN5A is a monomeric channel composed of 4 repeat domains (DI-DIV), each with 5 homologous hydrophobic transmembrane segments (S1-S3, S5, and S6) and 1 positively charged voltage-sensing segment (S4).23 Mutations map to all 4 domains, altering residues within membrane-spanning segments; 2 mutations occur within S4 voltage-sensing segments. The insertion or frameshift mutation severely truncates the protein by removing 13 of 24 transmembrane segments.
Sequencing
Genomic DNA samples selected for sequencing were reamplified by PCR, and products were treated with the PCR Product Pre-sequencing Kit (USB Corp, Cleveland, Ohio). The 2-bp insertion mutant allele was isolated from the wild-type allele following PCR on a 1× MDE gel (FMC Bioproducts, Rock-land, Md). Treated products were sequenced by the dye-terminator method in a core facility, using an ABI PRISM 377 DNA Sequencer (Applied Biosystems, Foster City, Calif). DNA sequence (Figure 2B) was viewed and analyzed using the Sequencher computer program (Gene Codes Corporation, Ann Arbor, Mich). Numbering of amino acids for SCN5A was based on inclusion of an alternatively spliced residue, Q1077.
Protein Structure Analyses
By aligning protein homologs and orthologs to SCN5A, we investigated the conservation of amino acid residues that were altered by the identified missense mutations. The complete sequence of SCN5A was submitted to the PredictProtein server (http://cubic.bioc.columbia.edu/predictprotein) yielding an alignment of the protein sequence against other known proteins in the database (Figure 2C). Mutations were mapped on a 2-dimensional schematic of SCN5A (Figure 2D).23
Genotype-Phenotype Correlation
Correlation between clinical phenotype and SCN5A genotype was determined for unrelated probands in whom an SCN5A mutation was identified, together with their relatives. Targeted sequencing of SCN5A was carried out to determine whether the identified mutation was present in each proband’s family members. The same markers on chromosome 3p used for refined mapping in DC-19 were used to construct haplotypes for these families (Figure 3).
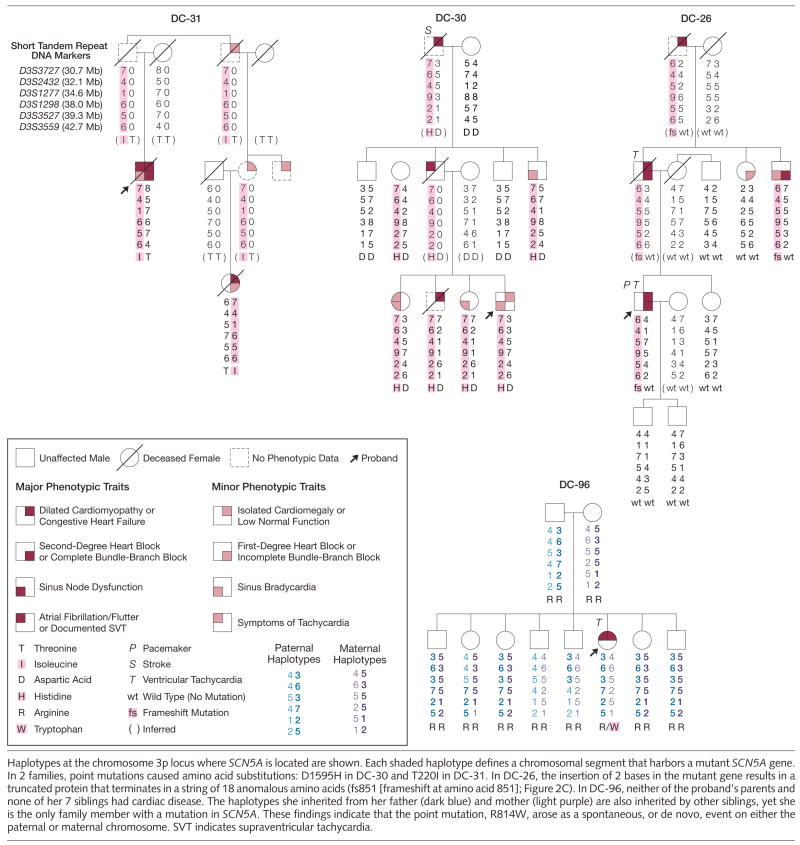
Figure 3.
Pedigree Structures and Clinical Features of Additional Families With SCN5A Mutations
Haplotypes at the chromosome 3p locus where SCN5A is located are shown. Each shaded haplotype defines a chromosomal segment that harbors a mutant SCN5A gene. In 2 families, point mutations caused amino acid substitutions: D1595H in DC-30 and T220I in DC-31. In DC-26, the insertion of 2 bases in the mutant gene results in a truncated protein that terminates in a string of 18 anomalous amino acids (fs851 [frameshift at amino acid 851]; Figure 2C). In DC-96, neither of the proband’s parents and none of her 7 siblings had cardiac disease. The haplotypes she inherited from her father (dark blue) and mother (light purple) are also inherited by other siblings, yet she is the only family member with a mutation in SCN5A. These findings indicate that the point mutation, R814W, arose as a spontaneous, or de novo, event on either the paternal or maternal chromosome. SVT indicates supraventricular tachycardia.
RESULTS
Phenotype of Index Family
Phenotypic data were updated for members of a large kindred (DC-19) who underwent cardiac evaluation since our previous reports (Figure 1).19,20 A 44-year-old woman had developed delayed-onset cardiac disease and was found to have a genetic recombination event that provided critical information for refined mapping of the disease gene locus. At age 29 years with symptoms of lightheadedness and palpitations, she had been classified as unaffected based on normal echocardiogram and electrocardiogram results. Fifteen years later, she developed paroxysmal atrial fibrillation and, after cardioversion, had an underlying rhythm of atrial standstill with junctional escape and incomplete right bundle-branch block.
Systematic evaluation of her family revealed variable, age-dependent penetrance, which resulted in a spectrum of disease manifestations including DCM, diagnosed in 6 family members (mean age, 46 years; range, 29–55 years). Left ventricular ejection fraction (EF) ranged from 25% to 47%, and 4 of the 6 had symptomatic congestive heart failure. Dilated cardiomyopathy was typically preceded by atrial fibrillation, sinus node dysfunction, and conduction block. Cardiac histopathology, available for 4 family members, revealed nonspecific degenerative changes in 2 and no abnormalities in 2 others. Morbidities included congestive heart failure, thromboembolic stroke attributed to atrial fibrillation and/or atrial standstill, and bradyarrhythmias necessitating cardiac pacemaker implantation. There was no history of sudden death and none had corrected QT-interval prolongation or ST-segment elevation, which are characteristic of SCN5A mutations in long QT syndrome4 or Brugada syndrome,5 respectively.
Refined Mapping
Genotyping was performed with polymorphic DNA markers within the 3p locus and haplotypes were constructed (Figure 1). The disease-associated hap-lotype was inherited by 22 clinically affected family members and an unaffected 10-year-old child (included to permit inference of her deceased father’s haplotype). Genetic linkage analysis revealed odds of more than 100 000:1 against spurious linkage to this locus (peak multipoint lod score, 5.8). Genetic recombination events defined a critical region of 12 megabases, flanked by markers D3S3727 and D3S3559. SCN5A was selected as a positional candidate gene based on high cardiac expression and similarity of the family’s cardiac automaticity and conduction abnormalities to other disorders caused by SCN5A mutations.9,24
Mutation Identification
Mutation scanning of all translated exons of SCN5A was carried out by PCR amplification of genomic DNA and denaturing high-performance liquid chromatography (D-HPLC) heteroduplex analysis. An anomalous chromatographic tracing (Figure 2A) identified a sequence variant in exon 21, present in all family members with the disease-associated haplotype from whom a DNA sample was available. Sequence analysis of the reamplified PCR fragment revealed a heterozygous point mutation, G3823A, resulting in a D1275N substitution (Figure 2B). The mutation was not present in 500 unrelated individuals (1000 chromosomes) and it altered a highly conserved amino acid in the SCN5A DIII/S3 transmembrane domain (Figure 2C and 2D). Both D-HPLC and sequence analysis of all remaining exons excluded additional SCN5A mutations in the gene harboring the mutation.
Mutation Scans In DCM Cohort
Comprehensive mutation scans of SCN5A were subsequently carried out in 156 unrelated patients with idiopathic DCM. Distinct mutations in 4 individuals were discovered (Figure 2A and 2B and Figure 3). Sequencing of the exon harboring an SCN5A mutation was then performed for relatives of each proband to determine their mutation-carrier status. Haplotypes were also constructed in each family demonstrating segregation of the mutation and the surrounding chromosomal region or, in the case of DC-96, showing that the mutation had arisen spontaneously in the proband (Figure 3).
None of the probands and their relatives had prolonged QT intervals or ST-segment elevation. To our knowledge, none had been exposed to procain-amide, which might have unmasked the electrocardiographic patterns of Brugada syndrome. Individuals with atrial fibrillation and DCM did not have rapid ventricular heart rates, excluding tachycardia-induced DCM. Similarly, the few family members with ventricular arrhythmia had nonsustained tachycardia based on Holter monitor or serial electrocardiographic recordings or, in 1 case, exhibited persistent DCM after medical control of tachycardia.
Genotype-Phenotype Correlation
In a 54-year-old man (DC-31) with DCM (EF, 15%), atrial fibrillation, and heart block, a heterozygous point mutation, C659T, was identified in exon 6 resulting in a T220I substitution. Coronary artery disease was excluded by angiography, and cardiac biopsy revealed moderate myocyte hypertrophy and marked interstitial fibrosis, consistent with idiopathic DCM. No ventricular tachycardia was observed on telemetry monitoring during a 6-day hospitalization. He died 13 years later in severe congestive heart failure. The mutation was also inherited by a female first cousin, once removed, who at age 55 years was diagnosed with DCM (EF, 10%) and incomplete bundle-branch block. Exercise testing revealed no evidence of ischemia or arrhythmia. She died 2 years later, also in severe congestive heart failure. Other relatives were reported to have enlarged hearts, but additional clinical information and DNA samples were unobtainable.
A second mutation was identified in a 23-year-old woman (DC-96) with sporadic DCM (EF, 30%), atrial flutter, and short runs of nonsustained ventricular tachycardia on 1 of 2 Holter monitor recordings. Cardiac biopsy revealed mild to moderate myocellular hypertrophy and mild interstitial fibrosis. Her parents and 7 siblings (aged from 19–36 years) had normal echo-cardiogram and electrocardiogram results. A heterozygous point mutation, C2440T, was identified in exon 16 resulting in an R814W substitution. The mutation arose de novo, because it was absent in the patient’s parents and siblings and biological paternity and maternity were verified (Figure 3).
A third mutation was identified in a man (DC-26) who at age 32 years developed DCM (EF, 35%) and monomorphic ventricular tachycardia (resolved on medical therapy). Third-degree heart block developed 7 years later, necessitating pacemaker implantation 7 years later. A chest computed tomography scan and stress echocardiography revealed no evidence for either coronary artery or is-chemic heart disease, respectively, and a cardiac biopsy was normal. A 2-bp insertion in exon 17, 2550-2551insTG, caused a frameshift at codon 851, resulting in a premature stop codon and truncation of the encoded protein. The mutation was inherited from his father, diagnosed with DCM (EF, 30%), left bundle-branch block, and monomorphic ventricular tachycardia at age 67 years, shortly before his death. The mutation was also present in his 63-year-old uncle who had sinus bradycardia, first-degree heart block, and complete left bundle-branch block. His paternal grandfather developed congestive heart failure at 50 years of age and died 6 years later, but DNA was unavailable.
A fourth mutation was discovered in a 7-year-old boy (DC-30) with sinus bradycardia, normal contractile function, and left ventricular dilation, an early manifestation of DCM.25,26 A heterozygous point mutation, G4783C, was identified in exon 27 resulting in a D1595H substitution. Analysis of DNA extracted from postmortem paraffin-embedded tissue confirmed presence of the mutation in his brother, who died at 34 years of age with an autopsy diagnosis of cardiomyopathy and only mild coronary artery disease. The mutation was also identified in 2 of his siblings (aged 17 and 24 years) and a paternal uncle (aged 45 years), all of whom had sinus bradycardia, and a paternal aunt (aged 53 years) with borderline left atrial enlargement. His father, an obligate mutation carrier, had atrial fibrillation and died at 49 years of age from pulmonary embolism following a surgical procedure. His paternal grandfather, a presumed mutation carrier, developed congestive heart failure at 70 years of age and died from a stroke 8 years later. Each identified mutation was absent in 500 unrelated individuals and altered highly conserved residues in transmembrane domains of SCN5A (Figure 2C and 2D).23 Mutation scans identified no additional mutations in the coding region of SCN5A in probands with a defined mutation.
In our study, 37 individuals in 5 families with available clinical data carried an SCN5A mutation. Isolated cardio-megaly and/or low-normal systolic function, considered early markers of DCM,25,26 were present in 27% of mutation carriers, with a mean age of 20.3 years at diagnosis. DCM was present in 38% of carriers, with mean age at diagnosis similar to that reported for patients with familial and sporadic forms of DCM (47.9 vs 45.2 years).3 Collectively, 65% of carriers had a cardiomyopathy phenotype. Atrial fibrillation was documented in 43% of individuals with an SCN5A mutation (mean age at diagnosis, 27.8 years). Sex was not observed to influence phenotypic expression.
COMMENT
Mutations in SCN5A have been previously discovered in 6 cardiac rhythm disorders: long QT syndrome,4 Brugada syndrome,5 idiopathic ventricular fibrillation,6 nonfamilial arrhythmia susceptibility,7 conduction system disease,24 and sick sinus syndrome.9 These conditions are characterized by ventricular tachyarrhythmia, heart block, or atrial bradyarrhythmia. We identified SCN5A mutations associated with atrial fibrillation, providing further evidence that ventricular and atrial tachyarrhythmias can be allelic disorders.10,27
Susceptibility to myopathic heart disease in patients with inherited and de novo heterozygous SCN5A mutations, now established in this report, may have gone unrecognized because of inherent selection bias in previous studies. Individuals and families with identified SCN5A mutations were usually studied based on diagnosis of an idiopathic rhythm disorder, with coexisting structural heart disease normally used as an exclusionary criterion.5,6,10,23 Moreover, studies have not typically reported cardiac dimensions and function in patients with rhythm disorders caused by SCN5A mutations.4,9,24 Systematic screening for SCN5A mutations in a DCM cohort has not been previously reported.
Our genetic data coupled with existing knowledge provide compelling evidence and a physiologic rationale for SCN5A dysfunction conferring risk for heart failure in DCM. First, intracellular sodium homeostasis is vital for myocellular function,28 including regulation of pH and intracellular calcium, the key mediator of excitation-contraction coupling. Indeed, high fidelity beat-to-beat regulation of intracellular calcium flux is essential for normal cardiac function, and altered calcium cycling due to phospholamban mutations has been established recently as a mechanism for familial DCM.17 We speculate that reduced sodium entry into myocytes could lead to compensatory increases in activities of the sodium-calcium exchanger (opposite the effect of digitalis) and sodium-hydrogen exchanger, resulting in decreased intracellular calcium and pH, respectively,28,29 and thus impairing calcium-mediated myocellular force production and/or mitochondrial energy production. Second, sodium channel agonists increase cardiac contractility,30 whereas certain class 1A and 1C antiarrhythmic drugs that block sodium channels can depress contractile function.31 Third, mutations in the homologous skeletal muscle sodium channel gene, SCN4A, cause a muscle disorder characterized by episodic weakness and, in some patients, progressive degenerative myopathy analogous to cardiac muscle disease in DCM.32 Fourth, 2 isolated cases of cardiomyopathy have been reported recently in small families with rhythm disorders who had heterozygous33 or compound heterozygous34 mutations in SCN5A.
Each of our identified mutations would be predicted to cause loss of function of the cardiac sodium channel, based on previous reports of identical or similar mutations. The same T220I and D1275N substitutions were reported as recessive allelesinsicksinussyndrome9 andfamilial atrial standstill,35 respectively. The de novo R814W substitution here identified in DII/S4 is exactly analogous in positiontoan R225W substitution in DI/S4, 34 identified in 2 siblings with compound heterozygosity (R225W and W156X mutations) and conduction system disease; one had cardiomyopathy on autopsy. The D1595H substitution is similar to a previously reported D1595N substitution in autosomal dominant conduction system disease.36 A frameshift and truncation mutation (V1397X) distal to the frameshift we identified was reported in Brugada syndrome.5 Collectively, in vitro biophysical analyses of these 5 mutations, identical or similar to those reported herein, resulted in loss of channel function with reduced sodium current.
The mechanisms by which similar or identical loss of function mutations in SCN5A lead to variable expression of heart disease remain unknown. Our data, together with previous reports, do not support mutation-specific genotype-phenotype correlations that predict either electrical or myopathic phenotypes. Indeed, it has been shown that the same mutation can cause either long QT syndrome, Brugada syndrome, or conduction abnormalities within the same family37 and that subtle in vitro changes in channel function can be associated with severe cardiac phenotypes.38 Extremes of heart rate have been proposed as a potential factor in variable phenotypic expression,38 raising the possibility that the bradycardia observed in many of our patients may have influenced development of DCM. In mice, heterozygous deletion of Scn5a leads to conduction system disease and ventricular arrhythmias, as observed in humans, at 8 to 10 weeks.39 If analogous to humans with SCN5A mutations, who develop DCM as middle-aged adults, ventricular dysfunction would not be expected to occur until much later in the 2-year lifespan of these mice. Targeted disruption of both Scn5a alleles causes severe defects in cardiac morphogenesis and embryonic lethality, precluding postnatal assessment of cardiac phenotype. Further mechanistic insight into the myopathic phenotype in humans associated with SCN5A mutations will require expression of these mutations in living animal models. The in vivo effects of SCN5A mutations leading to heart failure in DCM are likely modulated by additional genetic, environmental, and stochastic factors within the heterogeneous milieu of the cardiac myocyte.
Our findings uncover a novel molecular defect in DCM and further implicate impaired ion homeostasis in the patho-genesisofheartfailure.17,18 Together with LMNA15,16 and ABCC9,18 SCN5A may be a candidate for comprehensive genetic testing in the future, targeting patients with a syndrome of DCM and rhythm or conduction disturbances. Like patients with Brugada syndrome, avoidance of sodium channel–blocking drugs in patients with DCM and SCN5A mutations may prove to be prudent. Our findings provide a rationale for longitudinal studies of patients with isolated rhythm disorders caused by loss of SCN5A function, to determine whether they are also at risk for DCM.
Acknowledgments
Funding/Support: This work was supported by grant R01 HL71225 from the National Heart, Lung, and Blood Institute, National Institutes of Health; Miami Heart Research Institute; Marriott Program for Heart Disease Research; Siragusa Foundation; Mayo-Dubai Healthcare City Research Project; and the Mayo Foundation (Dr Olson).
Footnotes
Author Contributions: Dr Olson had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Olson, Ballew, Karst, Anderson.
Acquisition of data: Olson, Michels, Ballew, Reyna, Karst, Herron, Horton, Rodeheffer, Anderson.
Analysis and interpretation of data: Olson, Ballew, Karst, Anderson.
Drafting of the manuscript: Olson, Ballew, Reyna, Karst, Herron, Horton, Anderson.
Critical revision of the manuscript for important intellectual content: Olson, Michels, Ballew, Reyna, Rodeheffer.
Obtained funding: Olson.
Administrative, technical, or material support: Olson, Michels, Ballew, Reyna, Karst, Herron, Horton, Andersonf.
Study supervision: Olson, Anderson.
Role of the Sponsors: The sponsors had no role in the design and conduct of the study, in the collection, management, analysis, and interpretation of the data, or in the preparation, review, or approval of the manuscript.
References
- 1.Manolio TA, Baughman KL, Rodeheffer RJ, et al. Prevalence and etiology of idiopathic dilated cardiomyopathy (summary of a National Heart, Lung, and Blood Institute workshop) Am J Cardiol. 1992;69:1458–1466. doi: 10.1016/0002-9149(92)90901-a. [DOI] [PubMed] [Google Scholar]
- 2.Codd MB, Sugrue DD, Gersh BJ, Melton LJ., III Epidemiology of idiopathic dilated and hypertrophic cardiomyopathy: a population-based study in Olmsted County, Minnesota, 1975–1984. Circulation. 1989;80:564–572. doi: 10.1161/01.cir.80.3.564. [DOI] [PubMed] [Google Scholar]
- 3.Michels VV, Moll PP, Miller FA, et al. The frequency of familial dilated cardiomyopathy in a series of patients with idiopathic dilated cardiomyopathy. N Engl J Med. 1992;326:77–82. doi: 10.1056/NEJM199201093260201. [DOI] [PubMed] [Google Scholar]
- 4.Wang Q, Shen J, Splawski I, et al. SCN5A mutations associated with an inherited cardiac arrhythmia, long QT syndrome. Cell. 1995;80:805–811. doi: 10.1016/0092-8674(95)90359-3. [DOI] [PubMed] [Google Scholar]
- 5.Chen Q, Kirsch GE, Zhang D, et al. Genetic basis and molecular mechanism for idiopathic ventricular fibrillation. Nature. 1998;392:293–296. doi: 10.1038/32675. [DOI] [PubMed] [Google Scholar]
- 6.Akai J, Makita N, Sakurada H, et al. A novel SCN5A mutation associated with idiopathic ventricular fibrillation without typical ECG findings of Brugada syndrome. FEBS Lett. 2000;479:29–34. doi: 10.1016/s0014-5793(00)01875-5. [DOI] [PubMed] [Google Scholar]
- 7.Splawski I, Timothy KW, Tateyama M, et al. Variant of SCN5A sodium channel implicated in risk of cardiac arrhythmia. Science. 2002;297:1333–1336. doi: 10.1126/science.1073569. [DOI] [PubMed] [Google Scholar]
- 8.Splawski I, Shen J, Timothy KW, et al. Spectrum of mutations in long-QT syndrome genes. KVLQT1, HERG, SCN5A, KCNE1, KCNE2. Circulation. 2000;102:1178–1185. doi: 10.1161/01.cir.102.10.1178. [DOI] [PubMed] [Google Scholar]
- 9.Benson DW, Wang DW, Dyment M, et al. Congenital sick sinus syndrome caused by recessive mutations in the cardiac sodium channel gene (SCN5A) J Clin Invest. 2003;112:1019–1028. doi: 10.1172/JCI18062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Y-H, Xu S-J, Bendahhou S, et al. KCNQ1 gain-of-function mutation in familial atrial fibrillation. Science. 2003;299:251–254. doi: 10.1126/science.1077771. [DOI] [PubMed] [Google Scholar]
- 11.Towbin JA, Hejtmancik JF, Brink P, et al. X-linked dilated cardiomyopathy: molecular genetic evidence of linkage to the Duchenne muscular dystrophy (dystrophin) gene at the Xp21 locus. Circulation. 1993;87:1854–1865. doi: 10.1161/01.cir.87.6.1854. [DOI] [PubMed] [Google Scholar]
- 12.Olson TM, Michels VV, Thibodeau SN, Tai YS, Keating MT. Actin mutations in dilated cardiomyopathy, a heritable form of heart failure. Science. 1998;280:750–752. doi: 10.1126/science.280.5364.750. [DOI] [PubMed] [Google Scholar]
- 13.Schonberger J, Seidman C. Many roads lead to a broken heart: the genetics of dilated cardiomyopathy. Am J Hum Genet. 2001;69:249–260. doi: 10.1086/321978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Towbin JA, Bowles NE. The failing heart. Nature. 2002;415:227–233. doi: 10.1038/415227a. [DOI] [PubMed] [Google Scholar]
- 15.Fatkin D, MacRae C, Sasaki T, et al. Missense mutations in the rod domain of the lamin A/C gene as causes of dilated cardiomyopathy and conduction-system disease. N Engl J Med. 1999;341:1715–1724. doi: 10.1056/NEJM199912023412302. [DOI] [PubMed] [Google Scholar]
- 16.Taylor MR, Fain PR, Sinagra G, et al. Natural history of dilated cardiomyopathy due to lamin A/C gene mutations. J Am Coll Cardiol. 2003;41:771–780. doi: 10.1016/s0735-1097(02)02954-6. [DOI] [PubMed] [Google Scholar]
- 17.Schmitt JP, Kamisago M, Asahi M, et al. Dilated cardiomyopathy and heart failure caused by a mutation in phospholamban. Science. 2003;299:1410–1413. doi: 10.1126/science.1081578. [DOI] [PubMed] [Google Scholar]
- 18.Bienengraeber M, Olson TM, Selivanov VA, et al. ABCC9 mutations identified in human dilated cardiomyopathy disrupt catalytic KATP channel gating. Nat Genet. 2004;36:382–387. doi: 10.1038/ng1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greenlee PR, Anderson JL, Lutz JR, Lindsay AE, Hagan AD. Familial automaticity-conduction disorder with associated cardiomyopathy. West J Med. 1986;144:33–41. [PMC free article] [PubMed] [Google Scholar]
- 20.Olson TM, Keating MT. Mapping a cardiomyopathy locus to chromosome 3p22-p25. J Clin Invest. 1996;97:528–532. doi: 10.1172/JCI118445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olson TM, Kishimoto NY, Whitby FG, Michels VV. Mutations that alter the surface charge of alpha-tropomyosin are associated with dilated cardiomyopathy. J Mol Cell Cardiol. 2001;33:723–732. doi: 10.1006/jmcc.2000.1339. [DOI] [PubMed] [Google Scholar]
- 22.Olson TM, Illenberger S, Kishimoto NY, Huttel-maier S, Keating MT, Jockusch BM. Metavinculin mutations alter actin interaction in dilated cardiomyopathy. Circulation. 2002;105:431–437. doi: 10.1161/hc0402.102930. [DOI] [PubMed] [Google Scholar]
- 23.Tan HL, Bezzina CR, Smits JPP, Verkerk AO, Wilde AAM. Genetic control of sodium channel function. Cardiovasc Res. 2003;57:961–973. doi: 10.1016/s0008-6363(02)00714-9. [DOI] [PubMed] [Google Scholar]
- 24.Probst V, Hoorntje TM, Hulsbeek M, et al. Cardiac conduction defects associate with mutations in SCN5A. Nat Genet. 1999;23:20–21. doi: 10.1038/12618. [DOI] [PubMed] [Google Scholar]
- 25.Baig MK, Goldman JH, Caforio ALP, Coonar AS, Keeling PJ, McKenna WJ. Familial dilated cardiomyopathy: cardiac abnormalities are common in asymptomatic relatives and may represent early disease. J Am Coll Cardiol. 1998;31:195–201. doi: 10.1016/s0735-1097(97)00433-6. [DOI] [PubMed] [Google Scholar]
- 26.Michels VV, Olson TM, Miller FA, Ballman KV, Rosales G, Driscoll DJ. Frequency of development of idiopathic dilated cardiomyopathy among relatives of patients with idiopathic dilated cardiomyopathy. Am J Cardiol. 2003;91:1389–1392. doi: 10.1016/s0002-9149(03)00341-2. [DOI] [PubMed] [Google Scholar]
- 27.Wang Q, Curran ME, Splawski I, et al. Positional cloning of a novel potassium channel gene: KVLQT1 mutations cause cardiac arrhythmias. Nat Genet. 1996;12:17–23. doi: 10.1038/ng0196-17. [DOI] [PubMed] [Google Scholar]
- 28.Pieske B, Houser SR, Hasenfuss G, Bers DM. Sodium and the heart: a hidden key factor in cardiac regulation. Cardiovasc Res. 2003;57:871–872. doi: 10.1016/s0008-6363(02)00849-0. [DOI] [PubMed] [Google Scholar]
- 29.Bers DM, Barry WH, Despa S. Intracellular Na+ regulation in cardiac myocytes. Cardiovasc Res. 2003;57:897–912. doi: 10.1016/s0008-6363(02)00656-9. [DOI] [PubMed] [Google Scholar]
- 30.Flesch M, Erdmann E. Na+ channel activators as positive inotropic agents for the treatment of chronic heart failure. Cardiovasc Drugs Ther. 2001;15:379–386. doi: 10.1023/a:1013329203750. [DOI] [PubMed] [Google Scholar]
- 31.Zipes DP. Management of cardiac arrhythmias: pharmacological, electrical, and surgical techniques. In: Braunwald E, editor. Heart Disease: A Textbook of Cardiovascular Medicine. 5. Philadelphia, Pa: WB Saunders Co; 1997. pp. 597–610. [Google Scholar]
- 32.Plassart E, Reboul J, Rime CS, et al. Mutations in the muscle sodium channel gene (SCN4A) in 13 French families with hyperkalemic periodic paralysis and paramyotonia congenita: phenotype to genotype correlations and demonstration of the predominance of two mutations. Eur J Hum Genet. 1994;2:110–124. doi: 10.1159/000472351. [DOI] [PubMed] [Google Scholar]
- 33.Chen S, Chung MK, Martin D, Rozich R, Tchou PJ, Wang Q. SNP S1103Y in the cardiac sodium channel gene SCN5A is associated with cardiac arrhythmias and sudden death in a white family. J Med Genet. 2002;39:913–915. doi: 10.1136/jmg.39.12.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bezzina CR, Rook MB, Groenewegen WA, et al. Compound heterozygosity for mutations (W156X and R225W) in SCN5A associated with severe cardiac conduction disturbances and degenerative changes in the conduction system. Circ Res. 2003;92:159–168. doi: 10.1161/01.res.0000052672.97759.36. [DOI] [PubMed] [Google Scholar]
- 35.Groenewegen WA, Firouzi M, Bezzina CR, et al. A cardiac sodium channel mutation cosegregates with a rare connexin40 genotype in familial atrial standstill. Circ Res. 2003;92:14–22. doi: 10.1161/01.res.0000050585.07097.d7. [DOI] [PubMed] [Google Scholar]
- 36.Wang DW, Viswanathan PC, Balser JR, George AL, Benson DW. Clinical, genetic, and biophysical characterization of SCN5A mutations associated with atrioventricular conduction block. Circulation. 2002;105:341–346. doi: 10.1161/hc0302.102592. [DOI] [PubMed] [Google Scholar]
- 37.Grant AO, Carboni MP, Neplioueva V, et al. Long QT syndrome, Brugada syndrome, and conduction system disease are linked to a single sodium channel mutation. J Clin Invest. 2002;110:1201–1209. doi: 10.1172/JCI15570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Viswanathan PC, Balser JR. Inherited sodium channelopathies: a continuum of channel dysfunction. Trends Cardiovasc Med. 2004;14:28–35. doi: 10.1016/j.tcm.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 39.Papadatos GA, Wallerstein PM, Head CE, et al. Slowed conduction and ventricular tachycardia after targeted disruption of the cardiac sodium channel gene SCN5a. Proc Natl Acad Sci U S A. 2002;99:6210–6215. doi: 10.1073/pnas.082121299. [DOI] [PMC free article] [PubMed] [Google Scholar]