Abstract
The rodent malaria parasite Plasmodium yoelii has been an important animal model for studying malaria pathology and host-parasite interactions. Compared with other rodent malaria parasites such as Plasmodium chabaudi, however, genetic mapping studies on P. yoelii have been limited, partly due to the absence of genetic markers and the lack of well characterized phenotypes. Taking advantage of the available genome sequence, we initiated a project to develop a high-resolution microsatellite (MS) map for P. yoelii to study malaria disease phenotypes. Here we report screening the P. yoelii genome for simple sequence repeats and development of an inexpensive method (modified from a previously reported procedure) for typing malaria parasite MS: instead of labeling individual polymerase chain reaction primers, a single fluorescently labeled primer was used to type the MS markers. We evaluated various polymerase chain reaction cycling conditions and M13-tailed/labeled M13 primer ratios to establish a simple and robust procedure for typing P. yoelii MS markers. We also compared typing efficiencies between individually labeled primers and the M13-tailed single labeled primer method and found that the two approaches were comparable. Preliminary analyses of seven P. yoelii isolates deposited at MR4 with 77 MS showed that the markers were highly polymorphic and that the isolates belonged to two groups, suggesting potential common ancestry or laboratory contaminations among the isolates. The MS markers and the typing method provide important tools for genetic studies of P. yoelii. There is a good possibility that this method can be applied to type MS from other malaria parasites including important human pathogens Plasmodium falciparum and Plasmodium vivax.
Keywords: Genetic mapping, Genotyping, Rodent malaria parasite, Simple sequence repeat
1. Introduction
Microsatellites (MS) are simple sequence repeats (SSR) that have been shown to be highly variable and are widely used as genetic markers for genetic mapping and genotyping in many organisms, including malaria parasites [1–4]. In the Plasmodium falciparum parasite, MS are extremely abundant and are highly polymorphic, providing a powerful tool for genetic studies [2,5–8]. Although MS are present in the genome of the rodent malaria parasite Plasmodium yoelii [9], whether they are polymorphic or suitable for genetic markers have not previously been evaluated.
Generally, MS have 1–4 base pairs (bp) of repeat units and are typed using polymerase chain reaction (PCR) with primers flanking the repeat units. The PCR products are separated on DNA sequencing gels because of small sizes of the repeating units. To visualize the PCR products, one primer is usually labeled with fluorescent dyes or rarely with radioactive isotopes (typically P32); or the PCR products can be stained with reagents such as silver particles [10]. Labeling PCR products with fluorescently labeled primers has several advantages over the radioactive labeling or silver staining methods, including accurate product size calling and potential for high-throughput operation [11]; however, MS typing using fluorescently labeled primers can be expensive, requiring special equipment (DNA analyzers) to separate PCR products and fluorescently labeled primers that may prevent laboratories with limited budgets from typing large numbers of MS markers.
One alternative to individually labeled primers is to use a third primer labeled with a fluorescent dye [12–14]. In this method, one of the two PCR primers contains a ‘tail’—a unique sequence such as M13 universal primer sequence—in addition to the specific sequence matching the conserved sequence at one side of the MS repeat. PCR products from the amplification will contain the tail after the initial amplification cycle. A third primer with the tail sequence (M13 universal primer sequence) is then 5′ labeled with a fluorescent dye and included in the reaction, leading to incorporation of fluorescent labels into the PCR products (Supplemental Fig. 1). Unfortunately, this method does not always work well for all MS, and special cycling conditions are used for different markers [12]. Indeed, conflicting results have been reported using M13-tailed primers and individually labeled primer methods [15,16]. In one study comparing tailed primers and individually labeled primers, it was concluded that addition of the M13 tail greatly improved the specificity [15]; in another study, however, the M13-tailed method showed inconsistent performance in amplifying a plant genome (Pinus taeda) [16]. Although these differences could be attributed to the differences in genome sizes and genome compositions, it could not be ruled out that the discrepancy was due to differences in PCR setup and cycling conditions. It is clear, however, that some optimization may be necessary for obtaining consistent results using the M13-tailed primer method.
In our effort to develop a genetic map for the rodent malaria parasite P. yoelii, we have evaluated various methods for typing MS from the parasite. We compared a M13-tailed primer method with a method using individually labeled primers, evaluated different parameters in labeling PCR products with a single fluorescently labeled primer, and developed a reliable procedure for typing P. yoelii MS. Using the MS markers, we genotyped DNA from seven P. yoelii isolates and one Plasmodium berghei parasite and found that the P. yoelii parasites belonged to two groups. We conclude that the M13_tailed primer method is an economic and reliable method for typing MS from the AT-rich P. yoelii genome. To our knowledge, this work also represents the first study of MS from this rodent malaria parasite.
2. Materials and methods
2.1. Parasites and DNA extraction
P. yoelii parasites were obtained from MR4 (http://www.malaria.mr4.org/). P. yoelii parasites were thawed from frozen stocks and were injected directly into BALB/c mice under an NIH-approved animal usage protocol (#LMVR85). DNA samples were extracted from parasitized blood samples using a commercial blood sample extraction kit (Qiagen).
2.2. MS and oligonucleotide primer synthesis
MS were screened and identified by searching the P. yoelii genome sequences downloaded from the TIGR website (http://www.tigr.org/tdb/e2k1/pya1/). Potential MS such as TA(n), TAA(n), and other types of repeats were identified using the ‘find’ function in the Ward program and Repeat Finder [17] (http://tandem.bu.edu/trf/trf.html). Oligonucleotide primers based on the DNA sequences flanking the SSR were commercially synthesized. Because the P. yoelii genome is relatively AT rich [18], we selected primer sequences following some simple rules: Oligonucleotides of 20-mers with six GC were first selected. If no sequences at the specific flanking regions met the requirement, the primers could be lengthened (up to 25-mer if five GC or fewer) or shortened (18-mer if more than eight GC). The majority of primers designed following these rules had annealing temperatures of approximately 50°C (± 5°C).
2.3. PCR setups and cycling programs
The initial report on the M13-tailed primer method used various complicated PCR cycling programs for different primer pairs [12]. We compared several PCR cycling programs, taking into account the high AT content of our MS, and used an extension temperature of 60°C for all the cycling programs. The details of the cycling programs can be found in Supplemental Table 1. The simplest cycling program (cycling protocol 4) includes 94°C, 2 min for initial denaturation; 94°C, 20 sec; 50°C, 20 sec; 60°C, 30 sec for 40 cycles; and a final extension at 60°C for 2 min. PCR mixture in 15 μl volume contained 10 μl master PCR mix, 3 μl DNA (~50 pg), 1 μl reverse primer and M13-tailed primers (2 μM reverse primer and 1 μM M13-tailed primer ), and 1 μl D4-PA- or D3-PA-labeled M13 primer (1 μM, Sigma-Aldrich, St. Louis, MO). The master PCR mix contained 0.5 μl (1 U/μl) Tag polymerase, 0.3 μl dNTP (10 mM), 2.4 μl MgCl2 (25 mM), 1.5 μl 10× PCR buffer with (NH4)2SO4, and ddH2O to 10 μl. PCR setups for typing MS using individually labeled primers were the same as the procedures described except that equal amounts of labeled forward primer and unlabeled reverse primer (2 μM) were included in the reactions.
2.4. MS calling and MS data analysis
PCR products were either separated on a 2% agarose gel or resolved in a capillary gel matrix using a Beckman CEQ8000 DNA analyzer (Beckman Coulter, Fullerton, CA). For capillary gel electrophoresis, PCR product sizes were called using size standards provided by the company (Size Standard Kit 400). MS allele sizes were imported into an Excel sheet. Before analyses, the MS alleles were formatted according to instructions provided in MSAnalyzer [19] (http://i122server.vu-wien.ac.at/MSA/MSA_download.html). Genetic distance matrices were obtained with 1000 bootstrap replicates. The output files from MSAnalyzer were imported into ‘neighbor’ module in PHYLIP 3.6 (http://evolution.genetics.washington.edu/phylip.html) to generate an outtree file that was itself imported into ‘consense’ in PHYLIP 3.6 to obtain a consensus tree. To draw a tree, the consensus treefile was opened with ‘drawtree’ in PHYLIP 3.6 and plotfiles from ‘drawtree’ was displayed in Canvas 9.0.
3. Results
3.1. Screening MS from the P. yoelii genome
We screened and identified potential MS sequences from the P. yoelii genome sequence. Because the genome is relatively AT rich, we initially focused on searching for MS with AT(n) and other AT-related repeats and identified more than 600 candidate MS sequences (data not shown). We synthesized primer pairs based on the sequences flanking 77 P. yoelii MS that were identified in our initial screening. In addition, an individually fluorescent-labeled primer for each of 25 randomly selected MS from the 77 potential MS markers and a fluorescently labeled M13 primer (5′Dye-CACGACGTTGTAAAACGAC-3′) were also commercially synthesized. The primer sequences, allele sizes, numbers of alleles among tested parasites, and chromosomal localizations of these markers are listed in Table 1.
Table 1.
Microsatellite markers tested in this study and polymorphic alleles among seven Plasmodium yoelii isolates and Plasmodium berghei k173
| Marker | Forward primer | Reverse primer | Chr. | Size (bp) | Number of alleles |
|---|---|---|---|---|---|
| Py1003 | 5′AAATATAGAAACACCTCTGT3′ | 5′AAAAGAAATTCTGGTACTATT3′ | 9 | 158 | 4 |
| Py1006-1 | 5′AGCAAGATGAGTAGCTAC3′ | 5′CAGAGCTTAAGTAGTTGC3′ | 6 | 157 | 2(0) |
| Py1015 | 5′CAATAACTACTTATCCAGC3′ | 5′AGGAATGGAAGGAATGCT3′ | ? | 119 | 2(0) |
| Py1063 | 5′ATATCCAGCAGAAGGAGA3′ | 5′TCCATTCCATAAGCTTCC3′ | 9 | 127 | 4* |
| Py1084 | 5′GCATATCCAGTGACGCTA3′ | 5′CAATCGATTTGGGTGCTA3′ | 13 | 160 | 3 |
| Py1121 | 5′TGTTCTCTGACTACAGCA3′ | 5′CAACATACAAACATGTACC3′ | 6 | 222 | 2(0) |
| Py1188 | 5′ATTATTTCGAGGGATATACT3′ | 5′CAACAATTGCTGTTGCCA3′ | 4 | 185 | 1(0,0) |
| Py1193 | 5′AATGAATAAGGACGTGGTA3′ | 5′CTTTTCACACTAAGAAATCA3′ | ? | 184 | 3* |
| Py1199 | 5′TTACCTCTGTATAATGGCA3′ | 5′TTGCATAGATGAAGAAACAT3′ | 14 | 156 | 3 |
| Py1213 | 5′AGGAGGGAACAATAGGTA3′ | 5′TTGTCTGATAACATTGAATC3′ | 11 | 159 | 4 |
| Py1308 | 5′GAAATGAGCGCATTACCA3′ | 5′CAACTATAGTATGCTTACC3′ | 2 | 205 | 2(0) |
| Py1331 | 5′GGTCAATAATTCGGATTCT3′ | 5′TAGCACGTTGAATCTGGT3′ | 12 | 207 | 2(0) |
| Py1346 | 5′ACAGAACCGTCCTTCGAA3′ | 5′GATGTTACGAGTTGTGGA3′ | 1 | 125 | 3 |
| Py1356 | 5′TTTACGGTTACTTTGGGCA3′ | 5′TGTGTATCTAGCTATCTAAT3′ | 11 | 165 | 3 |
| Py1385 | 5′AGCCAAGCAAGAAACAGA3′ | 5′TTATGTACTTCTTCTCTGTA3′ | 9 | 105 | 2(0) |
| Py1414 | 5′AACAAAACAGGTAGCAACA3′ | 5′ATCATGGTTTATATTCACAG3′ | 4 | 207 | 2(0) |
| Py1418-1 | 5′TTACTGTCTATATATCACGT3′ | 5′AATAGACGTGCATATGGC3′ | 11 | 198 | 2(0) |
| Py1432 | 5′ACCCTTAATTAAAGGAAGC3′ | 5′AGAGAGAGAAATTGTAATTC3′ | 11 | 137 | 2 |
| Py1481 | 5′TCATAGGGTAAGAATAAAGT3′ | 5′TAGTCCATTGTGCAAAGG3′ | 9 | 130 | 1(0,0) |
| Py1516 | 5′TTAAAATGTCGTAAAGCAAC3′ | 5′AAGGGAACATTTCATGTGT3′ | 1 | 122 | 3 |
| Py1535 | 5′AACGGATCCTGATGGAAA3′ | 5′ACTGTTTGTGTTACTAATCA3′ | 9 | 238 | 3 |
| Py1556 | 5′AAGGAATTTTATATTCTCTGA3′ | 5′GTATATAGCAAGGTTATCG3′ | 9 | 165 | 1(0,0) |
| Py1615 | 5′GGACCAAATAAGTCACCA3′ | 5′CTATTGGATTAGGAATATCT3′ | 4 | 140 | 2(0) |
| Py1659 | 5′GAATCACGAAAGCAAAGAA3′ | 5′CCAAGTTTATATCCTTACTA3′ | 9 | 134 | 3 |
| Py1710 | 5′GAATAAGTTGATATCTAAGC3′ | 5′TAGTAATACTACCAAGCGT3′ | ? | 152 | 2(0) |
| Py174 | 5′AATTCAACGCCAATACCAA3′ | 5′GATCCCAGTAGCATTACA3′ | 7 | 104 | 2(0) |
| Py1836 | 5′CTTACTATTCGCAAAGTTG3′ | 5′ATCTTCTTGTAGGTTTCGA3′ | 10 | 172 | 2(0) |
| Py1841 | 5′TGGTAAATCATATGAAGAAC3′ | 5′CTATTCAAATATGCAGAGAT3′ | 14 | 145 | 3 |
| Py1858 | 5′GTCAAACAATAAAAACAGAG3′ | 5′CAAAACTAGCGAATGTTGA3′ | 13 | 171 | 3 |
| Py1861 | 5′GCATACACATATGTTGAGA3′ | 5′ACGGATGGAAACATTACAA3′ | ? | 112 | 3* |
| Py1873 | 5′CATTAAACCAACATGGGAT3′ | 5′GCAAATAAAACAGCTTGTC3′ | 4 | 195 | 2(0) |
| Py1885-1 | 5′TGAGCTAAACAAAAAGGCT3′ | 5′TTCTGTCTCAAGTGTGTC3′ | 9 | 115 | 1(0,0) |
| Py1902 | 5′TCGGGATTCCAATACTTG3′ | 5′TCGAGATTGTGCATGTATT3′ | ? | 222 | 1(0,0) |
| Py1914 | 5′GGAGACCTATAAATAAATAC3′ | 5′GGAGCAATAGAGAATGTAT3′ | 1 | 147 | 2(0) |
| Py1925 | 5′ATTATGTGATCTAGGATACA3′ | 5′GCTTAGTGGAGAACGTCA3′ | 6 | 124 | 2(0) |
| Py2081 | 5′AAGTATCACAATTTGCAAGA3′ | 5′CTAGAAATGTAGAGAATCTA3′ | 13 | 146 | 3 |
| Py2092 | 5′CATTTGTAAATCGTAGAAAG3′ | 5′GGCTTATCAAAAATAAGCAT3′ | 13 | 170 | 3 |
| Py2101 | 5′TAACACGAGCGCACCAAA3′ | 5′TTGTATGGACATATTCATCA3′ | 11 | 143 | 3 |
| Py2181 | 5′ATATTGTGTAAAAGTATTGCT3′ | 5′AATAAATGTATATGCAATTTGT3′ | 4 | 128 | 2* |
| Py2246 | 5′ATTTAGTGTACTTTCGGTG3′ | 5′TGTTGTTTTTACGCGACAA3′ | 4 | 146 | 3 |
| Py2355 | 5′GCAAAGCAAATATCAAGGA3′ | 5′AAAACGTATGTGGGCACA3′ | 11 | 192 | 2(0) |
| Py2370 | 5′ATTCATTTGTATATTCCCATT3′ | 5′TTATCCAACGGTGTTTTATT3′ | 8 | 176 | 2 |
| Py2391 | 5′ATTGACCTTAGCATAATAGT3′ | 5′TATCACGAGTTATGAGCAT3′ | 10 | 189 | 1(0,0) |
| Py2401 | 5′ATAATGGTAAAATAGAGGGT3′ | 5′AAAACGGTGAGAATAATGAA3′ | 13 | 218 | 3 |
| Py2423 | 5′CTTATTCAGACGGAACATT3′ | 5′GCAAACACTAGTATAGCAT3′ | 13 | 135 | 3* |
| Py2426 | 5′TGGGAACCTTAACATGATA3′ | 5′TCTCATTAATCATATTTGCTA3′ | 1 | 142 | 2(0) |
| Py2455 | 5′GAAAATATCGAATGGAGAAA3′ | 5′GAGAGCTAAACTAAACGC3′ | 9 | 125 | – |
| Py2572 | 5′TGAATTGTAGATTTGGAGTT3′ | 5′TATATGCATGACCATGCAT3′ | 10 | 158 | – |
| Py260 | 5′TGTACACATTGTGACATGT3′ | 5′TTCTCTCAATTATGATATACT3′ | 14 | 170 | 2(0) |
| Py2609 | 5′CTCATATTTAAGTTTTTCTCT3′ | 5′TAGGAAGACGATCTCGCT3′ | 13 | 183 | 3 |
| Py2673 | 5′CGTATACGAATATCCTTCA3′ | 5′CCAAAGGGAAAGAAACTG3′ | 2 | 140 | 2(0) |
| Py2742 | 5′TGCCAAGCAAATTCAGTG3′ | 5′TAACTGCACACAACATTAG3′ | 4 | 164 | 3 |
| Py279 | 5′ATGCATGTGTACGCACAT3′ | 5′TAGGGAATGCATATGCATC3′ | 13 | 160 | 3 |
| Py282 | 5′ATGAGAGGCACAGAATATT3′ | 5′ATTGTATACTTAAAATCCCTT3′ | 10 | 187 | 3* |
| Py2830 | 5′TTGGGGTTCCATTTTACAT3′ | 5′TAACAGCGGTCTCGCTAC3′ | 7 | 128 | 3(0) |
| Py348 | 5′GTATACGCGCAAAGTGGA3′ | 5′TCCTGGTTAGCTCTATGA3′ | 8 | 171 | 2(0) |
| Py353 | 5′TTGAAGGGGTTGACAGGT3′ | 5′GAATAAAGATAACTAACTGG3′ | 14 | 189 | 3(0) |
| Py378 | 5′CGAATGTTTTCAATCCCTT3′ | 5′TTCGTATTTGCTATTCATGT3′ | 2 | 214 | 3 |
| Py385 | 5′TCCAATTCCAAGTGTATAG3′ | 5′AAATGTGTTAATATGGCGAT3′ | 2 | 176 | 1(0,0) |
| Py453 | 5′GGTTGGAACACTAAAACG3′ | 5′AGCTGGAATTACCGACTG3′ | 10 | 89 | 2(0) |
| Py455 | 5′CCAAAACCCCAAAAAATAC3′ | 5′TGAATTCAATATCCATGCAA3′ | ? | 237 | 2*(0) |
| Py468 | 5′AAAGCAGCACAGTAGCTA3′ | 5′TGAAAATATAAGCATGGGAA3′ | 6 | 94 | 4 |
| Py477 | 5′ACTTGATAAATGCGATATGA3′ | 5′GAATTTCTTTACGTCCACA3′ | 3 | 122 | 3(0) |
| Py517-2 | 5′TTCAAACGTGATATCTCATT3′ | 5′CGATTAATGTGGAAATATGT3′ | 13 | 150 | 4 |
| Py523 | 5′AACTCTTATGTAATGTATGC3′ | 5′TCTTAACAAATTTCAAGAGC3′ | 13 | 161 | 3* |
| Py630 | 5′TACCCATACGATTTATGCT3′ | 5′TACTAACATGTACAGTTCAT3′ | 12 | 133 | 1(0,0) |
| Py658 | 5′CAATCGATCAATATCTCGA3′ | 5′GTATCAATCAAAATGTTCAC3′ | 8 | 140 | 2(0) |
| Py72 | 5′ATATATGTATCCTTGTATGTT3′ | 5′ATAACAACATATACAATGTTG3′ | 8 | 241 | – |
| Py747 | 5′AAGACTAAGACGGCTATG3′ | 5′TTTCCATTAATTGTGTGCAT3′ | 9 | 197 | – |
| Py762 | 5′ACAGGTTGGCTACATCAA3′ | 5′TAATGCCAATGATAGGCAA3′ | 9 | 221 | 2(0) |
| Py763 | 5′AAGAGTAGATAGATGACCA3′ | 5′TTTATGCTGATAAAGACCAT3′ | 14 | 111 | 1(0,0) |
| Py826 | 5′AATGTGCTTCATAAACATGT3′ | 5′TATACATGTAAAGACGTGTT3′ | 9 | 178 | 3 |
| Py845-3 | 5′GATGTGAATGGATGCGAA3′ | 5′ATTACATGACTTGCAACCA3′ | 3 | 153 | 2(0) |
| Py928 | 5′TTTTAGATTGACATTAGTCTT3′ | 5′CGCACACTATATATTACTG3′ | 11 | 173 | 3 |
| Py951 | 5′TCAAGCATAACACAATAGTA3′ | 5′TCACTTCCTGAATCAATCA3′ | 12 | 149 | 1(0) |
| Py952 | 5′TTGGTCCAAAAATATGTTGT3′ | 5′TTTTGTATGCACGTGATAG3′ | ? | 173 | 3 |
| Py984 | 5′TGGGTATGTCCATCACTT3′ | 5′TATCCTATGCCTTACTTAC3′ | 8 | 189 | 1(0) |
Note: Chr, chromosome; Size, allele sizes in base pairs based on Py-17XNL genome sequence; Number of alleles, numbers of alleles observed among the seven P. yoelii isolates and P. berghei; (0) indicates no product from P. berghei; (0,0), indicates no products from P. berghei and one of the two P. yoelii groups; * indicates more than one allele in one of the parasites when typed with individually labeled primers; ?, chromosomal location could not be determined. Markers in bold were also tested using individually labeled primers; and those underlined were markers that did not produce any PCR products in all cycling protocols.
Chromosome assignment of the markers was based on reference [26].
3.2. Optimization of a M13-tailed primer PCR method
Although a M13-tailed primer PCR method described previously [12] worked reasonably well, some of the procedures were long and complicated, containing multisegments in cycling programs; results from those studies did not always agree with each other [15,16]. The biggest advantage of the tailed-primer method compared with individually labeled primer is cost savings: only one fluorescently labeled primer is required for all the MS to be typed, rather than one labeled primer for each MS. Because we intended to develop hundreds of MS markers for genetic studies, it was critical for us to develop a simple, reliable, and affordable MS typing method. We therefore evaluated various PCR cycling conditions to improve and/or to simplify the M13-tailed MS typing assay. We reasoned that the critical elements for a successful tailed PCR would include properly designed primers with similar annealing temperatures, optimal cycling conditions, and a best labeled:tailed primer ratio. We followed simple rules in primer design and designed all the primers with similar annealing temperatures at approximately 50°C.
We tested and compared four different cycling programs (Supplemental Table 1), including those described previously [12,14] with some modifications to improve the amplification efficiency of the AT-rich MS in amplifying P. yoelii DNA. PCR products were separated in both agarose gels and capillary electrophoresis using a Beckman CEQ8000 DNA analyzer. In our hands, the four cycling conditions produced similar results (Fig. 1; Table 2). These results suggested that cycling programs (numbers and sequence of cycling steps) might not be critical as long as correct annealing temperatures and well-designed primers were used. Because our MS were mostly AT repeats, having a lower extension temperature at 60°C may be necessary for polymerase to extend through the AT rich regions [20]. Among the 52 pairs of primers, 4 did not produce any PCR products in the four cycling programs, suggesting that failure to amplify was likely due to problems in primer design rather than cycling programs (Table 1).
Fig. 1.

Agarose gels showing PCR products amplified using four different cycling protocols (Supplemental Table 1). Panel A are products using protocol 1; Panel B, protocol 2; Panel C, protocol 3; and Panel D, protocol 4. Names of different microsatellites are on top of the gel images. Four μl of the PCR products were loaded onto 2% agarose gels and stained with ethidium bromide.
Table 2.
Success rates of four different cycling programs in amplifying P. yoelii microsatellites
| Cycling methods | No. markers typed | No. markers w/bands | No. markers w/o bands | Success rate (%) |
|---|---|---|---|---|
| Protocol 1 | 52 | 48 | 4 | 92.3 |
| Protocol 2 | 52 | 48 | 4 | 92.3 |
| Protocol 3 | 52 | 48 | 4 | 92.3 |
| Protocol 4 | 52 | 48 | 4 | 92.3 |
We next compared different tailed:labeled primer ratios to improve fluorescent signals. Stronger bands were generally produced with increasing the amount of M13-tailed primer when PCR products were separated on agarose gels and stained with ethidium bromide (Fig. 2); however, better fluorescent signals were detected when the ratios of labeled M13 primer to M13-tailed primers were from 1:1 to 4:1 (Table 3). We therefore used a ratio of 1:1 for labeled M13 primer vs. M13-tailed primers in our reactions.
Fig. 2.
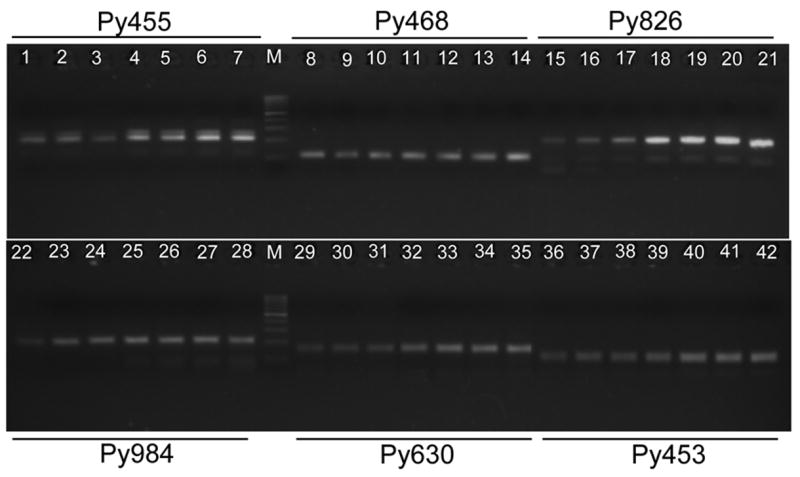
PCR products from six P. yoelii microsatellites amplified using different ratios of fluorescently labeled M13 primer and M13-tailed primer. For each microsatellite, products from fluorescently labeled M13 primer vs. M13-tailed primer ratios of 9:1 (lanes 1, 8, 15, 22, 29, 36), 4:1 (lanes 2, 9, 16, 23, 30, 37), 2:1 (lanes 3, 10, 17, 24, 31, 38), 1:1 (lanes 4, 11, 18, 25, 32, 39), 1:2 (lanes 5, 12, 19, 26, 33, 40), 1:4 (lanes 6, 13, 20, 27, 34, 41), and 1:9 (lanes 7, 14, 21, 28, 35, 42) were separated on 2% agarose gels. M, molecular weight markers (100-bp ladder). Generally, brighter bands were obtained in reactions with higher concentrations of M13-tailed primers.
Table 3.
Fluorescent signals obtained using various ratios of fluorescently labeled M13 primer:M13-tailed primer
| Ratio | |||||||
|---|---|---|---|---|---|---|---|
| Markers | 9:1 | 4:1 | 2:1 | 1:1 | 1:2 | 1:4 | 1:9 |
| Py455 | 28000 | 31500 | 13500 | 28000 | 14500 | 3500 | 400 |
| PY468 | 110000 | 135000 | 140000 | 132500 | 84000 | 46000 | 10200 |
| Py826 | 25000 | 60000 | 92000 | 130000 | 92000 | 38000 | 7200 |
| Py984 | 48000 | 30000 | 54000 | 13000 | 4200 | 2300 | 220 |
| Py630 | 24500 | 45000 | 40000 | 59000 | 43000 | 19000 | 8600 |
| Py453 | 37000 | 54000 | 47000 | 40000 | 24000 | 12000 | 4200 |
Numbers in bold indicate the highest signals. These MS were randomly selected from the 77 MS tested in this study.
3.3. Comparison of M13-tailed primer method with individually labeled primer method
To investigate how this cost-saving method compared with those using individually labeled primers in amplifying P. yoelii MS, we labeled the forward primers from 25 P. yoelii MS with D2-PA dye (Sigma-Aldrich) (Table 1; Supplemental Table 2). Comparison of the signals from PCR products using individually labeled primers and M13-tailed/labeled primers showed that the two methods were similar, although the signals from individually labeled primers were generally stronger than those from M13-tailed primers (Fig. 3). These results suggested that the M13-tailed primer method was as reliable as that using individually labeled primers in amplifying P. yoelli MS, except MS Py2830, which did not produce good signals using M13-tailed primer (Supplemental Table 2). Of interest, some MS (Py282, Py523, Py1861, and Py2423) had two alleles in some of the parasites when typed with individually labeled primers but only one allele each when typed with M13-tailed primers, suggesting improved specificity (also see Discussion). We concluded that the M13-tailed primer method is a reliable MS typing technique comparable to that using individually labeled primers. A simple PCR cycling program such as protocol 3 or 4 described in this paper with a PCR setup having a labeled M13 primer to M13-tailed primer ratio of 1:1, 2:1 or 4:1 should produce good results for the amplification of P. yoelii MS.
Fig. 3.
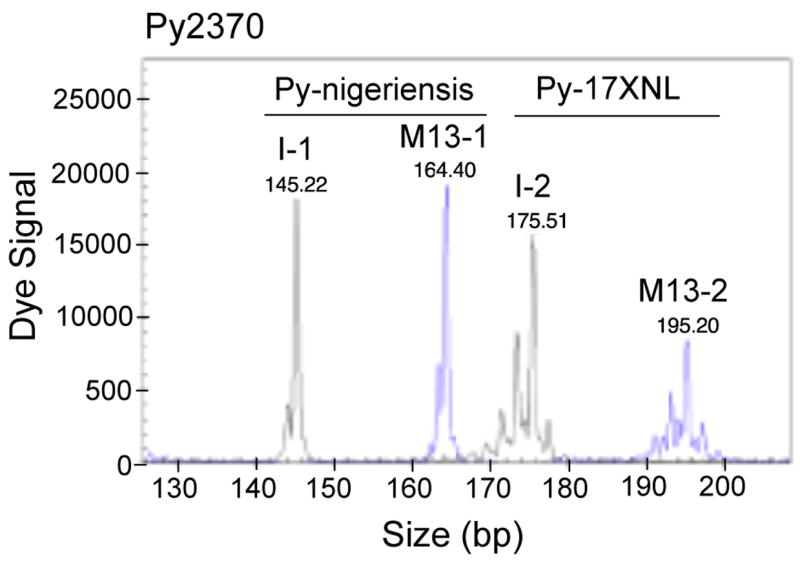
Comparison of dye signals from individually labeled primer and M13-tailed primer methods. PCR products of microsatellite Py2370 amplified using the two methods were mixed and loaded into the same capillary (0.2 μl PCR product for I-1, 0.6 μl for M13-1, 0.7 μl for I-2, and 1.6 μl for M13-2), and peaks of signals were printed directly from the DNA analyzer (Beckman CEQ8000). As expected, although signals from individually labeled primers were ~3 times stronger than those from M13-tailed/labeled primers, the signals from the M13-tailed primers were sufficient for allele calling. Weaker signals from the M13-tailed method can be compensated by loading a slightly larger amount of the PCR products. Peaks M13-1 and M13-2 were signals from M13-tailed primer method, and peaks I-1 and I-2 were from the individually labeled primer method. DNA samples were as labeled. Note that the patterns of the peaks were almost identical in both methods.
3.4. Highly polymorphic MS among P. yoelii isolates and P. berghei
To perform genetic studies, we needed to identify parasites that have different genetic backgrounds with various phenotypes. We therefore analyzed MS alleles from seven P. yoelii isolates deposited at MR4 (http://www.malaria.mr4.org/) and DNA from a P. berghei. Among the 77 MS typed, four (Py72, Py747, Py2572, and Py2455) did not produce any PCR products; two (Py951 and Py984) had only one allele among the P. yoelii isolates (monomorphic); nine produced PCR products in some isolates; and 62 had more than one allele (polymorphic, 80.5%) among the P. yoelii isolates (Table 1; Supplemental Table 2). Interestingly, all of the nine MS that produced PCR products in partial isolates had products from Py-17XNL, Py-17XL, and Py-312 only. These three parasites were closely related, but were quite different from the other four parasites (see below). The results suggested that no amplification in the other four parasites were likely due to polymorphism in primer sequences. If we consider those MS that did not produce PCR products in some parasites as polymorphic markers because of polymorphism at primer sequences, we have 92.2% of the MS being polymorphic among a limited number of isolates (basically two parasites; see below). For P. berghei, only 34 MS (44.2%) produced PCR products, with only 3 MS sharing the same alleles with some of the P. yoelii parasites. These results showed that MS from P. yoelii are highly polymorphic and can become useful markers for genetic studies.
3.5. P. yoelii isolates at MR4 belong to two major groups
Genetic distances between each pair of parasites typed with 25 MS markers (in bold, Table 1) were calculated using MSAnalyzer [19], and a distance matrix was produced and imported into ‘neighbor’ and ‘consense’ in PHYLIP 3.6 to generate a consense ‘tree’ (Fig. 4). Clearly, the parasites belonged to two major groups, with four parasites in one group (Py-Kenya, Py-NS, Py-238, and Py-nigeriensis) being almost identical. These parasites likely derive from a single laboratory-adapted parasite or simply contamination in the labs. The other group contains parasites Py-17XNL, Py-17XL, and Py-312. Although they were closely related, differences in some of the MS were observed. The P. berghei parasite was placed at the middle between the two groups, but the real distance of the P. berghei parasite should be larger than that shown in the tree, because approximately 56% of the markers could not be amplified using primers synthesized based on P. yoelii sequences.
Fig. 4.
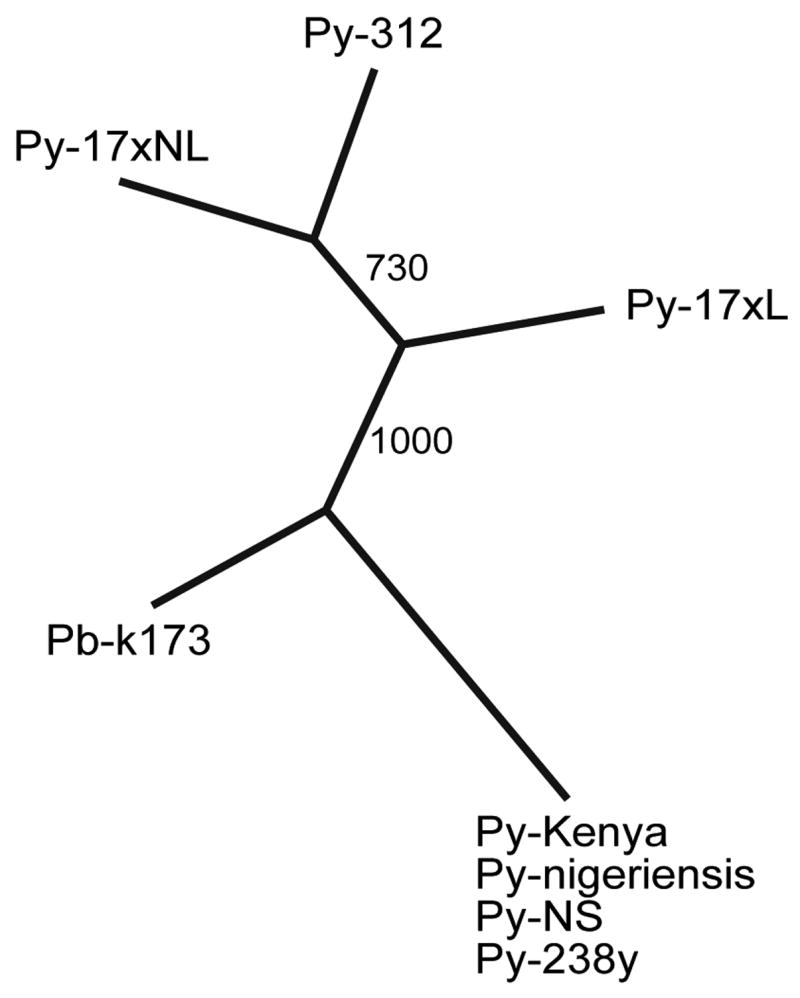
A neighbor joining ‘tree’ showing the relationship of seven Plasmodium yoelii isolates and one Plasmodium berghei parasite. The numbers are bootstrap values from 1000 replicates. The MR4 catalog numbers for each of the parasites are: Py-NS (MRA-418), Py-nigeriensis (MRA-427), Py-238y (MRA-642), Py-Kenya (MRA-428), Py-17XL (MRA-680), Py-17XNL (MRA-593), and Py-312 (MRA-312). Pb-k173 was a laboratory stock at Sun Yat-sen University, China, which was originally shipped from Dr. David Walliker’s laboratory at the University of Edinburgh, UK.
4.Discussion
P. yoelii was the first rodent malaria parasite whose genome was sequenced, because the parasite has been an important model for studies of host immunity against malaria parasites [9]. Like other rodent malaria parasites (Plasmodium chabaudi and P. berghei) [21,22], P. yoelii can become a useful malaria model for studying disease pathogenesis, virulence, and other traits using genetic crosses and linkage mapping if genetic markers and parasites with different phenotypes are available. Parasite isolates or strains that can cause different disease phenotypes —for example Py-17XNL (non-lethal), Py-17XL (lethal), or other isolates—can be exploited for studies of potential virulence factors. In this report, we describe preliminary results from our efforts to develop a genetic map to study important traits of P. yoelii parasites. Hundreds of MS have been identified from the parasite genome (data not known) and are being evaluated for use as genetic markers. Our results show that although MS are not as abundant as in the P. falciparum genome, reasonable numbers of polymorphic MS could be developed from the P. yoelii genome for genetic studies.
Our preliminary results from typing DNA samples of P. yoelii isolates deposited at MR4 showed that the majority of the P. yoelii isolates belonged to two groups (Fig. 4). The parasites in each group could be due to contamination while being adapted in the laboratories, as has been reported for P. falciparum [23–25]. Parasite Py-Kenya, Py-NS, Py-238, and Py-nigeriensis are basically identical, with 61 of the 62 (98.4%) MS typed being the same among the parasites (Supplemental Table 2). These parasites were likely derived from a single ancestor. Likewise, Py-17XNL, Py-17XL, and Py-312 are also very similar with only seven out of 73 (9.6%) MS showing differences in allele sizes. These differences are likely changes accumulated during in vivo passages after they were cloned and separated. Our results suggest that there is a need to genotype all the P. yoelii parasites being used in different laboratories so that experimental results can be properly evaluated or compared.
Typing MS requires labeling PCR products and separating the PCR products in high-resolution matrices such as DNA sequencing gels so that the small differences in PCR products (2–3 bp) can be detected. The common practice is to fluorescently label one PCR primer and detect the PCR product in a DNA analyzer. Unfortunately, fluorescent labeling a primer costs $70–$150 per oligonucleotide, which can be expensive if large numbers of primers are required in a study. Here we have refined a cost-saving method for typing large numbers of MS using a single fluorescently labeled primer. Because the procedure uses three PCR primers with different annealing temperatures, it has been difficult to develop a protocol that would work well for all the PCR primer pairs. In this study, we evaluated several PCR cycling programs and primer ratios, combined with primers of similar annealing temperatures, and developed a simple and robust method for typing P. yoelii MS.
Although the results were not always exactly the same, our M13-tailed primer method produced results comparable to those obtained using the individually labeled primer method (Supplemental Table 2). One interesting observation was that several MS produced two alleles in some parasites when typed with individually labeled primers, whereas only one allele was detected using the M13-tailed primer method. In particular, Py523 produced two alleles (159 and 132 bp) when Py-17XNL DNA was amplified with individually labeled primer but only one allele (180 bp) was present when amplified with M13-tailed primer method (Supplemental Table 2). Blast searches of the available P. yoelii genome sequences with the primer sequences for Py523 found only one match for each of the primers on contig MALPY00523, suggesting a single allele in the genome. Additionally, MS Py2423 had PCR products much smaller than the predicted size (39–45 bp smaller, possibly non-specific products) when amplified using the individually labeled primer method, but had a product with the predicted size when amplified using M13-tailed primer method. These results suggest that the M13-tailed primer method, in some cases, may improve specificity as suggested previously [15].
One potential problem associated with MS typing is the accuracy in estimating the sizes of PCR products, which are not always consistent with the sizes predicted from the genome sequences. The M13 primer was expected to add 19 nucleotides to the PCR products. Comparison of allele sizes from the predicted Py-17XNL genome sequences and those estimated from our Py-17XNL showed that the majority of 73 MS producing PCR products had alleles with added sizes of 15–22 bp (93%; average 17.4 bp), with two MS adding 14 bp, one adding 13 bp, one adding11 bp, one adding 6 bp, and one adding 4 bp using the M13-tailed method (Supplemental Table 2). The individually labeled primer method had allele size variation from −3 to 3 bp (mean −0.3 bp) when the estimated allele sizes were subtracted by the predicted allele sizes. The size variations could be due to inaccuracy in size estimates associated with the CEQ8000 machine and its software or real-size variations in the MS between parasite ‘strains’. If we consider the five MS that had differences 5 bp or larger between the predicted and estimated sizes representing real polymorphisms between the parasite ‘strains’, the accuracies of size estimates are very similar between the two methods. Indeed, this is the conclusion if the variations in size estimates from the 25 markers tested using both methods are compared. Nonetheless, absolute size estimate is not critical for applications such as typing progeny from a genetic cross or isolates from the field, because what is important in those cases is the relative, not the absolute, sizes of the markers. Despite the variation in PCR product size estimate, we believe that this M13-tailed method is a reliable and economic method for typing malaria parasite MS markers.
Supplementary Material
Acknowledgments
This work was supported by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health and by a special fund for graduate education from Xiamen University, People’s Republic of China. We thank Drs. David Walliker and Jane Carlton for advice and discussions on history and origins of various rodent malaria parasites, Dr. Deirdre Joy for advice on MSAnalyzer and PHYLIP, and NIAID intramural editor Brenda Rae Marshall for assistance.
Abbreviations
- bp
base pair(s)
- MS
microsatellite(s)
- PCR
polymerase chain reaction
- SSR
simple sequence repeat
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tautz D, Renz M. Simple sequences are ubiquitous repetitive components of eukaryotic genomes. Nucleic Acids Res. 1984;12:4127–38. doi: 10.1093/nar/12.10.4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Su X-z, Ferdig MT, Huang Y, et al. A genetic map and recombination parameters of the human malaria parasite Plasmodium falciparum. Science. 1999;286:1351–3. doi: 10.1126/science.286.5443.1351. [DOI] [PubMed] [Google Scholar]
- 3.Ferdig MT, Su X-z. Microsatellite markers and genetic mapping in Plasmodium falciparum. Parasitol Today. 2000;16:307–12. doi: 10.1016/s0169-4758(00)01676-8. [DOI] [PubMed] [Google Scholar]
- 4.Imwong M, Pukrittayakamee S, Renia L, et al. Novel point mutations in the dihydrofolate reductase gene of Plasmodium vivax: evidence for sequential selection by drug pressure. Antimicrob Agents Chemother. 2003;47:1514–21. doi: 10.1128/AAC.47.5.1514-1521.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson TJ, Haubold B, Williams JT, et al. Microsatellite markers reveal a spectrum of population structures in the malaria parasite Plasmodium falciparum. Mol Biol Evol. 2000;17:1467–82. doi: 10.1093/oxfordjournals.molbev.a026247. [DOI] [PubMed] [Google Scholar]
- 6.Roper C, Pearce R, Bredenkamp B, et al. Antifolate antimalarial resistance in southeast Africa: a population-based analysis. Lancet. 2003;361:1174–81. doi: 10.1016/S0140-6736(03)12951-0. [DOI] [PubMed] [Google Scholar]
- 7.Nair S, Williams JT, Brockman A, et al. A selective sweep driven by pyrimethamine treatment in southeast asian malaria parasites. Mol Biol Evol. 2003;20:1526–36. doi: 10.1093/molbev/msg162. [DOI] [PubMed] [Google Scholar]
- 8.Roper C, Pearce R, Nair S, et al. Intercontinental spread of pyrimethamine-resistant malaria. Science. 2004;305:1124. doi: 10.1126/science.1098876. [DOI] [PubMed] [Google Scholar]
- 9.Carlton JM, Angiuoli SV, Suh BB, et al. Genome sequence and comparative analysis of the model rodent malaria parasite Plasmodium yoelii yoelii. Nature. 2002;419:512–9. doi: 10.1038/nature01099. [DOI] [PubMed] [Google Scholar]
- 10.Bobba A, Marra E, Fathallah DM, Giannattasio S. Non-radioactive detection of five common microsatellite markers for ATP7B gene in Wilson disease patients. Mol Cell Probes. 2003;17:271–4. doi: 10.1016/s0890-8508(03)00065-3. [DOI] [PubMed] [Google Scholar]
- 11.Oda S, Oki E, Maehara Y, Sugimachi K. Precise assessment of microsatellite instability using high resolution fluorescent microsatellite analysis. Nucleic Acids Res. 1997;25:3415–20. doi: 10.1093/nar/25.17.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oetting WS, Lee HK, Flanders DJ, et al. Linkage analysis with multiplexed short tandem repeat polymorphisms using infrared fluorescence and M13 tailed primers. Genomics. 1995;30:450–8. doi: 10.1006/geno.1995.1264. [DOI] [PubMed] [Google Scholar]
- 13.Wildenberg SC, Oetting WS, Almodovar C, et al. A gene causing Hermansky-Pudlak syndrome in a Puerto Rican population maps to chromosome 10q2. Am J Hum Genet. 1995;57:755–65. [PMC free article] [PubMed] [Google Scholar]
- 14.Boutin P, Hani EH, Vasseur F, et al. Automated fluorescence-based screening for mutation by SSCP: use of universal M13 dye primers for labeling and detection. Biotechniques. 1997;23:358–62. doi: 10.2144/97233bm01. [DOI] [PubMed] [Google Scholar]
- 15.Boutin-Ganache I, Raposo M, Raymond M, Deschepper CF. M13-tailed primers improve the readability and usability of microsatellite analyses performed with two different allele-sizing methods. Biotechniques. 2001;31:24–8. [PubMed] [Google Scholar]
- 16.Zhou Y, Bui T, Auckland LD, Williams CG. Direct fluorescent primers are superior to M13-tailed primers for Pinus taeda microsatellites. Biotechniques. 2002;32:46–52. doi: 10.2144/02321bm05. [DOI] [PubMed] [Google Scholar]
- 17.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–80. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carlton J, Silva J, Hall N. The genome of model malaria parasites, and comparative genomics. Curr Issues Mol Biol. 2005;7:23–37. [PubMed] [Google Scholar]
- 19.Dieringer D, Schlötterer C. Microsatellite analyser (MSA): a platform independent analysis tool for large microsatellite data sets. Molecular Ecology Notes. 2003;3:167–9. [Google Scholar]
- 20.Su X-z, Wu Y, Sifri CD, Wellems TE. Reduced extension temperatures required for PCR amplification of extremely A+T-rich DNA. Nucleic Acids Res. 1996;24:1574–5. doi: 10.1093/nar/24.8.1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinelli A, Cheesman S, Hunt P, et al. A genetic approach to the de novo identification of targets of strain-specific immunity in malaria parasites. Proc Natl Acad Sci U S A. 2005 doi: 10.1073/pnas.0405097102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janse CJ, Franke-Fayard B, Mair GR, et al. High efficiency transfection of Plasmodium berghei facilitates novel selection procedures. Mol Biochem Parasitol. 2006;145:60–70. doi: 10.1016/j.molbiopara.2005.09.007. [DOI] [PubMed] [Google Scholar]
- 23.Wootton JC, Feng X, Ferdig MT, et al. Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature. 2002;418:320–3. doi: 10.1038/nature00813. [DOI] [PubMed] [Google Scholar]
- 24.Mu J, Awadalla P, Duan J, et al. Recombination hotspots and population structure in Plasmodium falciparum. PLoS Biol. 2005;3:e335. doi: 10.1371/journal.pbio.0030335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Volkman SK, Sabeti PC, DeCaprio D, et al. A genome-wide map of diversity in Plasmodium falciparum. Nat Genet. 2007;39:113–9. doi: 10.1038/ng1930. [DOI] [PubMed] [Google Scholar]
- 26.Kooij TW, Carlton JM, Bidwell SL, et al. A Plasmodium whole-genome synteny map: indels and synteny breakpoints as foci for species-specific genes. PLoS Pathog. 2005;1:e44. doi: 10.1371/journal.ppat.0010044. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


