Abstract
Cell surface oligosaccharides have been shown to play essential biological roles in such diverse biological phenomena as cellular adhesion, molecular recognition, and inflammatory response. The development of high-affinity ligands capable of selectively recognizing a variety of small motifs in different oligosaccharides would be of significant interest as experimental and diagnostic tools. As a step toward this goal we have developed DNA ligands that recognize the disaccharide cellobiose, whether in soluble form or as the repeating unit of the polymer, cellulose. These DNA “aptamers” bind with high selectivity to cellobiose with little or no affinity for the related disaccharides lactose, maltose, and gentiobiose. Thus, the DNA ligands can discriminate sugar epimers, anomers, and disaccharide linkages.
Keywords: systemic evolution of ligands by exponential enrichment, aptamer, cellulose, saccharide
Oligosaccharide antigens play essential biological roles in cellular adhesion, molecular recognition, and inflammatory response. Classical carbohydrate binding macromolecules are proteins. These include plant and animal lectins (1–3) and anticarbohydrate antibodies raised in animals and humans (4). The ability to discriminate the presence of different saccharide types with relatively subtle structural differences would be of benefit in both experimental applications and diagnostics. Many routine clinical screens and research applications use immunological methods for which there are currently no antibodies that can discriminate among similar sugar linkages. Possible applications for selective, high-affinity ligands include Western blots, in situ staining, “immuno” precipitation, and various types of “immuno” affinity in microtiter format.
High-affinity nucleic acid ligands, termed “aptamers,” can be selected from a random pool of nucleic acid sequences (refs. 5–12, reviewed in refs. 13–15). Aptamer selection is in theory much faster, cheaper, more versatile, and more reliable than antibody preparation. Such selection produces high-affinity ligands that can be propagated indefinitely with little risk of losing the stock, significantly lower cost than antibodies, and completely obviates the use of animals. In addition to possible ethical considerations, this also provides the opportunity to generate ligands to antigens that are toxic to humans and animals. The selection procedure, termed SELEX (systemic evolution of ligands by exponential enrichment, ref. 7) involves the iterative isolation of ligands out of the random sequence pool with affinity for a defined target molecule and PCR-based amplification of the selected RNA or DNA oligonucleotides after each round of isolation. This previously has been suggested as a route to drug discovery (7), with the possible use of nucleotide analogs to synthesize aptamers that are resistant to nucleolytic degradation (16). Targets for aptamer selection typically have been macromolecules with complex surfaces, but several cases of small molecule targets have been reported (17). Although SELEX has the potential to identify high-affinity ligands to a wide variety of molecular targets, use of the technology has been limited to date.
In developing such selective reagents for oligosaccharides, we have used the polysaccharide cellulose with a simple, uniform repeat unit, cellobiose, to identify highly selective DNA ligands. We have developed DNA ligands that bind tightly to cellulose. A subset of these aptamers recognize the cellobiose repeating units from cellulose with apparent Kd values from 10−5 to 10−7 M. These aptamers have little if any affinity for closely related disaccharides, suggesting that this type of ligand could be useful in differentiating among related biologically important oligosaccharides.
MATERIALS AND METHODS
Materials.
Random DNA oligonucleotides and primers were from the University of Michigan DNA Core. Taq DNA polymerase was prepared as described (18). T4 polynucleotide kinase was obtained from New England Biolabs. XbaI and SalI were from Boehringer Mannheim. γ-32P-ATP was from New England Nuclear and α-32P-ATP was from Amersham. Cellulose was purchased from W&R Balston, United Kingdom. Cellobiose was from Pfanstiehl Chemicals. Gentiobiose, lactose, and maltose were from Sigma. Cellotriose, cellotetraose, and cellopentaose were purchased from Seikagaku, Tokyo.
In Vitro Selection and Amplification.
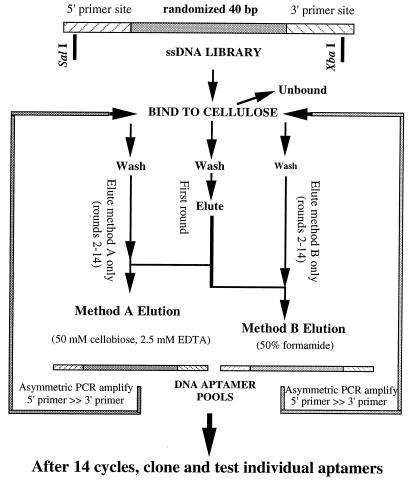
A pool of 86-nucleotide DNA oligomer containing 40 central nucleotides of random sequence flanked by defined primer-binding sites (Fig. 1) was synthesized. This resulted in an initial pool with estimated complexity of 1014–1016 different sequences: (5′-ATAGGAGTCGACCGACCAGAA [N]40 TATGTGCGTCTACATCTAGACTCAT).
Figure 1.
Selection of anticellulose DNA aptamers. Selection began with a library of 1015 DNA molecules containing known 5′ and 3′ sequences (for PCR priming) and 40 central nucleotide positions at which the sequence had been randomized during synthesis. In the first round, the library was bound to cellulose, washed extensively, and eluted first with method A (50 mM cellobiose competitor, 2.5 mM EDTA) and then with method B (50% formamide). DNA eluted from the first round of binding under these two conditions was kept separate in the subsequent 13 rounds of binding selection and amplification by PCR, eluting only with the original method. Pools of DNA aptamers from the 14th round of selection by each method were converted to double-stranded DNA, ligated into plasmid vectors to produce individual clones. DNA from more than 200 clones was reamplified to produce “monoclonal” aptamers, of which 80% were found to bind tightly to cellulose.
Short DNA oligonucleotides for amplifying selected sequences were: 5′-primer, 5′-ATAGGAGTCGACCGACCAGAA; 3′-primer, 5′-ATGAGTCTAGATGTAGACGCACATA.
The selection method is shown in Fig. 1. For the first-round selection, 1 mg of DNA library was passed through a 50-mg cellulose column 10 times. The column was washed with 4 × 400 μl of binding buffer (20 mM Tris, pH 7.5/100 mM NaCl/5 mM MgCl2), and sequentially eluted first with elution method A (50 mM cellobiose, 2.5 mM EDTA) and then with method B (50% formamide). Eluted aptamers were precipitated with ethanol and resuspended in 50 μl H2O. Asymmetric PCRs were used to amplify the recovered DNA sequences from 40 μl of the resuspended DNA eluate and recover single-stranded DNA. The 400-μl asymmetric PCR consisted of: 1× PCR buffer (10 mM Tris, pH 9.1/50 mM KCl/.5 mM MgCl2), 200 μM dNTPs, 18.5 μg 5′-primer, 0.64 μg 3′-primer, and 2.5 units Taq DNA. The cycling protocol was 30 cycles of: 94°C for 45 sec, 60°C for 1 min, 72°C for 2 min. A 350-μl aliquot of the PCR product was bound to the next 50-μl packed cellulose column and, the elution/amplification process was repeated 13 rounds. After the first round of elution by method A, then method B, rounds 2–14 kept the DNA selected by the two methods separate.
Obtaining Monoclonal Aptamers from Selected Pools.
Double-stranded DNA fragments were prepared from round 14 of method A and method B pools, cleaved at the terminal XbaI and SalI site, and ligated into those sites in plasmid pUC19. Transformation into Escherichia coli strain DH5α was followed by direct PCR screening of plasmid-containing bacterial colonies. The difference between the colony PCR and the PCR described above was that the cell samples were initially denatured at 94°C for 5–10 min, followed by 30–35 cycles using equal amount of 5′-primer and 3′-primer (0.5 μg) to produce double-stranded DNA (dsDNA) corresponding to unique sequences from the selected pool. Individual single-stranded aptamer DNAs was derived from an aliquot of this dsDNA stock by asymmetric PCR of the dsDNA amplification.
Cellulose Binding Assay.
α-32P-dATP was incorporated into single-stranded DNA by using asymmetric PCR and isolated after electrophoretic separation in 10% denaturing polyacrylamide gels. 32P-labeled single-stranded DNA was preincubated with binding buffer or 50 mM cellobiose (dissolved in binding buffer) for 10 min at room temperature. Next, 200 μl of the mixture was bound to 12–15 mg cellulose with gentle mixing for 20 min. The unbound fraction was saved, and the cellulose was washed with 200 μl of binding buffer. Bound aptamer was eluted with 200-μl elution method A or method B, depending on the conditions used in selection of that aptamer. Aliquots of 7.5 μl from the unbound, wash, or eluted fractions were analyzed by electrophoresis on denaturing 10% polyacrylamide gel and exposed to x-ray film.
DNA Sequencing.
Thirty-seven cellulose-binding aptamer clones were sequenced. They included all clones where binding was inhibited by cellobiose. Plasmid DNA was isolated by using Qiagen QIAprep-spin plasmid miniprep. DNA sequencing was performed by using primers flanking the plasmid insertion site and reagents from United States Biochemicals.
Hypochromicity Assay.
To quantitatively determine binding properties of three aptamers, milligram quantities were synthesized chemically. Only the internal variable region of three aptamers in were used: the 41 mer of Cel#16, the 40 mer of Cel#183, and the 36 mer of Cel#202 (see Table 1). A 41-mer control oligonucleotide also was used to show that the disaccharides generally did not cause hyperchromicity. The sequence of the 41 mer was 2-fold degenerate at four positions: 5′-CCGAATTCTGGAA(C or G)(A or T)CCC(T or A)(A or C)GCTTTCCTGATGAGTCCGTGA. To analyze binding of the DNAs to soluble disaccharides, hypochromicity was measured at 260 nm as a reflection of conformational change on binding the sugar. DNA aptamers at concentrations between 0.0006 and 0.0007 mM were preincubated with 0.00003 mM to 10 mM sugars (cellobiose, cellotetraose, gentiobiose, lactose, and maltose) at room temperature for 1 hr before reading absorbance at 260 nm. Higher concentrations of several sugars led to aberrantly large drops in absorbance, presumably caused by aggregation, and concentrations higher than 1 mM were not included in interpretations. Cellotriose and cellopentaose (not shown) gave data similar to that for cellotetraose. Readings were taken in triplicate, correcting for minor absorbance by the sugars, and each experiment was repeated three or more times. Hypochromicity is represented as the % change in absorbance from no sugar added (see Fig. 3); errors in the triplicate readings from one experiment are represented by bars.
Table 1.
Sequences of cellulose-binding DNA aptamers
| Aptamers | Elution method | # of clones | Cellobiose competition | Sequence | Nucleotide length |
|---|---|---|---|---|---|
| Cel#16 | A | 8 | + | GCGGGGTTGGGCGGGTGGGTTCGCTGGGCAGGGGGCGAGTG | 41 |
| Cel#183 | A | 2 | + | TAGCGGGTGTGGTGGGTGGGGGAGGCATGGTTTTTGGTAA | 40 |
| Cel#202 | A | 1 | + | GTCAAGGTGGGTGGGTGGGGTTGGTTGTTGTTTTGA | 36 |
| Cel#18 | A | 1 | − | CAGCACACAGGTAGAGAGGAGGGGGTGGGTGGGTGGTGCT | 40 |
| Cel#44 | A | 1 | + | TAGCGGGTGTGGTGGGTGGGGGAGGCATGGTTTTTGGTAG | 40 |
| Cel#75 | B | 1 | − | GTGAGCACATATGGGTAAAAGGAGGGTGGAGGGGTGGA | 38 |
| Cel#76 | B | 1 | − | GCACGAGGTGGTGGGTGGGGGAGGCATGGTTATACGAGTC | 40 |
| Cel#81 | B | 1 | − | GGGGGCGCGCATAGGGGTTGTAGTTCGTGGGTGGTGGTGA | 40 |
| Cel#88 | B | 1 | − | TGGATGGAGGTGGCAGCCCGAGGAGGGTCGGAGNNNA | 37 |
| Cel#89 | B | 1 | − | CTGGGAGGGAGACGCTTGAGGGAGAGAGTGGCTTAGGGAA | 40 |
| Cel#97 | A | 1 | − | TAACGGGTGTGGTGGGTGGGGGAGGCATGGTTTTTGGTAA | 40 |
| Cel#104 | A | 1 | − | GGGGCGCGCATAGGGGTTGTAGTACGTGGGTGGTGGTGA | 39 |
| Cel#105 | A | 1 | − | GGGGCGCGCATAGGGGTTGTAGTGCGTGGGTGGTGGTGA | 39 |
| Cel#124 | A | 1 | − | GTGAGCGAGGTGGAGGGGGTGGGTGGGTGGTAGGCCTGCT | 40 |
| Cel#125 | A | 1 | − | GGGCGCACGGCCAAGGAGTGGGGAGGGGGAGGGTGGTGGG | 40 |
| Cel#132 | A | 1 | − | GGGGCGGGGGGGAGGGGGAAGGTGTGGTCCGGTAATGGT | 39 |
| Cel#134 | A | 1 | − | TGCGATGTGAGAAGGTGTGTAGGATGGGGCGGTAGGTGGA | 40 |
| Cel#151 | A | 1 | − | CGGGCGACGACATCGCGTGAATCAACGCCAGATATGCCGG | 40 |
| Cel#154 | A | 1 | − | GGGCGACTCCAGTTTAATCGACACACAAGTTAGTGCTTCG | 40 |
| Cel#157 | A | 4 | − | GGGGGCGCGCATAGGGGTTGTAGTGCGTGGGTGGTGGTGA | 40 |
| Cel#169 | A | 3 | + | GACTAGATGGGAAGGGGTGGAGGGTGGGGTTGTAGGTTGT | 40 |
| Cel#184 | A | 1 | + | GGAGGATGCGGGTGGTTGGGCGGGGTTGCAGGTTTGTTGA | 40 |
| Cel#187 | A | 1 | + | GGAGGTAGAAGGCGGGTGGGTGGGGTCGGTGGTTGTAAGT | 40 |
| Cel#189 | A | 1 | + | GACTATATAAGAAGGGTTGGTGGGTTCGGTTGTAGTATGT | 40 |
Thirty-seven of the monoclonal aptamers that tightly bound to cellulose were sequenced, yielding 24 unique sequences. The sequences between the unique priming sites are shown, along with the number of times the sequence was identified, the elution method used, and whether cellobiose could effectively compete for binding with cellulose. The three aptamers shown in bold at the top (#16, #183, #202) were used in subsequent experiments to test their affinity and selectivity for binding to cellobiose in solution.
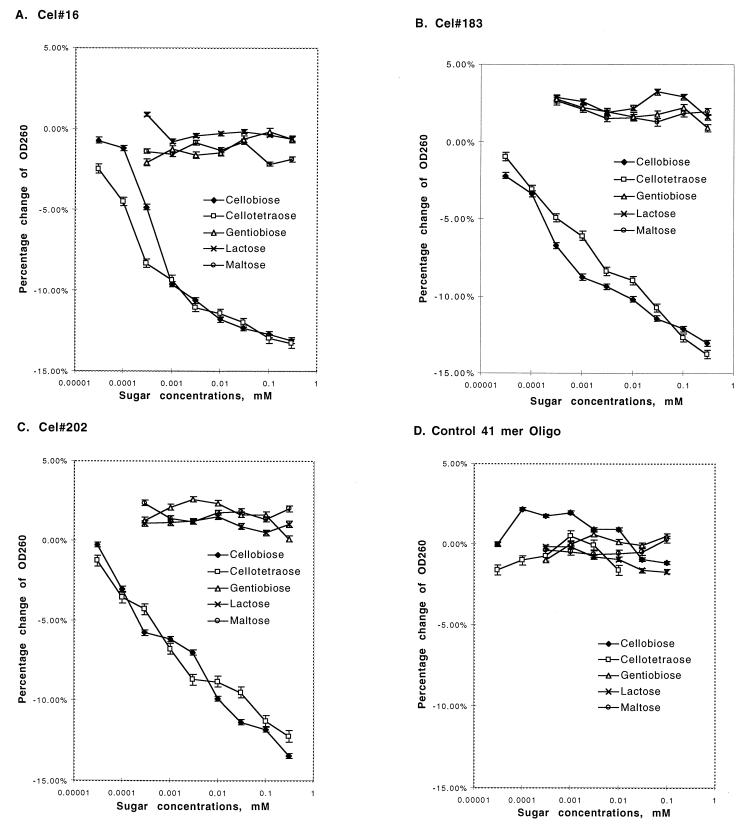
Figure 3.
Specificity of aptamer binding. Aptamers Cel#16, Cel#183, and Cel#202 were tested for the ability to bind four similar biose sugars by titrating the concentration of sugars and detecting DNA binding by measuring the resulting small decrease in absorbance of the solution at 260 nm (hypochromicity). The DNA concentration was constant within each experiment and was 0.6 to 0.7 μM. (A–C) For each aptamer, the cellobiose dimer and cellotetraose tetramer produced hypochromic shifts at similar concentrations (apparent Kd 10−7 to 10−5 M), but the related biose sugars did not produce significant hypochromicity. (D) A control 41-mer DNA oligonucleotide did not give a hypochromic response to any of the sugars tested.
RESULTS
In Vitro Selection and Amplification.
The iterative selection method used to obtain cellulose-binding DNA aptamers is shown schematically in Fig. 1. A random-sequence DNA library with a complexity of approximately 1015 different sequences was used. Only the center sequences of this library had been randomized during chemical synthesis, incorporating known sequences at the 5′ and 3′ termini to serve as primer binding sites for DNA amplification. One milligram of library was bound to cellulose under solution conditions that provided counterion shielding for the charged DNA backbone to stabilize folding. After washing extensively, bound DNA was eluted by using two different strategies. The first elution condition, termed method A, used only 2.5 mM EDTA (pH 8) and 50 mM cellobiose. This very low salt buffer and chelation of any residual divalent cation was intended to destabilize DNA structures by greatly reducing the charge shielding of the phosphate backbone. Cellobiose was used in addition to further compete for binding of DNA molecules capable of recognizing the disaccharide repeating unit of the cellulose polymer. As hoped, this led to aptamers from the method A elution capable of recognizing the disaccharide form. The second, more aggressive elution was performed also at room temperature with 50% formamide (method B) to denature the DNA and release it from the cellulose. Each of the eluted DNA pools was separately amplified by using asymmetric PCR to regenerate single-stranded “sense” DNA. These enriched pools of cellulose-binding DNAs were once again bound to cellulose and eluted with either method A or method B, depending on how they were originally removed in the first round. After 13 additional rounds of binding, elution, and amplification, the two DNA pools were end-labeled by kinasing with 32P-γ-ATP and tested for binding to cellulose under the same conditions used for selection. The majority (90%) of both DNA pools bound efficiently to cellulose and could be eluted with method A or method B.
Cellulose and Cellobiose Binding by Individual Aptamers.
The aptamer pools from the 14th rounds were converted into individual, “monoclonal” aptamers by shotgun cloning into a bacterial plasmid. Of the clones containing single inserts, 202 were converted back to single-stranded DNA by further asymmetric PCR and tested for the ability to bind cellulose. Eighty percent of the monoclonal aptamers bound cellulose tightly. These were further tested to identify a subset of aptamers with a sufficiently focused ligand recognition to bind tightly to the disaccharide unit of cellulose.
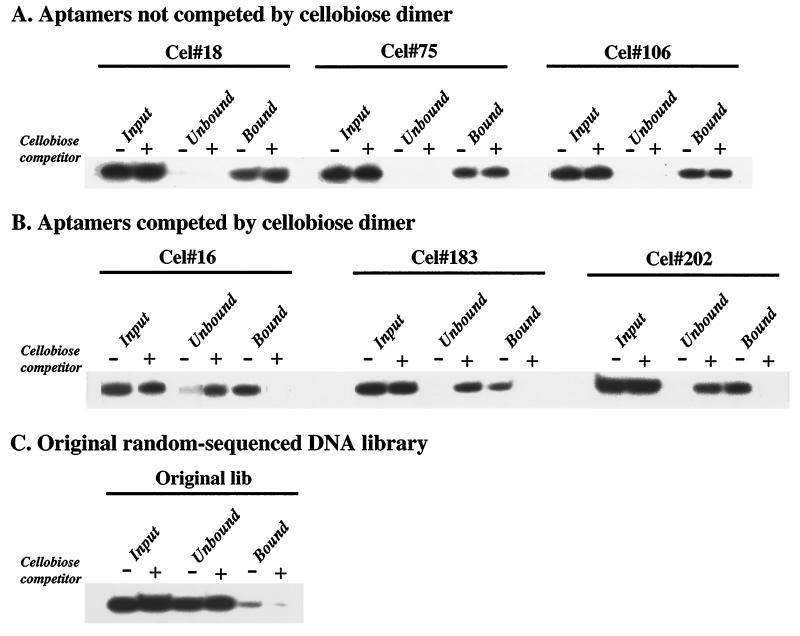
Labeled aptamers were preincubated with or without 50 mM cellobiose and then incubated with excess cellulose for 1 hr under selection conditions. Bound and unbound fractions were separated, and the bound aptamer fraction was eluted. Equal percentages of the input, unbound, and bound DNA fractions were visualized by electrophoresis on denaturing gels to assure the integrity of the DNA. Results are shown in Fig. 2 using the starting DNA library mixture, three aptamers that bound cellobiose and cellulose, and three aptamers that bound cellulose, but not cellobiose. Using this screen we identified 18 aptamer clones for which binding to cellulose could be reproducibly blocked by cellobiose.
Figure 2.
Blocking cellulose binding with cellobiose. The ability of the soluble repeating unit of cellulose, cellobiose, to inhibit cellulose binding was tested. Less than 1 nM 5′-32P DNA aptamer was incubated in 200 μl with 10 mg of insoluble cellulose in the presence (+) or absence (−) of 50 mM cellobiose. Equal percentages of the input DNA, unbound DNA, and the DNA eluted from the cellulose (with method A or method B, depending on aptamer) were visualized by using a PhosphorImager after denaturing PAGE. (A) Three aptamers (Cel#18, Cel#75, and Cel#106) not competed by cellobiose. (B) Three aptamers (Cel#16, Cel#183, and Cel#202) for which preincubation with cellobiose prevented most subsequent binding to cellulose. (C) Binding by the original random-sequence DNA library. Low levels of library DNA reproducibly bound to cellulose and could be eluted with method B (shown).
Sequence Analysis of Cellulose-Binding DNA Aptamers.
To examine common features of the DNA aptamers that bound cellulose, 37 of the clones were sequenced, including all 18 of the aptamers that bound stably to cellobiose. The insert sequences are listed in Table 1, along with the elution method used to select the aptamer, the number of clones containing the sequence, and whether cellobiose competed for its binding site(s). The three cellobiose-binding clones analyzed in subsequent experiments are listed in bold lettering at the top of the table, with others listed in the order they were identified. Although the length of sequence randomized in the original library was 40 nucleotides, the length of the clone inserts varied from 36 to 41 nucleotides, presumably because of insertions and deletions during PCR amplification. Several of the sequences, especially those identified as binding cellobiose, were identical, suggesting multiple identification of the same clone. It was expected that the aptamers that bound well to cellobiose would be drawn exclusively from the pool eluted with method A, because this method deliberately included cellobiose as part of the relatively mild elution buffer.
Examination of the sequences reveals a strong tendency toward G-richness, with most clones containing more than 50% G residues. A breakdown of average nucleotide of the general types of aptamers found in Table 1 is provided in Table 2, and shows that the G-richness does not reside in any particular subclass of the aptamers. In most instances the clusters of Gs have some tendency to be spaced by T residues. This tendency toward G clusters with T spacers is particularly pronounced in the clones that bind cellobiose well (Cel#16, Cel#183, Cel#202, Cel#169, Cel#184, and Cel#187) In the one aptamer that binds cellobiose but has only 37.5% G (Cel#189), there are central G clusters spaced by T residues. In the other two aptamers where the G content is below 50% (Cel#151 and Cel#154), there are only one or two small G clusters, suggesting that these may represent distinct binding strategies. Previous sequence characterization of a population of aptamers that bound nitrocellulose indicated that population was purine-rich, although the properties of the individual aptamers were not investigated in more detail (10). This population of cellulose-binding aptamers reported here appears to have properties quite distinct from the nitrocellulose aptamers.
Table 2.
Nucleotide makeup of different aptamer classes
| Percentage of nucleotides
|
||||
|---|---|---|---|---|
| dA | dC | dG | dT | |
| Method A aptamers | ||||
| Total | 12.1% | 10.3% | 55.4% | 22.2% |
| Competed by cellobiose | 10.1% | 8.9% | 57.4% | 23.6% |
| Not competed by cellobiose | 14.7% | 12.0% | 53.0% | 20.3% |
| Method B aptamers | 20.4% | 10.5% | 51.6% | 17.5% |
Secondary structure modeling of the G-rich aptamers using DNA Mfold (using Michael Zuker’s server at Washington University: http://www.ibc.wustl.edu/~zuker/dna/form1.cgi) did not produce any notable structure consensus for the inserts, but the superficial similarity of the G cluster/T spacer motif to repeated sequences found at the telomeres of eukaryotic chromosomes suggested that structures similar to “G-quartets” might somehow be involved (19). To test whether simple G-rich or telomere-like sequences might have an intrinsic affinity for cellulose, we synthesized oligodeoxynucleotides corresponding to dG10, dG4dT2dG4, and dG4dT2dG4dT2. Cellulose binding assays of the type used in Fig. 2 failed to detect higher affinity binding than found in the random sequence library (data not shown). We concluded that although some specialized form of DNA structure might be involved in the G-rich aptamers, any motif responsible for cellulose binding was not clearly defined by inspection of the DNA sequences.
Affinity and Selectivity of Disaccharide Binding.
Three of the aptamers that bound to cellobiose were selected for quantitative evaluation of the affinity and selectivity of binding. The internal DNA sequences of Cel#16, Cel#183, and Cel#202 minus the primer binding sites were synthesized in milligram quantities and found to bind tightly to cellulose as in Fig. 2 (not shown). Binding of soluble sugars to these three aptamers and an unrelated control 41-mer oligonucleotide was assayed indirectly by detecting the change in DNA structure on sugar binding. Transition from disordered polynucleotide structure to more ordered tertiary structures with greater base stacking can be detected through hypochromicity, a decrease in the absorbance at 260 nm. Maximum absorbance changes through this effect tend to be less than 25% of signal when going from completely single-stranded to completely double-stranded. Initial measurements of the three aptamers in the presence or absence of cellobiose suggested that the change in absorbance (corrected for a small sugar absorbance) was as high as 10–15%, consistent with a pronounced structural change. Careful titrations of sugar concentrations subsequently were undertaken with the three aptamers and control DNA 41-mer oligonucleotide of unrelated sequence. The results are shown in Fig. 3, with the percent change in the absorbance at 260 nm plotted against the concentration of sugar.
Both cellobiose and cellotetraose were used to test whether increased chain length would provide additional affinity for the aptamer. Aptamer Cel#16 had the highest affinity for both sugars, with a slightly higher affinity for cellotetraose than cellobiose. The apparent Kd for cellobiose is approximately 6 × 10−7, whereas the apparent Kd for cellotetraose is 3 × 10−7 or better. It is worth noting that this highest affinity aptamer is also the one displaying incomplete cellulose capture (Fig. 2). It is not clear whether this can be attributed to a nonhomogeneous DNA folding population, or whether the aptamer has a higher affinity for the soluble sugar than for the polymer. The observed increase in affinity for cellotetraose over cellobiose would argue that Cel#16 is unlikely to have lower affinity for the cellulose polymer than for cellobiose. Cellobiose and cellotetraose titrations for aptamers Cel#183 and Cel#202 had much lower affinities and shallower hypochromicity response curves. This gradual response suggests more complex binding events, possibly requiring multiple structural transitions. In contrast to the aptamer DNAs, the control oligonucleotide did not display significant hypochromicity (or hyperchromicity) with any of the sugars tested.
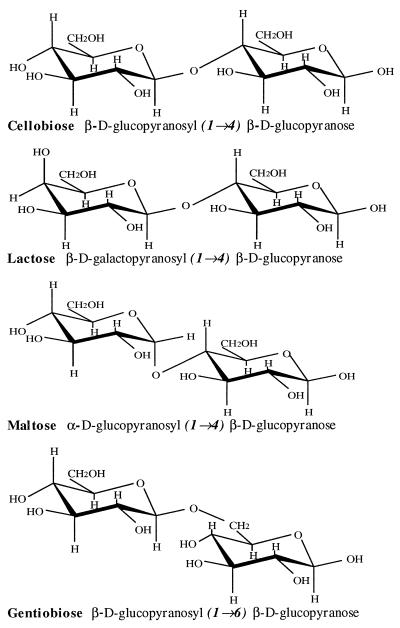
As a test of the specificity of the aptamer interactions with cellulose and its cellobiosyl units, several additional disaccharides were tested in the same assays. Lactose, maltose, and gentiobiose are closely related disaccharides that vary only slightly in structure from cellobiose, as shown in Fig. 4. Cellobiose and longer derivatives are made up of d-glucose monomers joined by β(1 → 4) linkages. Lactose differs from cellobiose only in containing a terminal galactosyl unit, a stereoisomer that differs at a single hydroxyl group (C-4′OH). Maltose provides a different isomerization, containing two glucose monomers with an α(1 → 4) linkage. Similar to cellobiose, gentiobiose consists of two d-glucose monomers, but contains a β(1 → 6) linkage. None of these three related sugars elicited a structural transition in any of the three DNA aptamers, indicating that the interaction of the aptamers with cellobiose and cellulose was highly selective.
Figure 4.
Structure comparison of cellobiose, lactose, maltose, and gentiobiose.
DISCUSSION
In this work high-affinity DNA ligands (aptamers) were selected that bound to cellulose, and further screening identified a subset of the aptamers that were capable of binding tightly to the minimal unique repeating unit in the polymer, cellobiose. Because aptamers to simple sugars had not previously been selected, it was not clear whether such aptamers could be selected or whether they would be highly specific in their oligosaccharide binding. Sugar groups have very few positions, primarily the hydroxyl groups, that are likely to be able to form noncovalent bonds with significant stability. These sugar “antigens” lack both charged interactions and aromatic ring structures with which to strongly direct nucleic acid interactions. It therefore is not surprising that the affinities of these aptamers for the disaccharides do not reach affinities equivalent to what can be obtained with more complex macromolecular targets, which can reach Kd values below 10−10 (15). However, the aptamer affinities compare favorably to characterized protein affinities for simple sugars. The association constants for lectin-carbohydrate interactions vary from 103 to 105 M−1 (1) and association constants of anticarbohydrate antibodies are reported to be 102 to 105 M−1 (4, 20).
The specificity of the aptamers for cellobiose, rather than closely related sugars, seems consistent with the paucity of possible interaction sites on the cellobiose. With no positively charged groups and no aromatic ring structures that could stack stably with the nucleic acid bases, the interactions are likely to be limited to hydrophobic sites and hydrogen bonding to hydroxyl and pyranose ring oxygens in the sugar. If any contact is lost through a change in the geometry of the target, it could constitute the loss of a large percentage of the binding energy. This points up one critical aspect to selection of antisaccharide aptamers for practical purposes. If the aptamer needs to recognize a particular sugar conformation in a specific context, for example as part of a glycoprotein surface, it is best to select the aptamers in that macromolecular context. Specificity for the sugar epitope is achieved by counterselecting for aptamers that bind to the macromolecule without the epitope of interest and screening monoclonal aptamers for the desired specificity.
The results of this study suggest it should be possible to develop specific aptamers against a wide array of carbohydrate antigens. A series of aptamers directed against different combinations of oligosaccharides might easily be able to discriminate among a mixture of macromolecular surfaces, including relatively minor differences in glycoprotein modifications. Simple sugars are not favored to provide high-affinity interactions with nucleic acids because of the limited opportunities for hydrogen bonding or aromatic ring stacking. More complex monomer could well provide significantly increased affinity while remaining selective for only particular combinations of monomer units. For example, there has been extensive development of RNA aptamers capable of interacting with aminoglycoside antibiotics (21–24), which contain both amino and hydroxyl groups for hydrogen bonding interactions and three or more ring structures. Many of the RNA aptamers selected against these targets have affinities in the range seen here, with Kd values between 10−4 and 10−7, although affinities have been reported as low as subnanomolar (25). The sequences reported for these RNA aptamers generally do not resemble the DNA G-rich sequences we have identified in this selection, although a subset in one study did have short G-rich sections (22). It is possible that the use of DNA in the present study restricted the structural motifs more than when RNA is used. Alternatively, the difference in sequence requirements could derive more from the fact that the potential interactions with the cellobiose target are far more limited than for aminoglycosides.
The increased affinity of aptamer ligands compared with protein ligands might make these nucleic acids attractive alternatives to lectin or mAb detection reagents for carbohydrate antigens. This advantage is multiplied by the practical advantages of aptamers over antibodies or lectins. Specific aptamers might be selected more easily and reliably than mAbs, and the artificial selection eliminates dependence on pre-existing lectins for a given carbohydrate antigen. Once selected and characterized, aptamers can be stored almost indefinitely, or can be resynthesized inexpensively from the known DNA or RNA sequence. Most types of diagnostic testing that depend on fluorescent or affinity-labeled ligands in microtiter plates could be easily adapted to the use of DNA or RNA ligands, in most cases with greater ease of labeling. Research applications that currently use antibodies for detection or immunoaffinity purification could also be undertaken with aptamers. Fluorescence-activated cell sorting, fluorescent in situ localization, and affinity purification methods all should be relatively straightforward with selective aptamer recognition. In addition, a number of applications are theoretically possible through fusing multiple aptamers directed against different antigens. For example, an anticellulose aptamer could be fused to an aptamer against a soluble toxin to provide an affinity purification (removal) for toxin-aptamer complexes on cellulose-based filters. A modified version of this might entail preparation of aptamers to membrane material used for human dialysis to provide a method to remove toxins from blood. Alternatively, aptamers directed against different cell-surface antigens might be fused to provide noncovalent crosslinking of the antigens.
This work demonstrates that relatively simple carbohydrate antigens are potential targets for highly selective DNA ligands, suggesting that it should be possible to select DNA, RNA, and modified polynucleotide aptamers from random sequence pools as recognition tools for a array of surface antigens.
Acknowledgments
This work was supported by a postdoctoral fellowship to Q.Y. from Parke-Davis Pharmaceutical Research. Oligonucleotide synthesis was partially defrayed by National Institutes of Health Grant P30 CA46592 to the University of Michigan Cancer Center, and support was available from National Institutes of Health Grant RO1 GM29470 to I.J.G.
References
- 1.Goldstein I J, Poretz R D. In: The Lectins: Properties, Function, and Applications in Biology and Medicine. Liener I E, Sharon N, Goldstein I J, editors. San Diego: Academic; 1986. pp. 33–247. [Google Scholar]
- 2.Weis W I, Drickamer K. Annu Rev Biochem. 1996;65:441–473. doi: 10.1146/annurev.bi.65.070196.002301. [DOI] [PubMed] [Google Scholar]
- 3.Gabius H-J. Eur J Biochem. 1997;243:543–576. doi: 10.1111/j.1432-1033.1997.t01-1-00543.x. [DOI] [PubMed] [Google Scholar]
- 4.Glaudemans C P J. Chem Rev. 1991;91:25–33. [Google Scholar]
- 5.Oliphant A R, Brandl C J, Struhl K. Mol Cell Biol. 1989;9:2944–2949. doi: 10.1128/mcb.9.7.2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Joyce G F. Gene. 1989;82:83–87. doi: 10.1016/0378-1119(89)90033-4. [DOI] [PubMed] [Google Scholar]
- 7.Tuerk C, Gold L. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 8.Ellington A D, Szostak J W. Nature (London) 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 9.Bartel D P, Zapp M L, Green M R, Szostak J W. Cell. 1991;67:529–536. doi: 10.1016/0092-8674(91)90527-6. [DOI] [PubMed] [Google Scholar]
- 10.Tuerk C, MacDougal S, Gold L. Proc Natl Acad Sci USA. 1992;89:6988–6992. doi: 10.1073/pnas.89.15.6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Famulok M, Szostak J W. J Am Chem Soc. 1992;114:3990–3991. [Google Scholar]
- 12.Ellington A D, Szostak J W. Nature (London) 1992;355:850–852. doi: 10.1038/355850a0. [DOI] [PubMed] [Google Scholar]
- 13.Ivine D, Tuerk C, Gold L. J Mol Biol. 1991;222:739–761. doi: 10.1016/0022-2836(91)90509-5. [DOI] [PubMed] [Google Scholar]
- 14.Burke J M, Berzal-Herranz A. FASEB J. 1993;7:106–112. doi: 10.1096/fasebj.7.1.8422956. [DOI] [PubMed] [Google Scholar]
- 15.Gold L, Polisky B, Uhlenbeck O, Yarus M. Annu Rev Biochem. 1995;64:763–797. doi: 10.1146/annurev.bi.64.070195.003555. [DOI] [PubMed] [Google Scholar]
- 16.Williams K P, Liu X H, Schumacher T N, Lin H Y, Ausiello D A, Kim P S, Bartel D P. Proc Natl Acad Sci USA. 1997;94:11285–11290. doi: 10.1073/pnas.94.21.11285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jenison R D, Gill S C, Pardi A, Polisky B. Science. 1994;263:1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- 18.Engelke D R, Krikos A, Bruck M E, Ginsburg D. Anal Biochem. 1990;191:396–400. doi: 10.1016/0003-2697(90)90238-5. [DOI] [PubMed] [Google Scholar]
- 19.Williamson J R, Raghuraman M K, Cech T R. Cell. 1989;59:871–880. doi: 10.1016/0092-8674(89)90610-7. [DOI] [PubMed] [Google Scholar]
- 20.Pavliak V, Nashed E M, Pozsgay V, Kovac P, Karpas A, Chu C, Schneerson R, Robbins J B, Glaudemans P J. J Biol Chem. 1993;268:25797–25802. [PubMed] [Google Scholar]
- 21.Wang Y, Rando R R. Chem Biol. 1995;2:281–290. doi: 10.1016/1074-5521(95)90047-0. [DOI] [PubMed] [Google Scholar]
- 22.Lato S M, Boles A R, Ellington A D. Chem Biol. 1995;2:291–303. doi: 10.1016/1074-5521(95)90048-9. [DOI] [PubMed] [Google Scholar]
- 23.Wallis M G, von Ahsen U, Schroeder R, Famulok M. Chem Biol. 1995;2:543–552. doi: 10.1016/1074-5521(95)90188-4. [DOI] [PubMed] [Google Scholar]
- 24.Jiang L, Suri A K, Fiala R, Patel D J. Chem Biol. 1997;4:35–50. doi: 10.1016/s1074-5521(97)90235-0. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, Killian J, Hamasaki K, Rando R R. Biochemistry. 1996;35:12338–12346. doi: 10.1021/bi960878w. [DOI] [PubMed] [Google Scholar]