Abstract
Many genes in parasitic nematodes are both cis- and trans-spliced. Previous studies have demonstrated that a 7nt element encoded in the first intron of the B. malayi 70 kDa heat shock protein (BmHSP70) gene was necessary to permit trans-splicing of transgenic mRNAs in embryos transfected with constructs encoding portions of the BmHSP70 gene. Here we demonstrate that this element (the B. malayi HSP70 trans-splicing motif, or BmHSP70 TSM) is necessary and sufficient to direct trans-splicing of transgenic mRNAs derived from two genes naturally containing this motif. Mutations introduced into any position of the BmHSP70 TSM abrogated its ability to direct trans-splicing. Transgenic mRNAs derived from embryos transfected with constructs containing promoters and associated downstream domains from two normally trans-spliced genes that lack a BmHSP70 TSM homologue (the B. malayi 12 kDa small subunit ribosomal protein (BmRPS12) gene and the B. malayi RNA binding protein (BmRBP1) gene), were not trans-spliced. Transfer of the BmHSP70 TSM into the first intron of the BmRPS12 gene rendered it competent for trans-splicing. Insertion of the BmHSP70 TSM into the single intron of the BmRBP1 gene did not render it trans-splicing competent. However, tagged constructs of the full-length BmRBP1 gene were trans-splicing competent. An analysis of the first exons and introns of over 200 trans-spliced B. malayi genes found homologues for the BmHSP70 TSM in roughly 25%. Thus, while the BmHSP70 TSM is necessary and sufficient to direct trans-splicing in some genomic contexts, independent trans-splicing signals are employed by other genes.
Keywords: filariasis, transfection, splicing, spliced leader
Introduction
mRNA 5′ trans-splicing is a process found in a variety of organisms, including kinetoplasts, many nematodes, flatworms and a few species in the phylum chordata [1]. It involves the addition of a small leader RNA sequence, known as the spliced leader, to the 5′ end of many pre-mRNAs. Trans-splicing appears to serve a variety of different functions, including processing of poly-cistronic pre-mRNAs [2], mRNA cap addition [3, 4], polishing and sanitizing the 5′ end of primary transcripts, resulting in optimal spacing for translation [5] and addition of an in-frame start codon [6].
In nematodes, two types of trans-splicing have been noted. The most conserved form involves the addition of a 22 nt sequence, known as the spliced leader 1 (SL1), to the 5′ end of the nascent mRNAs [7]. In the free living nematode Caenorhabditis elegans, the SL1 is added to pre-mRNAs transcribed from singlet genes and genes which are in the 5′ most position in operons. In C. elegans and some other nematodes, the downstream genes of operons are resolved through the addition of a distinct SL sequence, known as the SL2 [2]. However, in other nematodes, including the human filaria, SL2 trans-splicing appears not to exist, and all transcripts, including those located downstream in operon-like structures, contain SL1 [8].
The SL1 is encoded in the intragenic spacer domain of the 5S rRNA gene cluster of parasitic nematodes [7]. The SL RNA is transcribed from the intragenic spacer by RNA polymerase II, resulting in a pre-RNA which contains the SL sequence at its 5′ end, and binding sites for various components of the splicing machinery in its 3′ end [9, 10]. The SL sequence is then removed from the nascent SL transcript and is trans-spliced on to the 5′ end of the nascent mRNA, through a biochemical pathway that bears many similarities to the cis-splicing pathway [11], although certain proteins have been shown to be specifically required for trans-splicing [10, 11]
In vitro biochemical systems employing nuclear extracts have been used to extensively dissect the trans-splicing pathway in the intestinal parasite Ascaris suum [7, 10-13]. These studies have resulted in the identification of a number of factors that are involved in the trans-splicing process [9-11] and have also succeeded in identifying the structural factors in the SL pre-RNA that are necessary for correct processing of the nascent transcript [7]. However, because these studies have used synthetic templates and nuclear extracts, they could not be used to study trans-splicing in vivo, where transcription and post-transcriptional mRNA processing, including cis-and trans-splicing, are coupled.
Recently, the development of a transient transfection system for the parasitic nematode Brugia malayi was reported [14]. This system was subsequently employed to map the promoter domains in the sequences present upstream of the gene for the heat shock protein 70 (HSP70) homologue of B. malayi (BmHSP70) [15]. The native BmHSP70 message is trans-spliced in vivo, with the SL1 added at a site that is located 47 nt upstream of the translational start [16]. Surprisingly, mRNAs transcribed from B. malayi embryos transfected with a synthetic transgene consisting of the 659 nt upstream of the BmHSP70 ORF (including the native SL addition site) fused to a luciferase reporter gene were not trans spliced [17]. However, transgenes consisting of in frame fusions of the BmHSP70 659 nt upstream domain, exon 1, intron 1 and part of exon 2 were correctly cis-and trans-spliced [17]. Further studies demonstrated that downstream introns could not replace intron 1 in directing trans-splicing, and that a semi-conserved 7nt motif present in intron 1 was necessary for this process [18]. In the present manuscript, we have further explored the role that this conserved motif (designated the B. malayi HSP70 trans-splicing motif, or BmHSP70 TSM) plays in trans-splicing in transfected B. malayi embryos.
Materials and Methods
Preparation of parental constructs
Three parental plasmids, BmHSP70(-659 to 738)/luc, BmHSP70(-659 to 495)/luc and BmHSP70(-659 to 738; ∇98-489)/luc served as templates to prepare the BmHSP70 mutant constructs described below. The construction of these parental plasmids has been described in previous publications [17, 18].
A second gene containing the BmHSP70 TSM in it’s first intron was examined for it’s ability to support trans splicing in transfected embryos. This gene (BmATS) encodes an asparaginyl tRNA synthetase of B. malayi, and its mRNA was the first to be shown to be trans-spliced in this parasite [19]. To obtain a clone containing the promoter and downstream domains of the BmATS gene, the gene was identified by a BLAST search of The Institute for Genomic Research (TIGR) genomic sequence (available from the TIGR B. malayi genome project at http://www.tigr.org/tdb/e2k1/bma1/intro.shtml), using the full length mRNA sequence (Genbank Accession number J03266) as the query. The genomic sequence corresponding to the 5′ end of the gene was found in assembly BRSXP17TR of the B. malayi genomic sequence database. Primers corresponding to positions 23-47 in the coding orientation (5′ TCCATGTCCACTACCCGATCCTTTT 3′) and 759-778 in the non-coding orientation (5′ GCCAAGCTTGATAAAGCGTCCTGCAGTCA 3′) in this assembly were used to amplify the 544 nt upstream of the start of the open reading frame (ORF), the first exon, the first intron and 11nt of the second intron from B. malayi genomic DNA. The non-coding primer was designed with a synthetic Hind3 restriction site for subsequent sub-cloning and it was positioned so that the ORF of a correctly cis-spliced mRNA would be placed in frame with the luciferase reporter gene in the reporter vector pGL3 basic. If, as expected, translation began at the initiation codon of the BmATS gene, this arrangement would permit translation of the luciferase ORF, albeit as a chimeric protein, fused to the peptide encoded by the residual upstream ORF. The resulting amplicon was cloned into the TA cloning vector (Invitrogen, Carlsbad, CA) and its sequence verified. The cloned amplicon was then excised from the TA cloning plasmid by digestion with EcoR1 and Hind3 and cloned into the reporter vector pGL3 basic, as previously described [15].
Two trans-spliced genes lacking a homologue of the BmHSP70 TSM in their first introns were also examined. These included the B. malayi 12 kDa ribosomal small subunit protein (BmRPS12) gene and the B. malayi RNA binding protein (BmRBP1) gene. The clone of the promoter for the BmRPS12 gene was prepared as previously described [15]. To obtain a clone containing the promoter and downstream domains of the BmRPS12 gene, the BmRPS12 gene sequence was first identified by a BLAST search of TIGR genomic sequence using the putative full length EST sequence of the BmRPS12 mRNA (available from NEMBASE2 (http://www.nematodes.org/nematodeESTs/nembase.html)) as the query sequence. The BmRPS12 genomic sequence was found to reside in TIGR assembly 12,701. Two primers corresponding to positions 4255-4273 (coding orientation; 5′ GGGCTCGAGCAATAACACGCAAACACAC 3′) and 4812-4830 (non-coding orientation; 5′ GGGAAGCTTTTCCTCCTGCTTCCATGTTCA 3′) in this sequence were then used to amplify the corresponding locus from B. malayi genomic DNA, essentially as previously described [15]. This resulted in the amplification of a sequence consisting of the 562 nt upstream of the initiating codon of the ORF, the first exon, first intron and 8 nt of the second exon of the BmRPS12 gene. The primers were developed with a synthetic Xho1 site on the 5′ end of the upstream primer and a Hind3 site at the 5′ end of the downstream primer to facilitate subsequent sub-cloning. The primers were also engineered to ensure that a correctly cis-spliced mRNA produced from this construct would place the residual upstream BmRPS12 ORF in frame with the luciferase reporter gene, as described above. No attempt was made to determine the actual start site of translation of this construct. The resulting amplicon was cloned into the TA cloning vector (Invitrogen, Carlsbad, CA) and its sequence verified. The cloned amplicon was then excised from the TA cloning plasmid by digestion with Xho1 and Hind3 and cloned into the reporter vector pGL3 basic, as previously described [15].
The constructs derived from the BmRBP1 gene were prepared in essentially the same manner as those for the BmRPS12 gene. In this case, a comparison of a putative full length EST sequence to the TIGR database resulted in the localization of the BmRBP1 gene to TIGR assembly 14,972. Two primers developed from this sequence, representing nucleotides 2,050,904-2,050,923 in the coding orientation (5′ GGGCTCGAGACATTCTCTTCCTCTTCTGC 3′) and 2,051,958 - 2,051,976 in the non-coding orientation (5′ GGGAAGCTTGAGGCAAGTTTCCAACTGGA 3′), were used to amplify the sequence consisting of the 463 nt upstream of the ORF, the first exon, the single intron and 15 nt of the second exon of the BmRBP1 gene. Similar to the BmRPS12 primer pair, these primers were designed with synthetic Xho1 and Hind3 restriction sites to facilitate subsequent sub-cloning. The resulting amplicon was cloned as described above and the DNA sequence of a representative clone was confirmed. The amplicon was excised with Xho1 and Hind3 and sub-cloned into pGL3 basic as described above, resulting in the clone designated BmRBP1 E1-I-E2-luc. As before, the primers were designed so that a correctly cis-spliced product would place the residual BmRBP1 ORF in frame with the downstream luciferase reporter gene.
To amplify the complete BmRBP1 gene, a coding orientation primer corresponding to positions 2,050,923 - 2,050,941 in TIGR assembly 14,972 (5′ CACCCACACCCACCCTTCC 3′) was used in conjunction with a non-coding primer derived from positions 2,052,421-2,052,442 (5′ TCGTAGCTTCACAAAATCATTC 3′). This resulted in the amplification of a product of 1520 nt containing the 443 nt upstream of the BmRBP1 ORF, the complete ORF and 252 nt downstream of the ORF, extending 34 nt downstream of the poly A addition site for the gene. The amplicon was cloned as described above and its DNA sequence confirmed. This clone (still in the TA vector) was used as the parental construct for preparation of the tagged version of the BmRBP1 gene described in the text.
The TIGR sequencing effort is part of the International Brugia Genome Sequencing Project and is supported by an award from the National Institute of Allergy and Infectious Diseases, National Institutes of Health. Preliminary sequence data resulting from this effort are deposited regularly into the GSS division of GenBank.
In vitro mutagenesis of the parental constructs
Mutants were prepared from the parental constructs using the GeneTailor™ site-directed mutagenesis system (Invitrogen), following the protocols described by the manufacturer. In brief, plasmid DNA from each of the parental constructs was methylated with DNA methylase. The methylated DNA was used as a template in a PCR amplification reaction that employed a coding and non-coding primer set that overlapped by 15-20 nt (depending upon the construct) at a position that was located just 5′ to the mutagenesis site. The primer in the coding orientation was designed to encode the mutation to be introduced (insertion or substitution) just 3′ of this overlap zone. The resulting amplicons were transformed directly into DH5α-T1 competent cells (Invitrogen). Plasmid DNA from the resulting transformants was recovered and their identity confirmed by DNA sequence analysis.
Transfection and analysis of transgene expression
Isolated B. malayi embryos were biolistically transiently transfected as previously described [17]. In brief, embryos were isolated from gravid female parasites and transfected with the experimental DNA driving the expression of firefly luciferase mixed with a constant amount of an internal standard, consisting of the BmHSP70 promoter fragment (positions -659 to -1) driving the expression of renilla luciferase. Transfected embryos were maintained in culture for 48 hours before being assayed for transgene activity. Firefly luciferase activity was normalized to the amount of renilla luciferase activity in each sample to control for variations in transfection efficiency. Firefly/renilla activity ratios for each sample were further normalized to the activity ratio found in embryos transfected in parallel with a control construct (BmHSP70(-659 to -1)/luc). This permitted comparisons of data collected in experiments carried out on different days. All constructs were assayed in triplicate.
RNA prepared from 48-hour transfected embryos was used as the template in individual hemi-nested RT-PCRs to detect evidence of cis- and trans-splicing, as previously described [17]. To detect trans-splicing, total RNA was purified from each batch of transfected embryos using Trizol (Invitrogen) following the manufacturer’s instructions. A total of 1μg of this purified RNA was used as a template in a single tube RT-PCR amplification reaction, employing the Qiagen One Step reagent (Qiagen, Valencia, CA), following the manufacturer’s protocol. The one step reaction employed primers derived from the SL1 sequence (5′ GGTTTAATTACCCAAGTTTGAG 3′) and the luciferase gene encoded in the pGL3 basic vector (luc nc490; 5′ TTTGCAACCCCTTTTTGGAA 3′). Amplification conditions consisted of an initial incubation at 50°C for 30 minutes, followed by heating at 95°C for 15 minutes to denature the reverse transcriptase and activate the polymerase. This was followed by 40 cycles consisting of 94°C for 30 sec, 52°C for 30 sec and 72°C. A total of 5ul of this amplification reaction was used as a template in a 50 μl hemi-nested PCR reaction, employing Pfu Turbo DNA polymerase (Invitrogen, Carlsbad, California) using the buffer and conditions recommended by the manufacturer. The hemi-nested PCR reaction employed the SL primer and a nested primer derived from the luciferase gene (luc nc238; 5′ CGACGATTCTGTGATTTGTA 3′). Amplification conditions consisted of 35 cycles of 30 sec at 94°C, 30 sec at 52°C and 1 min at 72°C. The amplification reactions were analyzed by agarose gel electrophoresis. The identity of all amplicons were confirmed by DNA sequence analysis.
Cis-splicing in the BmHSP70 transfected embryos was assayed using a nested RT-PCR assay that specifically targeted the transgenic mRNA. RNA was prepared from transfected embryos using the method described above. It was then treated with DNAse-free RNAse (Qiagen) following the manufacturer’s instructions, and purified using the RNAeasy spin column kits (Qiagen). The resulting treated purified RNA was then used as a template in a nested PCR amplification reaction using Pfu Turbo DNA polymerase, as described above. In the case of RNA purified from embryos transfected with the construct BmHSP70 E1-E2-I2-E3-luc, the first amplification employed a primer derived from exon 1 of the BmHSP70 gene (HSP70c1200; 5′ TATTCCTGCGTGGGTGTGT 3′) and the luc nc490 primer. Amplification conditions consisted of 40 cycles of 30 sec at 94°C, 30 sec at 52°C and 1 min at 72°C. A total of 5 μl of the first reaction was used as a template in a 50 μl nested PCR amplification reaction employing a primer spanning the deletion site of intron 1 (BmHSP70 ex1-Xho-ex2; 5′ ATCAAGCTCGAGGTAATCGT 3′) and the luc nc238 primer. The amplification products were analyzed by agarose gel electrophoresis. Any RNA sample producing a detectable amplicon (indicative of residual DNA contamination) was subjected to further rounds of DNAse treatment and purification until no visible products were produced in the nested PCR. The purified RNA was then analyzed using a nested RT-PCR, employing the Qiagen one step RT-PCR kit in the first step and Pfu Turbo DNA polymerase in the nested step, as described above.
Cis-splicing in embryos transfected with the substitution mutants of the BmHSP70 TSM were assayed following a similar nested PCR procedure. In this case, the assay employed BmHSP70c1169 (5′ GAATGCCATTGGTATCGAC 3′) and luc nc490 as primers in the first step and BmHSP70c1200 and luc nc238 in the nested step. All amplification reactions were analyzed by agarose gel electrophoresis and the identity of amplicons confirmed by DNA sequencing.
In the case of the full length hemagglutinin (HA) tagged constructs of the complete BmRBP1 gene, luciferase primers could not be used to assay for trans-splicing, as the constructs did not contain a luciferase reporter gene. In this case, a hemi-nested RT-PCR assay was used that employed the SL primer together with a primer spanning the 3′ end of the inserted HA tag sequence and a portion of the 3′ flanking domain (5′ CTGCTGAAAACATTCAAGCGT 3′) in the first step. The second step used the SL primer and a primer containing the 5′ end of the inserted HA tag sequence and a portion of the associated 5′ region (5′ GTAATCTGGTACGTCGTATCG 3′). The resulting amplicons were analyzed by agarose gel electrophoresis and DNA sequence analysis as described above.
Bioinformatic analyses
A database of 1,314 expressed sequence tags from the JHU93SL-BmL3 library [20] was searched by BLASTN against the B. malayi coding sequence database (BMA1.cds; available at http://www.tigr.org/tdb/e2k1/bma1/; also available at http://www.wormbase.org/db/seq/gbrowse/brugia/). The JHU93SL library was prepared by PCR amplification of cDNA prepared from B. malayi infective larvae mRNA using oligo dT and a primer corresponding to the SL sequence, thus specifically amplifying mRNAs containing the SL. Significant hits with an E-value below e-5 matching from position 1 of the B. malayi CDS were selected, assuring that only full length mRNA sequences were included in the analysis. For the 218 genes meeting these criteria, we excerpted from the appropriate assembly sequence the string of bases ranging from the beginning of the first exon to the beginning of the second exon. These excerpts thus comprised the gene’s first exon and first intron. A regular expression search for the motif “RGATRAA” was performed on the multifasta file of exons and introns. The number of occurrences and the location within the excerpt of each specific match to the ambiguous motif were tabulated.
Genes in which a TSM homologue were identified were further analyzed to determine if the sequences flanking the TSM were conserved. To accomplish this, the 20 nt upstream and downstream from each TSM were extracted. Multiple alignments of these sequences were prepared and the results graphically displayed with the WebLogo algorithm [21].
Results
The BmHSP70 TSM is necessary and sufficient to direct trans-splicing
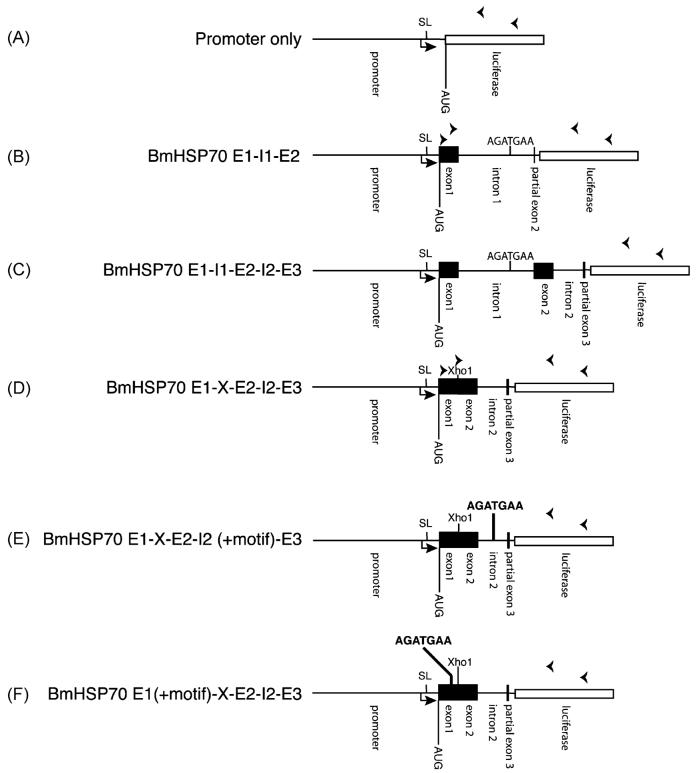
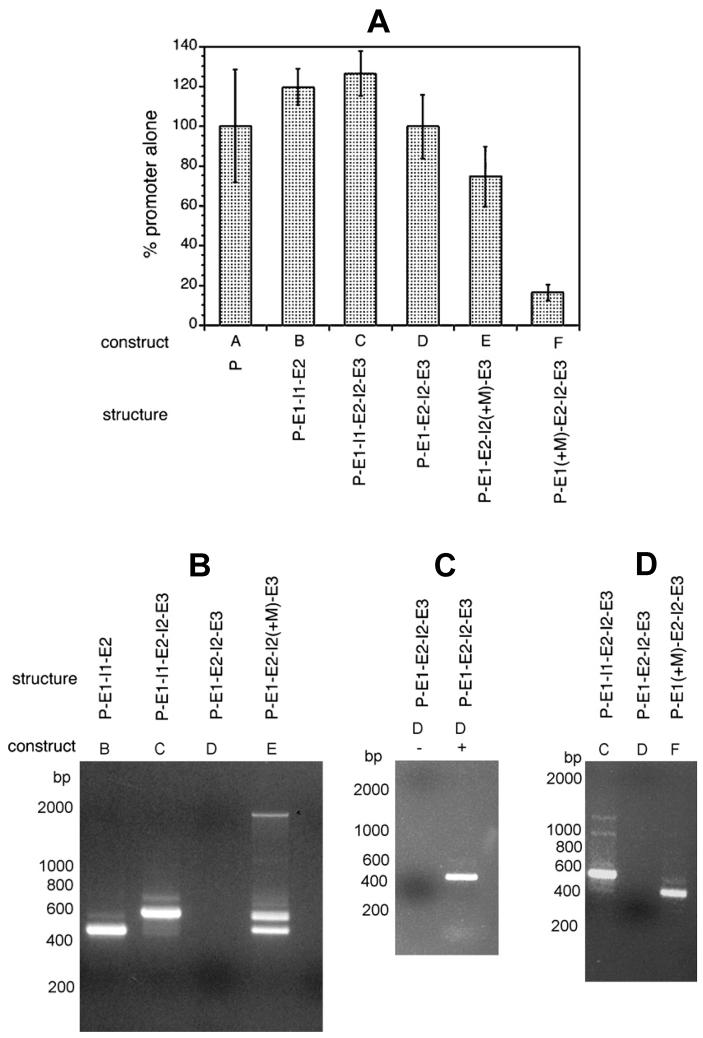
Previous studies had demonstrated that sequences present in the first intron of the BmHSP70 gene were necessary to direct trans-splicing of transgenes in B. malayi transfected embryos, and that the second intron of this gene could not substitute for the first intron in this process [18]. Furthermore, in vitro deletion mutagenesis studies demonstrated that a semi-conserved sequence present in the first intron (the BmHSP70 TSM) was necessary for this process [18]. However, these studies did not address the question of whether or not the BmHSP70 TSM was sufficient to direct trans splicing. To address this question, a number of constructs were prepared, transfected into B. malayi embryos and trans-splicing of the resulting transgenic mRNAs assayed as described in Materials and Methods. These constructs are outlined schematically in Figure 1 and the results of these studies presented in Figure 2. As previously shown [17, 18], embryos transfected with a construct containing the BmHSP70 promoter, exon 1, intron 1, and part of exon 2 (BmHSP70 E1-I1-E2-luc) or a construct containing the promoter and exon 1, intron 1, exon 2, intron 2 and part of exon 3 (BmHSP70 E1-I1-E2-I2-E3-luc) produced RT-PCR products demonstrating that transgenic mRNAs derived from these constructs were trans-spliced (Figure 2, Panel B, constructs B and C). DNA sequence analysis of these amplicons confirmed that they were derived from transgenic RNA molecules which were correctly cis-and trans-spliced. In contrast, a construct derived from BmHSP70 E1-I1-E2-I2-E3-luc in which intron 1 was deleted and replaced by a synthetic Xho1 site (BmHSP70 E1-X-E2-I2-E3-luc) was not trans-spliced (Figure 2, Panel B, construct D). The luciferase activity from embryos transfected with this construct was not significantly different from that seen in the parental construct, demonstrating the presence of transgenic mRNA (Figure 2, Panel A, constructs B, C and D). Furthermore, RT-PCR assays using a set of internal primers targeting the transgenic mRNA from embryos transfected with BmHSP70 E1-X-E2-I2-E3-luc demonstrated that the transgenic mRNA in these samples was correctly cis-spliced (Figure 2, Panel C).
Figure 1. Schematic diagram of the structure of the BmHSP70 clones.
The elements representing each feature are proportional to their size in the construct (with the exception of the luciferase ORF, which is truncated for clarity). Black boxes indicate exon sequences, and lines indicate non-coding sequences (e.g. promoter domains, introns and spacers). The open box represents the luciferase reporter ORF. In each panel, the bent arrow pointing to the right indicates the start site of transcription of the BmHSP70 gene [17]. “SL” indicates the position of the SL1 addition site. “AUG” indicates the first methionine codon in the long open reading frame present in a correctly cis-spliced message, presumably representing the start site of translation. “Xho1” indicates the Xho1 site introduced during the procedure used to delete intron 1. The sequence “AGATGAA” indicates the position of the BmHSP70 TSM. This is indicated in normal type when in its native position, and in bold type when introduced into a different position. Arrowheads over the luciferase gene and the exons in the schematic diagrams indicate the position and orientation of the primers used in the cis- and trans-splicing assays described in the text.
Figure 2. Cis- and trans-splicing of transgenic mRNAs from embryos transfected with BmHSP70 constructs.
In each panel, the “construct” refers to the designation of each construct in the detailed schematic diagrams found in Figure 1, while “structure” provides a shorthand description of the structure of each construct. In the latter P=promoter, E1=exon 1, I1=intron 1, E2=exon 2, I2=intron 2 and E3=exon 3. “+M” indicates that the BmHSP70 TSM was inserted into the element to the left. For example the structure P-E1-E2-I2(+M)-E3 represents the construct containing the promoter, exon 1, exon 2 (i.e. intron 1 has been deleted), intron 2 (into which the BmHSP70 TSM has been inserted), and exon 3. Panel A: Luciferase activity in embryos transfected with the different BmHSP70 constructs. Bars represent means and error bars standard deviations of triplicate determinations. Panel B: SL mediated hemi-nested RT-PCR analysis of trans-splicing in transgenic mRNAs from constructs containing and lacking the BmHSP70 TSM in intron sequences. Panel C: Analysis of cis-splicing in transgenic mRNA from embryos transfected with construct D, which was not trans spliced. The lane labeled “D+” represents an cis-splicing RT-PCR assay carried out in the presence of reverse transcriptase, while the lane labeled “D-” represents an parallel assay carried out without reverse transcriptase. Panel D: SL mediated hemi-nested RT-PCR analysis of trans-splicing in transgenic RNA collected from embryos transfected with the construct in which the BmHSP70 TSM was inserted into the first exon of the BmHSP70 gene (construct F). Constructs C and D served as positive and negative controls respectively for this experiment.
To test if the BmHSP70 TSM was sufficient to induce trans-splicing, a construct was prepared in which the motif was inserted into the remaining intron of BmHSP70 E1-X-E2-I2-E3-luc (Figure 1, construct E). When assayed using the RT-PCR assay described in Materials and Methods, this construct (BmHSP70 E1-X-E2-I2(+M)-E3-luc) produced two amplicons (Figure 2, Panel B, construct E). DNA sequence analysis of these two amplicons demonstrated that the larger was derived from a transgenic mRNA that was correctly cis- and trans-spliced; thus it had the structure SL-E1-X-E2-E3-luc. The smaller of the two products was derived from a transgenic mRNA that was correctly trans-spliced, but in which the synthetic Xho1 site and adjacent flanking sequences inserted during the deletion of intron had served as a de novo cis-splice donor, resulting in a transgenic mRNA with the structure SL-E1-X-E3-luc.
Context and primary sequence requirements of the BmHSP70 TSM
The studies described above indicated that the BmHSp70 TSM was sufficient to drive trans-splicing of the BmHSP70 transgenic mRNA regardless of its intron context, functioning within either intron 1 or intron 2. However, the makeup of introns and exons is generally different, so it was of interest to determine if the activity of the BmHSP70 TSM was dependent upon the functional context in which it appeared, i.e. was it necessary for the motif to be present in an intron sequence, or was it also functional when placed in an exon? To answer this question a construct was prepared in which the BmHSP70 TSM was inserted as an in frame substitution mutation into the first exon of BmHSP70 E1-X-E2-I2-E3-luc (Figure 1, construct F). RT-PCR analysis of RNA isolated from embryos transfected with this construct (BmHSP70 E1(+M)-X-E2-I2-E3-luc) resulted in the production of an amplicon of roughly the predicted size (Figure 2, Panel D, construct F). DNA sequence analysis of this product revealed that it was correctly trans-spliced. However, as was seen with the smaller product in the experiments involving the construct E1-X-E2-I2(+M)-E3-luc, the synthetic Xho1 site added during the deletion of intron 1 had created a splice donor, resulting in a transgenic mRNA with the structure SL-E1(+M)-X-E3-luc.
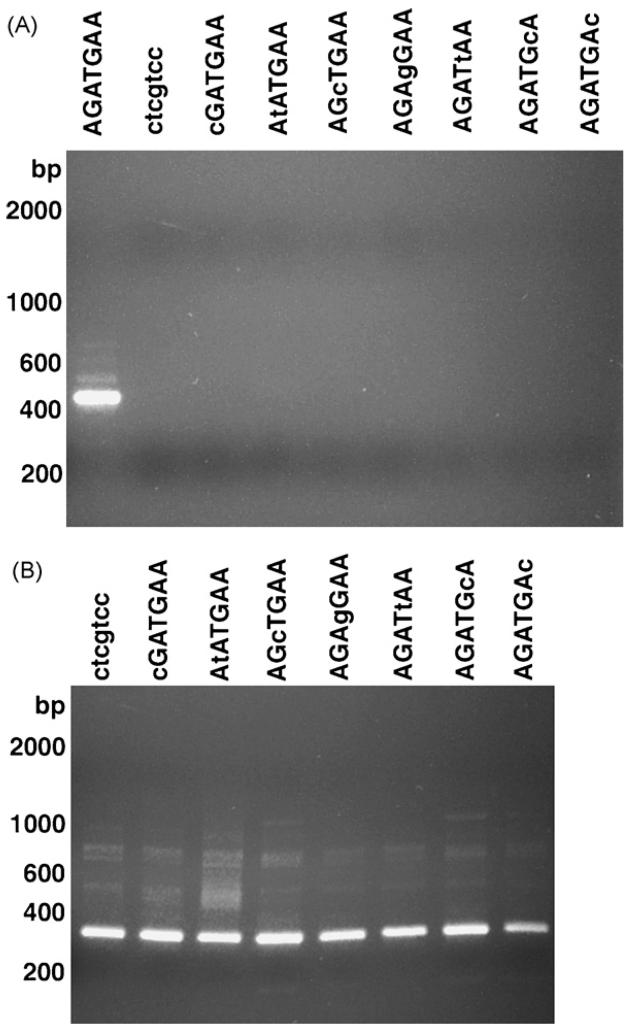
To determine which nucleotides in the BmHSP70 TSM were essential to support trans-splicing, a series of constructs were prepared in which each of the individual nucleotides in the motif present in the construct E1-I1-E2-luc were replaced by in vitro mutagenesis. These constructs were then transfected into isolated B. malayi embryos and the transgenic mRNAs examined for evidence of cis- and trans-splicing. Replacement of any or all of the nucleotides in the BmHSP70 TSM eliminated trans-splicing (Figure 3, Panel A). As expected, mutagenesis of the motif did not affect cis-splicing in the transgenic mRNAs (Figure 3, Panel B).
Figure 3. Analysis of cis- and trans-splicing in transgenic mRNAs isolated from embryos transfected with mutants of the BmHSP70 TSM.
Panel A: SL mediated hemi-nested RT-PCR analysis of trans-splicing of transgenic mRNAs isolated from embryos transfected with constructs containing replacement mutations of the BmHSP70 TSM. Panel B: RT-PCR analysis of cis-splicing in the non-trans-spliced samples analyzed in Panel A. In each panel, the lanes are labeled with the sequence of the BmHSP70 TSM contained in the construct analyzed. Capital letters represent the wild type sequence and small letters the mutation introduced in each construct. In panel A, the first lane (labeled AGATGAA) contains RT-PCR amplicons produced from mRNA isolated from the parental construct (BmHSP70 E1-I1-E2-luc).
Activity of the BmHSP70 TSM in other genes
Analysis of the first introns of several B. malayi genes indicated that sequences similar to the BmHSP70 TSM are present in many, but not all, of the first introns of trans spliced B. malayi genes. It was therefore of interest to examine if downstream sequences were capable of mediating trans-splicing in other genes both containing and lacking this motif. To answer the first question, a portion of the B. malayi asparaginyl tRNA synthetase (BmATS) gene was isolated and tested for its ability to direct trans-splicing. The BmATS gene encodes the first mRNA shown to be trans-spliced in B. malayi [19] and it contains a perfect homologue of the BmHSP70 TSM in its first intron (Figure 4, panel A). A construct (BmATS E1-I1-E2-luc) containing the 544 nt upstream of the ORF, the first exon, the first intron and 11 nt of the second intron (Figure 4, Panel A) was cloned upstream from the luciferase reporter gene of pGL3 basic and transfected into B. malayi embryos. mRNA was isolated from these embryos and assayed for evidence of trans-splicing in the transgenic transcripts using the SL mediated RT-PCR assay described above. This produced two products of approximately 400 nt and 320 nt (Figure 4, Panel B). DNA sequence analysis of these two products revealed that the larger was derived from a correctly cis- and trans-spliced transgenic mRNA. The lower product was found to be a spurious amplicon produced from a genomic sequence found in TIGR assembly BRSOZ87TR. This assembly did not contain an identifiable long open reading frame and did not match any predicted gene in the TIGR database (data not shown), and was therefore not characterized further.
Figure 4. Trans-splicing in transgenic mRNAs isolated from embryos transfected with BmATS E1-I1-E2-luc.
Panel A: Schematic diagram of the construct. The elements representing each feature are proportional to their size in the construct (with the exception of the luciferase ORF, which is truncated for clarity). Black boxes indicate exon sequences and lines non-coding sequences (e.g. promoter domains, introns and spacers). The open box represents the luciferase reporter ORF. “SL” indicates the position of the SL1 addition site. “AUG” indicates the first methionine codon in the long open reading frame present in a correctly cis-spliced message, presumably representing the start site of translation. The sequence “AGATGAA” indicates the position of the BmHSP70 TSM homologue found in the first intron of the BmATS gene. Arrowheads over the luciferase gene indicate the position and orientation of the primers used in the trans-splicing assays described in the text. Panel B: SL mediated hemi-nested RT-PCR analyses of RNA prepared from embryos transfected with BmATS E1-I1-E2-luc. Lane A = positive control (RNA extracted from embryos transfected with BmHSP70 E1-I1-E2-luc). Lane B = SL mediated hemi-nested RT-PCRs carried out on mRNA extracted from embryos transfected from BmATS E1-I1-E2-luc carried out in the presence of reverse transcriptase. Lane C = SL mediated hemi-nested RT-PCRs carried out on mRNA extracted from embryos transfected from BmATS E1-I1-E2-luc carried out in the absence of reverse transcriptase.
Two normally trans-spliced genes lacking homologues of the motif in their first intron were then examined. The first of these encodes the B. malayi homologue of the 12 kDa protein of the small subunit of the ribosome (BmRPS12), while the second encodes an abundant RNA binding protein homologue (BmRBP1). The upstream domains of both of these genes are capable of driving transgene expression in B. malayi embryos [15], and EST evidence suggested that the native mRNAs derived from both of these genes were trans-spliced. This was confirmed experimentally by SL mediated RT-PCR for both native genes (data not shown). The predicted gene structure of the downstream domains of both genes was determined by comparison of the genomic sequences present in the TIGR database to those derived from predicted full length ESTs. The structure of the BmRPS12 gene was fairly typical of most B. malayi genes [22] containing 4 exons and 3 introns, which ranged from 128 nt to 156 nt in size. The RBP gene was less typical, containing two exons and a single intron of 578 nt.
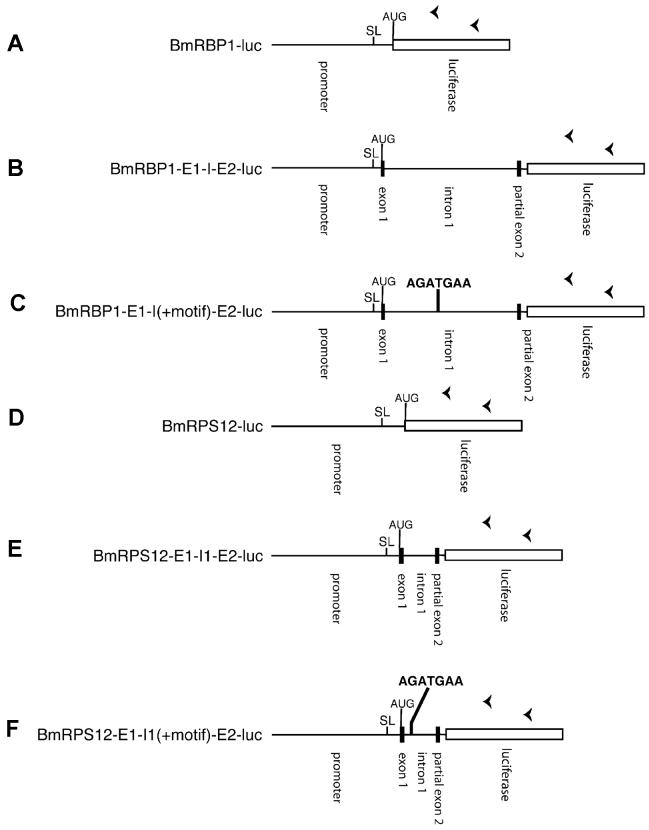
The regions upstream of the open reading frame of both genes, as well as the regions consisting of the upstream domains, the first exon, the first intron and a portion of the second exon were cloned into the pGL3 Basic reporter vector as described in Materials and Methods. These constructs are depicted schematically in Figure 5. All constructs were transfected into embryos and assayed for luciferase reporter gene activity and trans-splicing. Constructs containing the upstream domains alone, while producing luciferase activity and therefore transgenic mRNA (Figure 6, Panel A, constructs A and D), were not trans-spliced (Figure 6, Panel B, constructs A and D). Similarly, constructs containing the downstream domains of both genes including the first exon, the first intron and a portion of the second exon were also not trans-spliced (Figure 6, Panel B, constructs B and E). These data suggested that first introns lacking a sequence similar to the BmHSP70 TSM were incapable of supporting trans-splicing of a transgenic transcript.
Figure 5. Schematic diagram of the BmRBP1 and BmRPS12 constructs.
The elements representing each feature are proportional to their size in the construct (with the exception of the luciferase ORF, which is truncated for clarity). Black boxes indicate exon sequences and lines non-coding sequences (e.g. promoter domains, introns and spacers). The open box represents the luciferase reporter ORF. “SL” indicates the position of the SL1 addition site. “AUG” indicates the first methionine codon in the long open reading frame present in a correctly cis-spliced message, presumably representing the start site of translation. The bold sequence “AGATGAA” indicates the position of the inserted BmHSP70 TSM. Arrowheads over the luciferase gene indicate the position and orientation of the primers used in the SL mediated hemi-nested RT-PCR trans-splicing assays described in the text.
Figure 6. Analysis of trans-splicing in transgenic mRNA isolated from embryos transfected with BmRBP1 and BmRPS12 constructs.
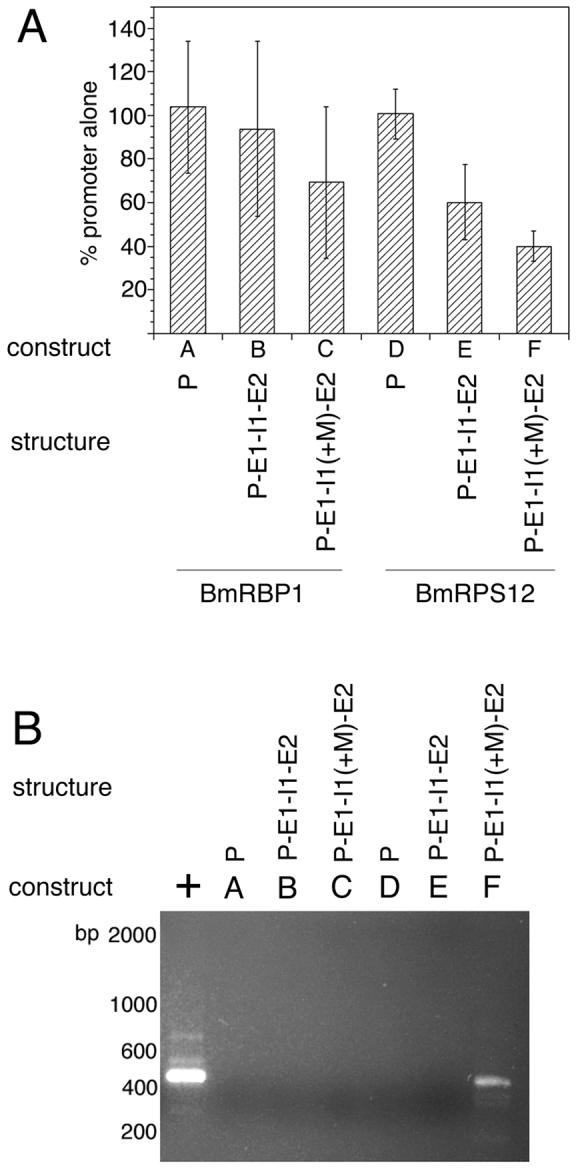
In each panel, the “construct” refers to the designation of each construct in the detailed schematic diagrams found in Figure 4, while “structure” provides a shorthand description of the structure of each construct. In the latter, P=promoter, E1=exon 1, I1=intron 1, E2=exon 2, I2=intron 2 and E3=exon 3. “+M” indicates that the BmHSP70 TSM was inserted into the element to the left. Panel A: Luciferase activity in embryos transfected with the different BmRBP1 and BmRPS12 constructs. Bars represent means and error bars standard deviations of triplicate determinations. Panel B: SL mediated hemi-nested RT-PCR analysis of trans-splicing in transgenic RNAs isolated from embryos transfected with these constructs. The lane labeled “+” in Panel B represents a positive control for the trans-splicing RT-PCR assay (RNA isolated from embryos transfected with BmHSP70 E1-I1-E2-luc).
The finding that the first introns of BmRBP1 and BmRPS12 were incapable of supporting trans-splicing offered the opportunity of exploring whether the BmHSP70 TSM was capable of directing trans-splicing when placed in a genomic context in which it is not normally found. Constructs were prepared in which the BmHSP70 TSM was inserted into the first intron of BmRBP1 and BmRPS12. These constructs were then transfected into B. malayi embryos and assayed as described above. RT-PCR analysis of transgenic RNA isolated from embryos transfected with the construct in which the BmHSP70 TSM was inserted into the first intron of the BmRPS12 gene (BmRPS12 E1-I1(+M)-E2-luc) produced an amplicon of the expected size for a correctly cis- and trans-spliced product (Figure 6, Panel B, construct F). DNA sequence analysis confirmed that this product was derived from a correctly cis-and trans-spliced transgenic mRNA. In contrast, no product was detected in similar assays conducted on RNA obtained from embryos transfected with BmRBP1 E1-I(+M)-E2-luc (Figure 6, Panel B, construct C).
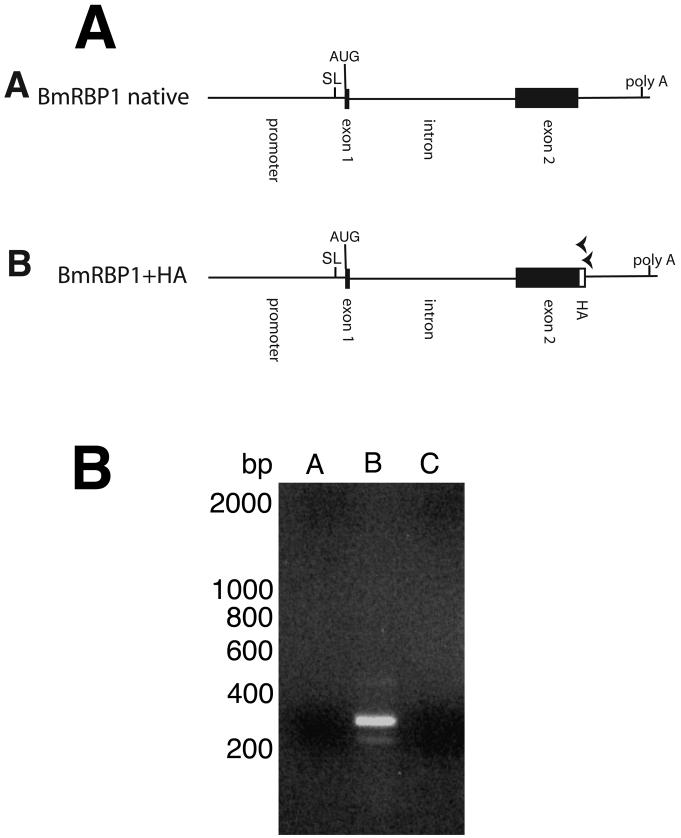
The inability of the HSP70 TSM to support trans-splicing when inserted into the single intron of the BmRBP1 gene was surprising, given the fact that the BmHSP70 TSM was active when inserted into either intron 2 or exon 1 of the BmHSP70 gene, as well as in intron 1 of the BmRPS12 gene. One possibility was that the rather unusual structure of the BmRBP1 gene, which contains only a single large intron, might prevent transgenic mRNAs produced from this locus from being trans-spliced when expressed from an exogenously introduced plasmid. To determine if the unusual gene structure was precluding trans-splicing of BmRBP1 transgenic pre-mRNAs, the complete BmRBP1 gene (consisting of the 378 nt upstream from the SL addition site, continuing through the coding region to a point located 34nt downstream of the poly A addition signal) was isolated by PCR amplification (Figure 7, Panel A, construct A). This amplicon was cloned and mutated to insert a 21nt sequence encoding for a synthetic hemagglutinin (HA) tag into the 3′ end of the open reading frame (Figure 7, Panel A, construct B). The plasmid containing this HA tagged version of BmRBP1 was transiently transfected into B. malayi. RT-PCR analysis of the resulting RNA isolated from these embryos targeting the HA tagged transgenic mRNA produced a product of the size expected for a correctly cis-and trans-spliced mRNA (Figure 7, Panel B, lane B). DNA sequence analysis of the product confirmed that it was derived from a correctly cis- and trans-spliced transgenic mRNA. A similar product was not obtained in reactions carried out in the absence of reverse transcriptase, or from RNA prepared from un-transfected embryos (Figure 7, Panel B, lanes A and C).
Figure 7. Analysis of trans-splicing in mRNA isolated from embryos transfected with an HA tagged full length BmRBP1 gene.
Panel A: Schematic diagram of the native BmRBP1 gene and the HA tagged construct. Black boxes indicate exons, and lines non-coding sequences. The HA tag is indicated by a white box. “SL” indicates the position of the splice leader addition site and “poly A” the position of the poly A addition site. The arrowheads over the schematic diagram of the HA tagged construct indicate the position and orientation of the primers used in the trans-splicing assays described in the text. Panel B: SL mediated hemi-nested RT-PCR analysis of trans-splicing in transgenic RNA isolated from embryos transfected with the BmRBP1-HA construct. A = amplicons produced from RT-PCR analysis of RNA isolated from untransfected embryos (containing native BmRBP1 mRNA). B = amplicons produced from RT-PCR analysis of RNA isolated from embryos transfected with the BmRBP1-HA construct. C = sham RT-PCR of the RNA from embryos transfected with the BmRBP1-HA construct carried out in the absence of reverse transcriptase.
The HSP70 TSM was initially identified by a search of a limited number of B. malayi genomic sequences looking for sequences that were conserved in the first introns of genes producing a trans-spliced mRNA, but which were not conserved in downstream intron sequences of the same genes [17]. An analysis of six such genes revealed the presence of a semi-conserved 7 nt sequence (RGATRAA) which was found in 5/6 of the first introns but which was not present in the downstream intron sequences [17]. However, as described above, it is clear that some trans-spliced gene sequences lack a BmHSP70 homologue. To conduct a more thorough investigation of the proportion of trans-spliced genes containing a homologue of the BmHSP70 TSM, the first exon and first intron sequences of 218 genes where experimental evidence suggested that their transcripts were trans-spliced were examined. Both the first intron and first exon sequences were examined, as the data above suggested that the BmHSP70 TSM was functional in both of these genomic contexts. The results of these analyses are summarized in Table 1. A total of 56/218 (26%) of the genes analyzed were found to contain TSM homologues. Of these, 48% contained a BmHSP70 TSM homologue in their first exon, and 61% contained a homologue in their first intron. Just 5% of the genes contained more than one copy of the motif.
Table 1.
Presence of homologues of the BmHSP70 TSM (RGATRAA) in trans-spliced genes of B. malayi
| Sequence of motif | |||||
|---|---|---|---|---|---|
| AGATAAA | GGATAAA | GGATGAA | AGATGAA | ||
| Total # genes analyzed | 218 | ||||
| # genes with at least one motif | 56 | ||||
| # genes with two or more motifs | 11 | ||||
| # exons with motif | 27 | 6 | 3 | 9 | 9 |
| # introns with motif | 34 | 18 | 7 | 5 | 11 |
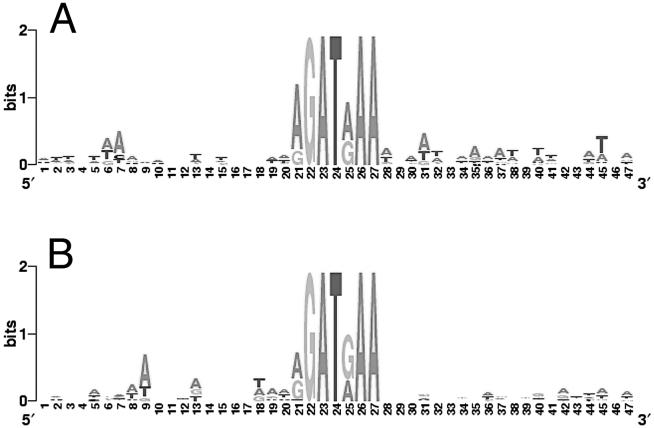
The 20 nt upstream and downstream of the motif homologues were then extracted and examined for evidence of local sequence conservation. No evidence of local sequence conservation outside of the motif sequences themselves was detected (Figure 8).
Figure 8. Analysis of regions flanking the TSM homologues.
Sequence conservation in the 20 nt upstream and downstream of the TSM homologues in the genes found to contain them was analyzed using WebLogo [21], as described in Materials and Methods. The height of the letters indicates the conservation of the nucleotides at each position. Panel A = sequence conservation in the regions surrounding the TSM homologues found in first introns. Panel B = sequence conservation in the regions surrounding the TSM homologues found in first exons.
Discussion
Previous studies had demonstrated that the first intron of the BmHSP70 gene was necessary to permit trans-splicing of transgenic mRNAs and that a downstream intron could not substitute for intron 1 in mediating trans-splicing [18]. Furthermore, deletion mutagenesis had demonstrated that a semi-conserved 7 nt motif found in the first intron (the HSP70 TSM) was necessary to support trans-splicing [18]. We have extended these observations, showing that the BmHSP70 TSM is sufficient to mediate trans-splicing. Thus, insertion of the BmHSP70 TSM into the trans-splicing incompetent construct BmHSP70-E1-X-E2-I2-E3-luc permitted trans-splicing of the mRNAs produced from this construct. Interestingly, the BmHSP70 TSM was capable of directing trans-splicing when inserted into either the intron or first exon of BmHSP70-E1-X-E2-I2-E3. Thus, the exact functional environment of the sequence in which the BmHSP70 TSM is located (i.e. an intron versus exon sequence) appears not be a determining factor in whether it is capable of mediating trans-splicing.
The hypothesis that the BmHSP70 TSM is sufficient to direct trans-splicing was also supported by the experiments involving the BmRPS12 gene. This gene lacks the motif sequence in its first intron, and constructs containing the BmRPS12 promoter alone or the promoter plus the first exon, first intron and part of the second exon were not trans-spliced. However, RNA derived from a construct in which the BmHSP70 TSM was inserted into the first intron of the BmRPS12 gene was trans-spliced. Thus, the BmHSP70 TSM was sufficient to direct trans-splicing when placed into an unrelated genomic environment.
In contrast, the BmHSP70 TSM was not capable of supporting trans-splicing when inserted into the intron of the BmRBP1 gene. However, previous studies have shown that intron secondary structure can affect cis-splicing in some circumstances [23, 24]. It is thus possible that the secondary structure or local sequence environment surrounding the BmHSP70 TSM may play a role in its activity in the trans-splicing process. This hypothesis is currently under investigation.
The mechanism through which the BmHSP70 TSM acts to direct trans-splicing is still unknown. Presumably, the BmHSP70 TSM is serving as a binding site for a trans-acting factor that is responsible for assisting the trans-spliceosomal machinery in binding to the pre-mRNA and then carrying out the trans-splicing reaction. By analogy to what is currently known concerning the process of cis-and trans-splicing, it is tempting to speculate that this factor might be a small ribonuceloprotein complex (snRNP) or an accessory protein that binds to the existing spliceosome. Experiments to identify such a factor are currently underway.
Although the BmHSP70 TSM is both necessary and sufficient to direct the trans-splicing reaction in some circumstances, it is apparently not the exclusive signal used by B. malayi in this process. Just over 25% of the trans-spliced genes examined here contained a homologue of the BmHSP70 TSM in their first exon or first intron. This result suggests that as yet unidentified sequences in these genes may substitute for the BmHSP70 TSM in directing trans-splicing. More experiments will be necessary to determine the identity of these alternative signals.
Acknowledgements
We would like to thank Dr. Naomi Lang-Unnasch for critical reading of the manuscript prior to publication and Dr. Julian Rayner for critical reading and helpful discussions. We also acknowledge the use of the B. malayi genomic sequence produced and maintained by TIGR. TIGR’s sequencing effort is part of the International Brugia Genome Sequencing Project and is supported by an award from the National Institute of Allergy and Infectious Diseases, National Institutes of Health. Preliminary sequence data from this project are deposited regularly into the GSS division of GenBank. This work was supported by a grant from National Institute of Allergy and Infectious Diseases to TRU (Project#R01AI048562).
Abbreviations
- ATS
Asparaginyl tRNA synthetase
- BmHSP70 TSM
B. malayi HSP70 trans-splicing motif
- HSP70
70 kDa heat shock protein
- luc
luciferase
- nt
nucleotides
- ORF
open reading frame
- RBP1
RNA binding protein
- RPS12
ribosomal small subunit 12 kDa protein
- SL
spliced leader
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hastings KE. SL trans-splicing: easy come or easy go? Trends in Genetics. 2005;21:240–47. doi: 10.1016/j.tig.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Blumenthal T, Evans D, Link CD, Guffanti A, Lawson D, Thierry-Mieg J, Thierry-Mieg D, Chiu WL, Duke K, Kiraly M, Kim SK. A global analysis of Caenorhabditis elegans operons. Nature. 2002;417:851–54. doi: 10.1038/nature00831. [DOI] [PubMed] [Google Scholar]
- 3.Maroney PA, Denker JA, Darzynkiewicz E, Laneve R, Nilsen TW. Most mRNAs in the nematode Ascaris lumbricoides are trans-spliced: A role for spliced leader addition in translational efficiency. RNA. 1995;1:714–23. [PMC free article] [PubMed] [Google Scholar]
- 4.Lall S, Friedman CC, Jankowska-Anyszka M, Stepinski J, Darzynkiewicz E, Davis RE. Contribution of trans-splicing, 5′ -leader length, cap-poly(A) synergism, and initiation factors to nematode translation in an Ascaris suum embryo cell-free system. Journal of Biological Chemistry. 2004;279:45, 573, 85–45. doi: 10.1074/jbc.M407475200. [DOI] [PubMed] [Google Scholar]
- 5.Blumenthal T. Trans-splicing and polycistronic transcription in Caenorhabditis elegans. Trends Genet. 1995;11:132–36. doi: 10.1016/s0168-9525(00)89026-5. [DOI] [PubMed] [Google Scholar]
- 6.Cheng G, Cohen L, Ndegwa D, Davis RE. The flatworm spliced leader 3′-terminal AUG as a translation initiator methionine. Journal of Biological Chemistry. 2006;281:733–43. doi: 10.1074/jbc.M506963200. [DOI] [PubMed] [Google Scholar]
- 7.Nilsen TW. Trans-splicing of nematode premessenger RNA. Ann Rev Microbiol. 1993;47:413–40. doi: 10.1146/annurev.mi.47.100193.002213. [DOI] [PubMed] [Google Scholar]
- 8.Guiliano DB, Blaxter ML. Operon conservation and the evolution of trans-splicing in the phylum Nematoda. Public Library of Science-Genetics. 2006;2:e198. doi: 10.1371/journal.pgen.0020198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maroney PA, Yu YT, Jankowska M, Nilsen TW. Direct analysis of nematode cis- and trans-spliceosomes: a functional role for U5 snRNA in spliced leader addition trans-splicing and the identification of novel Sm snRNPs. RNA. 1996;2:735–45. [PMC free article] [PubMed] [Google Scholar]
- 10.Denker JA, Zuckerman DM, Maroney PA, Nilsen TW. New components of the spliced leader RNP required for nematode trans-splicing. Nature. 2002;417:667–70. doi: 10.1038/nature00783. [DOI] [PubMed] [Google Scholar]
- 11.Denker JA, Maroney PA, Yu YT, Kanost RA, Nilsen TW. Multiple requirements for nematode spliced leader RNP function in trans-splicing. RNA. 1996;2:746–55. [PMC free article] [PubMed] [Google Scholar]
- 12.Hannon GJ, Maroney PA, Denker JA, Nilsen TW. Trans splicing of nematode pre-messenger RNA in vitro. Cell. 1990;61:1247–55. doi: 10.1016/0092-8674(90)90689-c. [DOI] [PubMed] [Google Scholar]
- 13.Hannon GE, Hannon GJ, Maroney PA, Nilsen TW. Transcription of a Nematode U1 Small Nuclear RNA in vitro - 3′-End Formation Requires Cis-Acting Elements Within the Coding Sequence. J Biol Chem. 1994;269:12, 387, 90–12. [PubMed] [Google Scholar]
- 14.Higazi TB, Merriweather A, Shu L, Davis R, Unnasch TR. Brugia malayi: Transient transfection by microinjection and particle bombardment. Exp Parasitol. 2002;100:95–102. doi: 10.1016/S0014-4894(02)00004-8. [DOI] [PubMed] [Google Scholar]
- 15.Higazi TB, DeOliveira A, Katholi CR, Shu L, Barchue J, Lisanby M, Unnasch TR. Identification of elements essential for transcription in Brugia malayi promoters. J Mol Biol. 2005;353:1–13. doi: 10.1016/j.jmb.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 16.Rothstein N, Rajan TV. Characterization of an HSP70 gene from the human filarial parasite, Brugia malayi (Nematoda) Mol Biochem Parasitol. 1991;49:229–37. doi: 10.1016/0166-6851(91)90066-f. [DOI] [PubMed] [Google Scholar]
- 17.Shu L, Katholi C, Higazi T, Unnasch TR. Analysis of the Brugia malayi HSP70 promoter using a homologous transient transfection system. Mol Biochem Parasitol. 2003;128:67–75. doi: 10.1016/s0166-6851(03)00052-5. [DOI] [PubMed] [Google Scholar]
- 18.Higazi TB, Unnasch TR. Intron encoded sequences necessary for trans splicing in transiently transfected Brugia malayi. Mol Biochem Parasitol. 2004;137:181–84. doi: 10.1016/j.molbiopara.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 19.Takacs AM, Denker JA, Perrine KG, Maroney PA, Nilsen TW. A 22-nucleotide spliced leader sequence in the human parasitic nematode Brugia malayi is identical to the trans-spliced leader exon in Caenorhabditis elegans. Proc Nat Acad Sci USA. 1988;85:7932–36. doi: 10.1073/pnas.85.21.7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blaxter ML, Raghavan N, Ghosh I, Guiliano D, Lu W, Williams SA, Slatko B, Scott AL. Genes expressed in Brugia malayi infective third stage larvae. Mol Biochem Parasitol. 1996;77:77–93. doi: 10.1016/0166-6851(96)02571-6. [DOI] [PubMed] [Google Scholar]
- 21.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: A sequence logo generator. Genome Res. 2004;14:1188–90. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghedin E, Wang S, Foster JM, Slatko BE. First sequenced genome of a parasitic nematode. Trends Parasitol. 2004;20:151–53. doi: 10.1016/j.pt.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 23.McAlinden A, Havlioglu N, Liang L, Davies SR, Sandell LJ. Alternative splicing of type II procollagen exon 2 is regulated by the combination of a weak 5′ splice site and an adjacent intronic stem-loop cis element. J Biol Chem. 2005;280:32, 700, 11–32. doi: 10.1074/jbc.M505940200. [DOI] [PubMed] [Google Scholar]
- 24.Glazov EA, Pheasant M, Nahkuri S, Mattick JS. Evidence for Control of Splicing by Alternative RNA Secondary Structures in Dipteran homothorax pre-mRNA. RNA Biol. 2006;3:1. doi: 10.4161/rna.3.1.2719. [DOI] [PubMed] [Google Scholar]