Abstract
Nonhistone chromosomal protein HMG-14 is a nucleosomal binding protein that unfolds the higher-order chromatin structure and enhances the transcriptional potential of chromatin, but not that of DNA. Both the transcriptional enhancement and the chromatin unfolding activities of HMG-14 are mediated through the C-terminal region of the protein. Here we study the molecular interactions of both this region and the N-terminal region of HMG-14 with nucleosome cores. By protein photocrosslinking we demonstrate that the N-terminal domain of HMG-14 targets a restricted region in histone H2B, whereas the C-terminal chromatin unfolding domain of HMG-14 targets a restricted region in the N terminus of histone H3. The N-terminal regions of the core histones are involved in the folding of oligonucleosomes and are the target of various activities associated with chromatin unfolding and transcriptional activation. We suggest that specific interactions between the C-terminal domain of HMG-14 and the N-terminal tail of histone H3 reduce the compaction of chromatin. These findings provide insights into the molecular mechanism whereby HMG-14/-17 proteins reduce the repressive effect of chromatin, and they also broaden the scope of the molecular interactions involving the N termini of the core histones in nucleosomes.
In eukaryotic cells, the chromatin fiber serves not only to package the DNA into the confines of the nucleus but also to suppress and regulate various cellular processes involving DNA. Transcriptional activation is associated with rearrangements in the structure of chromatin, both at the level of the nucleosome—i.e., at the primary unit of chromatin organization—and at the higher-order chromatin levels—i.e., the folding of nucleosomal arrays into condensed, compact, structures (1–5). The higher-order chromatin structure is stabilized by the linker histone H1 (1, 2, 6) and by internucleosomal interactions involving the N termini of the core histones (7–11), which are relatively exposed (11) and protrude beyond the nucleosomal surface (12, 13). It has been suggested that nonhistone proteins such as the high mobility group proteins (HMG) could induce structural changes and affect various processes such as transcription and replication occurring in chromatin (reviewed in ref. 14). In this study we demonstrate that HMG-14 protein specifically targets the N-terminal region of a core histone, suggesting a molecular mechanism for decompaction of chromatin by HMG-14/-17 proteins.
Nonhistone chromosomal protein HMG-14, and its close homologue HMG-17, are the only nuclear proteins that bind specifically to the 146-bp nucleosome core particle (reviewed in ref. 14). The proteins bind to the nucleosome core cooperatively to form complexes containing either two molecules of HMG-14 or two molecules of HMG-17 (15). Both proteins contact the DNA 25 bp from the end of the DNA and in the two major grooves flanking the nucleosomal dyad axis (16–18). Results from several laboratories indicate that HMG-14/-17 can stimulate both RNA polymerase II and RNA polymerase III transcription from chromatin templates but not from deproteinized “naked” DNA (19–24). Likewise, HMG-17 increases the replication efficiency of a chromatin template but not of protein-free DNA (25). These findings suggest that the proteins act as modifiers of chromatin structure rather than polymerase-specific factors. Indeed, sedimentation analysis and nuclease digestion studies revealed that the HMG-14/-17-mediated transcriptional stimulation is associated with decompaction of the nucleosome array in the chromatin template (21, 23). Therefore, we suggested that the binding of HMG-14/-17 to nucleosomes induces an extended chromatin conformation and enhances the utilization of chromatin templates (23, 26).
What is the mechanism whereby these proteins decompact chromatin? Both transcriptional activation and decompaction of the chromatin template are mediated by the negatively charged C-terminal domain of HMG-14 (21, 24). Because HMG-14 binds specifically to the nucleosome core particle, we assume that its C-terminal domain decompacts nucleosome arrays by interacting with a component of the nucleosome core. Indeed, contacts between the C terminus of HMG-14/-17 and nucleosome cores have been detected (27); however, the nucleosomal target of the C terminus was not identified. In this study, we use site-specific photocrosslinking (28) to study interactions between either the N-terminal domain or the C-terminal domain of HMG-14 and the nucleosome core. We find that each HMG-14 domain targets a restricted region in a specific histone. The results provide insights into the mechanism whereby nonhistone chromosomal proteins facilitate various DNA-dependent activities in the context of chromatin.
MATERIALS AND METHODS
Nucleosome Cores.
Cores were prepared from HeLa cells, grown in either the presence or the absence of 10 mM sodium butyrate, according to Ausio and van Holde (29).
Generation of Mutant Proteins.
The HMG-14 mutant proteins were generated by site-directed mutagenesis using standard PCR procedures and were expressed as described in ref. 30.
Labeling of the Crosslinker with 125I and Coupling to Mutant HMG-14.
The crosslinking reagent S-[N-(4-azidosalicyl)cysteaminyl]-2-thiopyridyl (28) was purchased from Molecular Probes. Borosilicate glass tubes (12 × 75 mm) containing 0.1 mg of Iodo-Gen (Pierce) were prepared by evaporating 100 μl of a 1 mg/ml solution of Iodo-Gen in dichloromethane under a very gentle stream of nitrogen. Fifty microliters of a reaction mixture containing 0.35 mM crosslinker, 1 mCi (1 mCi = 37 MBq) of Na125I, and 100 mM sodium borate (pH 8.4), was transferred to the Iodo-Gen-coated borosilicate tubes. After 1 min at 22°C the labeling reaction was terminated and the coupling reaction was started by transfer of the solution to a vial containing 30 μg of mutant HMG-14 protein in 50 μl of 100 mM sodium borate, pH 8.4. This vial was incubated at 22°C for 30 min. The modified HMG protein was purified by chromatography on a Bio-Gel P6DG spin column (Bio-Rad).
UV-Crosslinking.
We followed the procedure of Chen et al. (28). The modified HMG proteins were bound to nucleosome core particles in 40 mM Tris⋅HCl, pH 8.0/50 mM NaCl/3 mM MgCl2/5% (vol/vol) glycerol, and the reaction mixture was incubated for 15 min on ice. The mixtures contained 1 μg of cores in 10 μl total volume. In the peptide mapping experiments the reaction mixture contained 50 μg of cores in 500 μl total volume. The Eppendorf reaction vials were placed inside borosilicate glass tubes (13 × 100 mm) and UV-irradiated (360 nm) for 30 sec from a distance of 20 cm, by using a Black-Ray long-wave UV lamp. If the samples were to be analyzed on polyacrylamide gels, 1 vol of 2× SDS-sample buffer containing 4% (vol/vol) 2-mercaptoethanol was added. For purification of the histone proteins by reverse-phase HPLC, 10 mM MnCl2 was added, followed by digestion of the nucleosomal DNA with 20 units of DNase I (Worthington) for 15 min at 37°C. After addition of 35% (vol/vol) acetonitrile and 2% 2-mercaptoethanol the samples were heated to 95°C for 5 min, cooled on ice, made 0.1% in trifluoroacetic acid (TFA), and loaded on the HPLC column.
Protein and Peptide Purification by HPLC.
The histone proteins were purified on a C4 reverse-phase HPLC column (2.1 mm inner diameter × 250 mm; Vydac) using a 35–55% acetonitrile/0.1% TFA gradient at a flow rate of 0.25 ml/min. The peptides derived from histones H2B and H3 were purified on a C18 reverse-phase HPLC column (2.1 mm inner diameter × 250 mm; Vydac) using a 3–55% acetonitrile/0.1% TFA gradient at the same flow rate.
Cleavage of Histone Proteins.
The proteins were cleaved with endoproteinase Glu-C (Boehringer Mannheim) in 25 mM ammonium carbonate buffer (pH 7.8) at 25°C for 18 h, at a protein-to-enzyme ratio of 10:1 or 100:1.
Proteins were cleaved with endoproteinase Asp-N (Boehringer Mannheim) in 50 mM sodium phosphate buffer (pH 8.0) at 37°C for 5 h, at a protein-to-enzyme ratio of 1000:1.
Gel Electrophoresis.
Proteins were separated on 15% polyacrylamide gels containing SDS according to Laemmli (31). Peptides were separated by Tricine/SDS/polyacrylamide gel electrophoresis as described by Schagger and Jagow (32). The composition of these gels was 16.5% T, 6% C. After staining, the gels were scanned on a Molecular Dynamics computer densitometer, dried, and exposed to Kodak X-Omat AR film at −70°C. Electrophoresis in Triton/acid/urea gels was performed as described elsewhere (33).
Protein Sequencing.
The amino acid sequence was determined on either an Applied Biosystems model 476 liquid-phase protein sequencer or a Beckman LF3000 gas-phase protein sequencer, both of which equipped with on-line PTH (phenylthiohydantoin) amino acid analyzers. Samples were loaded (in HPLC solvent) directly onto Polybrene-treated glass fiber filters for the Applied Biosystems instrument or on Beckman peptide disks for either instrument. Mass spectral analysis were accomplished by using a PerSeptive Biosystems (Framingham, MA) matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometer according to the manufacturer’s instructions. Samples were spotted onto the sample plate in 50% acetonitrile/water containing 0.1% TFA along with α-cyano-4-hydroxycinnamic acid as the matrix.
RESULTS
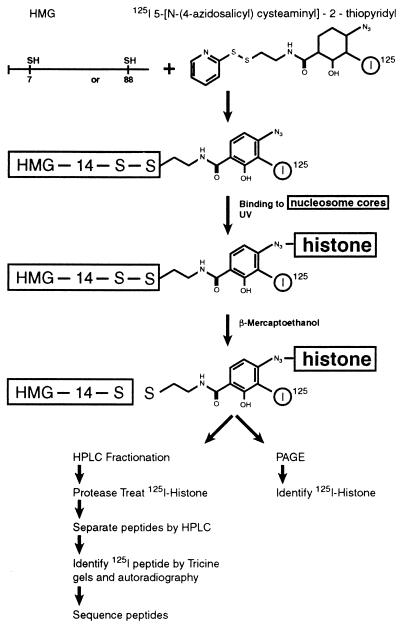
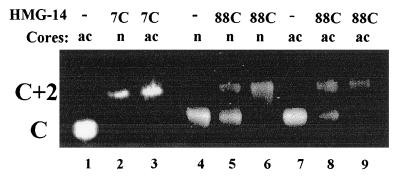
The procedure outlined in Fig. 1 was used to identify the nucleosomal component targeted by either the N-terminal or the C-terminal region of HMG-14. To attach a photoactivable crosslinking reagent to a particular site in the HMG-14 molecule, we constructed two mutant HMG-14 proteins in which either the 7th (a serine, located in the N-terminal domain) or the 88th (a serine, located in the C-terminal activation domain) amino acid residue was replaced by cysteine. Radioactively labeled S-[N-(4-azidosalicyl)cysteaminyl]-2-thiopyridyl, which was previously used to map the targets of specific amino acids in nucleoprotein complexes (28), was covalently attached to the cysteine through a disulfide linkage. The modified proteins retain their ability to bind specifically to nucleosome core particles, as revealed by mobility-shift assays (Fig. 2). The mutant HMG-14 proteins, carrying the crosslinker at either position 7 or position 88, bound cooperatively to the nucleosome cores, forming complexes containing two molecules of HMG per core particle. The level of acetylation of the nucleosomal histones did not significantly affect the binding of the HMG-14 mutants to the core particles.
Figure 1.
Procedure for mapping nucleosome core–HMG-14 interactions. Radioactively labeled S-[N-(4-azidosalicyl)cysteaminyl]-2-thiopyridyl is attached to either 7C or 88C HMG-14 point mutants. The labeled point mutants are complexed with nucleosome cores and the phenylazide moiety is activated by exposure to UV. The bond between the HMG protein and the crosslinking agent is disrupted by 2-mercaptoethanol, and the target of crosslinking is identified by polyacrylamide gel electrophoresis and autoradiography. The 125I-labeled histones are purified by HPLC and proteolyzed, and the resulting peptides are fractionated by HPLC and polyacrylamide gel electrophoresis. The targeted peptides are detected by autoradiography and identified by their amino acid sequence. See text for further details.
Figure 2.
Mobility-shift assays indicate that modified HMG-14 point mutants bind to nucleosome cores. Nucleosome cores containing either nonacetylated (n) or acetylated (ac) histones were incubated either without (lanes 1, 4, and 7) or with a 4-fold molar excess (lanes 2, 5, and 8) or an 8-fold molar excess (lanes 3, 6, and 9) of the modified HMG-14 point mutant indicated on the top of the lanes. C, position of free core particle; C+2, position of core containing two molecules of HMG-14 protein (see also Fig. 3).
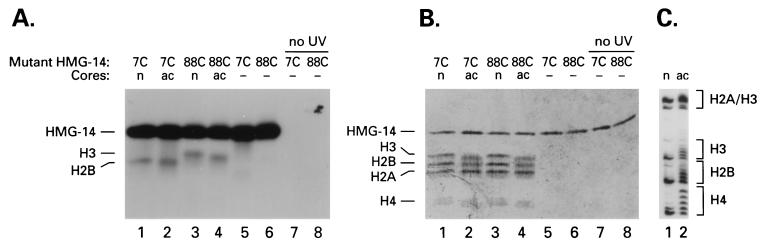
The HMG–nucleosome complex was irradiated with UV light, causing the photoactivable phenylazide moiety of the crosslinker to form a highly reactive nitrene intermediate that forms a covalent bond with any protein or DNA in close vicinity. The linker arm is 15 Å long and suitable for analysis, occurring at or just beyond side-chain contact distance (28). The disulfide bond between the HMG protein and the crosslinker is subsequently cleaved with 2-mercaptoethanol, thereby transferring the radiolabel to the target. No crosslinking to DNA was observed. The protein target in nucleosome cores containing either normal or hyperacetylated histones (Fig. 3C) was identified by SDS gel electrophoresis and autoradiography (Fig. 3 A and B). Crosslinking from amino acid residue 88C occurred exclusively with histone H3, independent of the acetylation state of the core particles (Fig. 3 A and B, lanes 3 and 4). Crosslinking from amino acid residue 7C occurred exclusively with histone H2B, also independent of the acetylation state (Fig. 3 A and B, lanes 1 and 2). As expected, because of intramolecular crosslinking a significant amount of radioactivity was retained in the HMG-14 mutant proteins. The intramolecular crosslinking lowers the yield, but not the specificity, of the targeted crosslinking. In the absence of UV the HMG-14 proteins were not labeled (Fig. 3A, lanes 7 and 8). In the absence of nucleosome cores all the radioactivity was confined to the mutant HMG-14 proteins (Fig. 3A, lanes 5 and 6). These results suggest that in the nucleosome core particle, the C-terminal domain of HMG-14 specifically targets histone H3, whereas the N-terminal region of HMG-14 specifically targets histone H2B. The specificity of the reaction was identical at all the HMG-to-core ratios examined.
Figure 3.
Identification of the targets of the HMG-14 point mutants. Acetylated (ac) or nonacetylated (n) cores containing the 125I-labeled HMG-14 point mutants indicated at the top of the lanes were exposed to UV, treated with 2-mercaptoethanol, and analyzed by SDS/PAGE. (A) Autoradiograph. (B) Coomassie blue stain. Lanes 5–8 contained only the labeled HMG-14 mutants. Lanes 7 and 8 were not exposed to UV and therefore the radioactive label runs with the ion front. (C) The degree of acetylation of the histones in the nucleosome cores was assessed by Coomassie blue staining of Triton/acid/urea polyacrylamide gels (33).
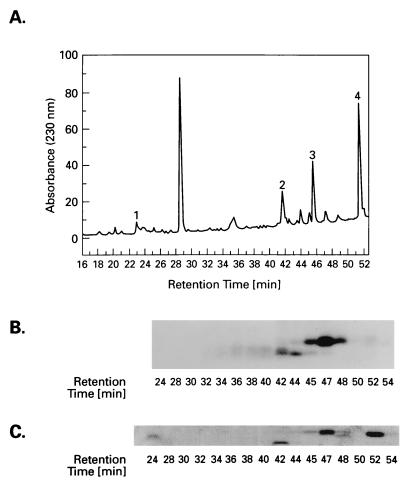
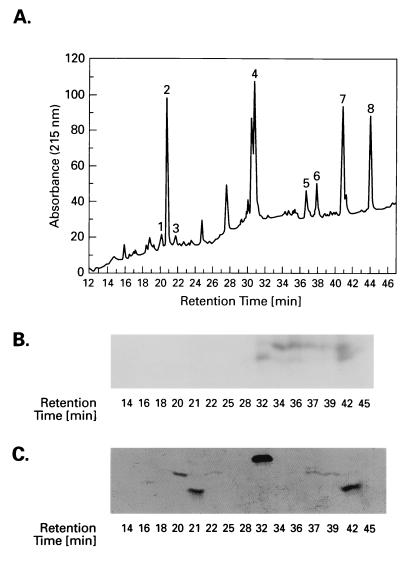
To map the HMG-14 target sites within the histones, the labeled histones, obtained from either acetylated or nonacetylated nucleosome cores, were first separated by HPLC and then proteolyzed with either endoproteinase Glu-C (histone H3) or endoproteinase Asp-N (histone H2B). The digests were fractionated on HPLC (Fig. 4A and 5A), and each of the HPLC fractions was examined by electrophoresis in a second dimension on polyacrylamide gels. The peptides within each fraction were visualized by staining the gels with Coomassie blue (Figs. 4C and 5C) and the targeted peptides were identified by autoradiography (Figs. 4B and 5B). The matching of the radioactivity with the Coomassie blue-stained peptides is complicated by the fact that the crosslinker is a hydrophobic compound that causes a shift in the HPLC-elution time of hydrophilic peptides. Therefore, the labeled fraction of a hydrophilic peptide will elute later in an HPLC gradient than the unlabeled fraction. The more hydrophilic the peptide is, the greater the difference in elution time will be. However, by comparing the mobility of the radioactive and the unlabeled Coomassie blue-stained peptides in SDS/polyacrylamide gels we were able to unambiguously identify the labeled peptides. All the peptides were purified and identified by sequence analysis and, in some cases, additionally by mass spectroscopy. For both histone H2B and histone H3 the sum of the peptides accounted for the entire protein sequence.
Figure 4.
Identification of the region in histone H3 targeted by the HMG-14 88C point mutant. A proteolytic digest of the 125I-labeled histone was fractionated by HPLC (A) and the peptides present in the fractions were analyzed by PAGE. The targeted peptides were identified by autoradiography (B) of Coomassie blue-stained gels (C).
Figure 5.
Identification of the region in histone H2B targeted by the HMG-14 7C point mutant. A proteolytic digest of the 125I-labeled histone was fractionated by HPLC (A) and the peptides present in the fractions were analyzed by PAGE. The targeted peptides were identified by autoradiography (B) of Coomassie blue-stained gels (C).
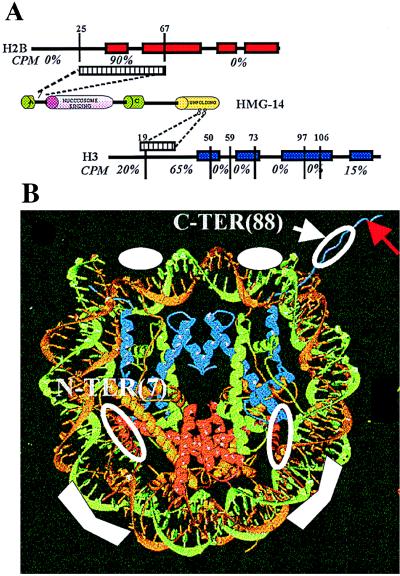
Fig. 6A summarizes the distribution of the radioactivity in the various regions of the histones targeted by amino acid residue 7 or 88 of HMG-14. Amino acid 88, located in the chromatin unfolding domain of HMG-14, targets histone H3. Most of the label (≈65%) is found in the peptide spanning amino acid residues 20–50. When not labeled, this peptide elutes at 31 min. (Fig. 4A, peak 4), whereas when labeled, it elutes between 32 and 42 min (Fig. 4B). The broadened elution peak of the labeled peptide could be due to differential elution time, depending on the location of the label in the peptide. Toward the N terminus, 20% of the label is found in the peptide spanning amino acid residues 1–19. This peptide when unlabeled elutes in two peaks at 20 and 22 min (Fig. 4A, peaks 1 and 3), and when labeled it elutes between 32 and 36 min (Fig. 4B). Toward the C-terminal region, the abutting peptide, spanning amino acid residues 51–59 (Fig. 4A, peak 2), is not labeled. Taken together, these results indicate that the C-terminal activation domain of HMG-14 targets a region close to the N terminus of the peptide spanning amino acid residues 20–50 in the N-terminal tail of histone H3 (see Fig. 6A). This region is close to the C terminus of the tail region, which is removed by limited trypsin digestion of nucleosome cores (residue 1–27) (34). The distribution of radioactivity among the peptides was independent of the acetylation level of the histones (not shown), perhaps because most of the contact area is located outside the region containing the major acetylation sites (K9, K14, K18, K23). A small amount of label (≈15%) is found near the C terminus of H3, in the peptide spanning amino acid residues 106–133, eluting when unlabeled in two peaks (Fig. 4A, peaks 5 and 6) at 36.5 and 38 min, when labeled, between 39 and 42 min (Fig. 4B). This peptide contains the L2 loop of H3 (residues 115–120), which contacts the nucleosomal DNA on either side of the dyad axis (12). In hydroxy radical footprinting experiments, this area of the DNA was protected by HMG-17 (16).
Figure 6.
Organization of HMG-14 in nucleosome cores. (A) Specific crosslinks between HMG-14 and histones in nucleosome cores. The α-helical regions in the histones are depicted as boxes. The evolutionarily conserved domains in HMG-14 are depicted as cylinders. The amino acid positions at which the proteases cleaved the histones are indicated above their sequence. The radioactive counts present in each of the peptides is indicated, expressed as percent of total. The regions targeted in H2B and H3 by amino acid residues 7 and 88 of HMG-14, respectively, are indicated by the striped boxes. (B) Model of the HMG-14 binding sites. The ribbon traces for the DNA and histones in the core particles were reproduced by permission from the recent article in Nature by Luger et al. (12), copyright 1997, Macmillan Magazines Ltd. The solid white symbols, in the two major grooves flanking the dyad axis and approximately 25 bp from the end of the DNA, indicate the regions where HMG-14/-17 protect the DNA from hydroxyl radical cleavage. The open white circles represent the approximate location of the crosslinks identified in the present study. The red arrow points to the N-terminal region of histone H3. Histones H2B and H3 are represented by red and blue ribbons, respectively.
The HMG-14 mutant carrying the crosslinker at amino acid residue 7 specifically targets histone H2B. Almost all the label is found in the peptide eluting at 46 min (Fig. 5A, peak 3). Sequencing and mass spectroscopy revealed that this peptide spans residues 25–66 of histone H2B, which includes the N-terminal half of the histone fold domain and some residues preceding it, but not the N-terminal tail proper (residues 1–24) as defined by limited trypsin digestion (34)(see also ref. 11 for references).
A model depicting the known sites of interactions between HMG-14 and the nucleosome core particles is presented in Fig. 6B. The nucleosome model is reproduced from the recently published article by Luger et al. (12). The main contacts of HMG-14 with DNA, as mapped by hydroxyl radical and DNase I footprinting (16–18) and by crosslinking (35), are indicated by solid white symbols. They occur in the two major grooves flanking the dyad axis and approximately 25 bp from the end of the nucleosomal DNA. The open circles depict the protein–protein contacts identified in the present study. The N-terminal region of HMG-14 crosslinks in the N-terminal half of the H2B histone fold domain. This region is located inside the nucleosomal DNA approximately 15–25 bp from its end—i.e., near one of the main HMG–DNA contacts (16–18, 35). The C-terminal domain of HMG-14 contacts the N-terminal domain of histone H3 in the region where it exits from the nucleosome—i.e., near the ends of the nucleosomal DNA (12). The detailed structure and location of the N terminus of H3 are not known.
DISCUSSION
In view of the recent evidence that the acidic C-terminal domain of HMG-14 facilitates transcription and replication of chromatin by unfolding the higher-order chromatin structure (21, 23–25), it was important to identify the target of this domain in the nucleosome core. We have used site-specific crosslinking to map the contacts between the HMG-14 and the nucleosome core, by identifying the targets of a specific amino acid located in either the N-terminal or the C-terminal chromatin unfolding domain of HMG-14. Earlier studies on the location of HMG-14/-17 in nucleosomes used crosslinkers that reacted with all the lysines in the proteins and therefore were not suitable for identifying the nucleosomal targets of specific regions in the protein (36–38). These studies identified multiple contacts between HMG-14/-17 and all of the core histones. Our findings that the site-specific crosslinkers target distinct regions in specific histones suggest that the mobility of the HMG-14 on the nucleosome is limited and argue for specific placement of the HMG-14 protein in the nucleosome cores.
The main finding of the present study is that the chromatin unfolding domain of HMG-14 targets the N-terminal tail of histone H3 in the nucleosome core. We also find that the N–terminal domain of HMG-14 crosslinks to a specific protein domain in H2B. Both of the histone domains that crosslink to HMG-14 are located in the peripheral part of the nucleosome (12, 13). The location of the crosslinks between HMG and the nucleosomal histones in our study is consistent with earlier crosslinking and footprinting studies, which also placed the HMG molecule at the periphery of the nucleosome core (16, 27, 35, 37, 38).
We have documented that the C-terminal region of HMG-14 is necessary for transcriptional activation, whereas the functional significance of the N-terminal domain of HMG-14 is not known. A mutant HMG-14 protein lacking the first 10 amino acids in the N-terminal region binds to nucleosome cores and is not impaired in its ability to either unfold chromatin or stimulate transcription from chromatin templates (21, 24). The interaction between the C-terminal domain of HMG-14 and the tail domain of histone H3 provides clues regarding the molecular mechanism whereby HMG-14/-17 may unfold the higher-order chromatin structure. It is well documented that the N termini of the core histones are involved in various processes leading to rearrangements in the structure of chromatin and stimulation of DNA-dependent activities (2, 11). The histone N termini are necessary for the activity of the nucleosomal rearrangement factor NURF (39). Removal of the termini prevents nucleosomal arrays from folding into a higher-order structure (8). The tail domains undergo numerous posttranslational modifications, such as acetylation, phosphorylation, methylation, ubiquitination, and ADP-ribosylation (reviewed in ref. 11). The functional significance of most of the modifications is not known, but a link between transcriptional activation and acetylation has been firmly established (40–42).
Some of the interactions between the N termini of histones and nonhistone proteins lead to chromatin inactivation. For example, in yeast, the tail domains of histones H3 and H4 interact with the nonhistone proteins SIR 3, SIR 4, and TUP1 (reviewed in refs. 11 and 40), all of which are involved in establishing repressive chromatin structures. The present study extends the range of known specific interactions between nonhistone chromosomal proteins and the tail domain of histone H3. Competitive interactions at the H3 N terminus between proteins that compact chromatin and those that decompact it could affect the structure and function of a given chromatin region.
What is the mechanism whereby specific interactions between the C-terminal domain of HMG-14 and the N-terminal domain of histone H3 unfold the higher-order chromatin structure? One obvious possibility is that these interactions disrupt H3-mediated internucleosomal contacts. The crystal structure of the nucleosome core (12) shows that residues 39–43 of histone H3 pass between the gyres of the DNA superhelix, through channels formed by the minor grooves of the DNA terminus and the central turn near the dyad axis. Although the tail domain of H3 is not visible in the crystal structure, Richmond and his co-workers (12) suggested that it protrudes from the nucleosome and interacts with neighboring nucleosomes in the chromatin fiber. Thus, HMG-14/-17 may disrupt the higher-order chromatin structure by interfering with these interactions. A second possibility stems from data suggesting that in the nucleosome core, the H3 (and H4) tail domains have a defined structure and are bound to DNA (43). Because the H3 tails are located at the end of the nucleosome, it is conceivable that the interaction between histone H3 and HMG-14/-17 changes the entry/exit angle of the nucleosomal DNA, thereby unfolding the 30-nm fiber (44, 45). A third possibility stems from the finding that the position of HMG-14/-17 in nucleosome cores is close to that of histone H1 (16) and that HMG-14 can counteract H1-mediated transcriptional repression and chromatin compaction (21). It is well documented that H1 compacts chromatin and stabilizes higher-order chromatin structure. Interactions between histone H1 and the N termini of core histones have been observed (46). By affecting these interactions HMG-14 could unfold chromatin. All these possibilities are consistent with the proposed location of HMG-14 in the nucleosome core and with the proposed role of H3 N termini in stabilizing the higher-order chromatin structure.
What is the size of the chromatin domain unfolded by HMG-14? We have recently reported that in chromatin, nucleosomes containing HMG-14 are clustered into distinct domains that on the average contain six contiguous nucleosomes (47). It is possible that one of the mechanisms described above is responsible for unfolding the chromatin structure in such an HMG-14-containing chromatin domain. To further elucidate the molecular mechanism whereby HMG-14/-17 unfolds chromatin it will be necessary to study the interactions between nucleosomes and HMG in nucleosomal arrays, and to map the location of the histone tail domains.
Acknowledgments
We thank Ms. Anke Rickers for help in generating the HMG mutants, Dr. Terry Sumpter for mass spectroscopy analysis, and Drs. J. Herrera, K. Marsh, Y. Postnikov, and J. Wagner for critical comments on the manuscript.
ABBREVIATIONS
- HMG
high mobility group
- TFA
trifluoroacetic acid
References
- 1.van Holde K E. Chromatin. New York: Springer; 1988. [Google Scholar]
- 2.Wolffe A P. Chromatin Structure and Function. London: Academic; 1995. [Google Scholar]
- 3.Kornberg R D, Lorch Y. Curr Opin Cell Biol. 1995;8:371–375. doi: 10.1016/0955-0674(95)80092-1. [DOI] [PubMed] [Google Scholar]
- 4.Adams C C, Workman J L. Cell. 1993;72:305–308. doi: 10.1016/0092-8674(93)90109-4. [DOI] [PubMed] [Google Scholar]
- 5.Paranjape S M, Kamakaka R T, Kadonaga J T. Annu Rev Biochem. 1994;63:265–297. doi: 10.1146/annurev.bi.63.070194.001405. [DOI] [PubMed] [Google Scholar]
- 6.Thoma F, Koller T. Cell. 1977;12:101–107. doi: 10.1016/0092-8674(77)90188-x. [DOI] [PubMed] [Google Scholar]
- 7.Hansen J C, Ausio J. Trends Biochem Sci. 1992;17:187–191. doi: 10.1016/0968-0004(92)90264-a. [DOI] [PubMed] [Google Scholar]
- 8.Garcia-Ramirez M, Dong F, Ausio J. J Biol Chem. 1992;267:19587–19595. [PubMed] [Google Scholar]
- 9.Krajewski W A, Ausio J. Biochem J. 1996;316:395–400. doi: 10.1042/bj3160395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tse C, Hansen J C. Biochemistry. 1997;36:11381–11388. doi: 10.1021/bi970801n. [DOI] [PubMed] [Google Scholar]
- 11.Hansen J. Chemtracts: Biochem Mol Biol. 1997;10:56–69. [Google Scholar]
- 12.Luger K, Mader A W, Richmond R K, Sargent D F, Richmond T J. Nature (London) 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 13.Arents G, Moudrianakis E N. Proc Natl Acad Sci USA. 1993;90:10489–10493. doi: 10.1073/pnas.90.22.10489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bustin M, Reeves R. Prog Nucleic Acid Res Mol Biol. 1996;54:35–100. doi: 10.1016/s0079-6603(08)60360-8. [DOI] [PubMed] [Google Scholar]
- 15.Postnikov Y V, Trieschmann L, Rickers A, Bustin M. J Mol Biol. 1995;252:423–432. doi: 10.1006/jmbi.1995.0508. [DOI] [PubMed] [Google Scholar]
- 16.Alfonso P J, Crippa M P, Hayes J J, Bustin M. J Mol Biol. 1994;236:189–198. doi: 10.1006/jmbi.1994.1128. [DOI] [PubMed] [Google Scholar]
- 17.Mardian J K, Paton A E, Bunick G J, Olins D E. Science. 1980;209:1534–1536. doi: 10.1126/science.7433974. [DOI] [PubMed] [Google Scholar]
- 18.Sandeen G, Wood W I, Felsenfeld G. Nucleic Acids Res. 1980;8:3757–3778. doi: 10.1093/nar/8.17.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crippa M P, Trieschmann L, Alfonso P J, Wolffe A P, Bustin M. EMBO J. 1993;12:3855–3864. doi: 10.1002/j.1460-2075.1993.tb06064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding H F, Rimsky S, Batson S C, Bustin M, Hansen U. Science. 1994;265:796–799. doi: 10.1126/science.8047885. [DOI] [PubMed] [Google Scholar]
- 21.Ding H F, Bustin M, Hansen U. Mol Cell Biol. 1997;17:5843–5855. doi: 10.1128/mcb.17.10.5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paranjape S M, Krumm A, Kadonaga J T. Genes Dev. 1995;9:1978–1991. doi: 10.1101/gad.9.16.1978. [DOI] [PubMed] [Google Scholar]
- 23.Trieschmann L, Alfonso P J, Crippa M P, Wolffe A P, Bustin M. EMBO J. 1995;14:1478–1489. doi: 10.1002/j.1460-2075.1995.tb07134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trieschmann L, Postnikov Y, Rickers A, Bustin M. Mol Cell Biol. 1995;15:6663–6669. doi: 10.1128/mcb.15.12.6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vestner, B., Bustin, M. & Gruss, C. (1998) J. Biol. Chem., in press. [DOI] [PubMed]
- 26.Bustin M, Trieschmann L, Postnikov Y V. Semin Cell Biol. 1995;6:247–255. doi: 10.1006/scel.1995.0033. [DOI] [PubMed] [Google Scholar]
- 27.Cook G R, Minch M, Schroth G P, Bradbury E M. J Biol Chem. 1989;264:1799–1803. [PubMed] [Google Scholar]
- 28.Chen Y, Ebright Y W, Ebright R H. Science. 1994;265:90–92. doi: 10.1126/science.8016656. [DOI] [PubMed] [Google Scholar]
- 29.Ausio J, van Holde K. Biochemistry. 1988;25:1421–1428. doi: 10.1021/bi00354a035. [DOI] [PubMed] [Google Scholar]
- 30.Bustin M, Becerra P S, Crippa M P, Lehn D A, Pash J M, Shiloach J. Nucleic Acids Res. 1991;19:3115–3121. doi: 10.1093/nar/19.11.3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 32.Schagger H, Jagow G. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 33.Lennox R W, Cohen L H. Methods Enzymol. 1989;170:532–548. doi: 10.1016/0076-6879(89)70063-x. [DOI] [PubMed] [Google Scholar]
- 34.Bohm L, Crane-Robinson C. Biosci Rep. 1984;4:365–386. doi: 10.1007/BF01122502. [DOI] [PubMed] [Google Scholar]
- 35.Shick V V, Belyavsky A V, Mirzabekov A D. J Mol Biol. 1985;185:329–339. doi: 10.1016/0022-2836(85)90407-3. [DOI] [PubMed] [Google Scholar]
- 36.Espel E, Bernues J, Perez-Pons J A, Querol E. Biochem Biophys Res Commun. 1985;132:1031–1037. doi: 10.1016/0006-291x(85)91910-2. [DOI] [PubMed] [Google Scholar]
- 37.Cook G R, Yau P, Yasuda H, Traut R R, Bradbury E M. J Biol Chem. 1986;261:16185–16190. [PubMed] [Google Scholar]
- 38.Brawley J V, Martinson H G. Biochemistry. 1992;31:364–370. doi: 10.1021/bi00117a008. [DOI] [PubMed] [Google Scholar]
- 39.Georgel P T, Tsukiyama T, Wu C. EMBO J. 1997;16:4717–4726. doi: 10.1093/emboj/16.15.4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grunstein M. Nature (London) 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 41.Brownell J E, Allis C D. Curr Opin Genet Dev. 1996;6:176–184. doi: 10.1016/s0959-437x(96)80048-7. [DOI] [PubMed] [Google Scholar]
- 42.Turner B M, O’Neill L P. Semin Cell Biol. 1995;6:229–236. doi: 10.1006/scel.1995.0031. [DOI] [PubMed] [Google Scholar]
- 43.Baneres J L, Martin A, Parello J. J Mol Biol. 1997;273:503–508. doi: 10.1006/jmbi.1997.1297. [DOI] [PubMed] [Google Scholar]
- 44.Woodcock C L, Horowitz R A. Trends Cell Biol. 1995;5:272–277. doi: 10.1016/s0962-8924(00)89038-8. [DOI] [PubMed] [Google Scholar]
- 45.van Holde K, Zlatanova J. Proc Natl Acad Sci USA. 1996;93:10548–10555. doi: 10.1073/pnas.93.20.10548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Juan L J, Utley R T, Adams C C, Vettese-Dadey M, Workman J L. EMBO J. 1994;13:6031–6040. doi: 10.1002/j.1460-2075.1994.tb06949.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Postnikov Y V, Herrera J E, Hock R, Scheer U, Bustin M. J Mol Biol. 1997;274:454–465. doi: 10.1006/jmbi.1997.1391. [DOI] [PubMed] [Google Scholar]