SUMMARY
Circadian clocks are widely distributed in mammalian tissues, but little is known about the physiological functions of clocks outside the suprachiasmatic nucleus of the brain. The retina has an intrinsic circadian clock, but its importance for vision is unknown. Here we show that mice lacking Bmal1, a gene required for clock function, had abnormal retinal transcriptional responses to light and defective inner retinal electrical responses to light, but normal photoreceptor responses to light and retinas that appeared structurally normal by light and electron microscopy. We generated mice with a retina-specific genetic deletion of Bmal1, and they had defects of retinal visual physiology essentially identical to those of mice lacking Bmal1 in all tissues and lacked a circadian rhythm of inner retinal electrical responses to light. Our findings indicate that the intrinsic circadian clock of the retina regulates retinal visual processing in vivo.
INTRODUCTION
Circadian clocks are endogenous oscillators that drive daily rhythms of physiology and behavior. In mammals, the circadian clock mechanism is built upon a molecular feedback loop in which the CLOCK-BMAL1 transcription factor drives expression of its PER and CRY inhibitors (Gekakis et al., 1998; Ko and Takahashi, 2006). The clock generates circadian rhythms cell-autonomously, and it is thought to generate rhythms of physiology in large part by driving rhythms of transcription of output genes (Panda et al., 2002; Storch et al., 2002; Akhtar et al., 2002; Duffield et al., 2002). The adaptive significance of circadian clocks likely lies in their ability to allow anticipatory responses to predictable daily variations in the environment (Ouyang et al., 1998).
It has long been known that the circadian clock regulating behavior in mammals is located in the suprachiasmatic nucleus (SCN) of the brain (Ko and Takahashi, 2006). Recently it has become clear that circadian clocks are distributed in mammalian tissues, present at sites such as the retina (Tosini and Menaker, 1996), multiple brain regions (Abe et al., 2002), and in many peripheral tissues (Balsalobre et al., 1998; Yamazaki et al., 2000; Damiola et al., 2000). It is thought that clocks outside the SCN have physiological functions, but to date few studies have addressed this question (Durgan et al., 2006; McDearmon et al., 2006). In Drosophila there is compelling evidence that a clock in the antenna drives rhythms of olfactory sensitivity (Tanoue et al., 2004).
Retinas from a wide range of vertebrates, including amphibians (Besharse and Iuvone, 1983), birds (Pierce et al., 1993), and mammals (Tosini and Menaker, 1996), contain a circadian clock. In Xenopus retina, photoreceptors are the circadian clock cells (Cahill and Besharse, 1993; Hayasaka et al., 2002), whereas in the mammalian retina circadian clock cells are found in the inner retina (Witkovsky et al., 2003; Gustincich et al., 2004; Ruan et al., 2006) (Supplemental Figure 1).
Fundamental retinal processes are under circadian control, including photoreceptor disc shedding (LaVail and Ward; 1978), release of melatonin and dopamine (Doyle et al., 2002), and retinal electrical responses to light, manifested as a circadian rhythm of one or more components of the electroretinogram (ERG) (Manglapus et al., 1998; Barnard et al., 2006). At present little is known about the biochemical pathways under circadian control in the mammalian retina or the molecular mechanisms that modulate retinal physiological responses to light. A circadian rhythm of melatonin release is driven autonomously from the retina (Tosini and Menaker, 1996), but it is not yet known to what extent circadian rhythms of retinal electrical activity in response to light reflect the action of a local retinal clock or the action of a remote clock, such as the SCN. Evidence from non-mammalian vertebrates suggests that circadian rhythms of retinal electrical responses to light are driven at least in part from the brain (Miranda-Anaya et al., 2002) or pineal (McGoogan and Cassone, 1999).
RESULTS
Daily rhythms of retinal gene expression
To gain a view of molecular regulation in the mammalian retina by a circadian clock, light, or both, we performed whole-genome microarray studies in mice to identify genes with a ∼24-hr rhythm of expression during a three-day interval in constant darkness (DD) or in a 12:12-hr light-dark cycle (LD) (Supplemental Figure 2). Because of the role of melatonin in retinal function (Doyle et al., 2002), we used CBA/CaJ mice, a strain that makes melatonin (Goto et al., 1989) and does not have retinal degeneration. To optimize the efficiency of tissue collection, we used whole eyes for RNA extraction (Supplemental Note 1).
To identify rhythmic variations in gene expression, we computed the best-fit function that models the expression of a gene across the three-day, 18-time-point microarray profile as a ∼24-hr rhythmic pattern; we did not assume a simple waveform. For some genes the best-fitting rhythmic function makes a good fit, for most a poor fit. Next we randomly permuted the order of the 18 time-points for each gene 50,000 times. With 18 time-points, random permutation of the time-series will degrade the fit of a truly rhythmic profile but will have little or no effect on the fit of noisy or flat profiles (Supplemental Figure 2). This procedure allows all or any subset of the 45,101 probe sets on the array to be ranked quantitatively for rhythmicity, and one of its key advantages is that a statistical threshold for rhythmicity can be set according to any desired false discovery rate, defined as the percentage of genes ranking above the threshold that can be accounted for by noise (Supplemental Note 2).
In DD, we identified 277 genes with a circadian rhythm of expression at a moderate threshold corresponding to a 15% false discovery rate, (Figure 1A, top, DD). Phases of peak expression around-the-clock were represented roughly equally (Figure 1A, top, DD), and the dataset included genes with known circadian regulation in other tissues, including clock components (Supplemental Table 1). Overall, genes expressed in the retina made a major contribution to the dataset (Supplemental Note 1; Supplemental Figure 3), and the genes represent a wide range of functions, including synaptic transmission, photoreceptor signaling, intercellular communication, and regulation of the cytoskeleton and chromatin (Supplemental Figure 4, Supplemental Table 1). For some genes, expression was limited to photoreceptors (Supplemental Figure 3), likely reflecting non-cell autonomous regulation that depends on clock cells in the inner retina or elsewhere. The rhythmic dataset included secreted factors expressed exclusively in the inner retina, candidates for circadian signals from inner retinal clock cells to photoreceptors (Supplemental Fig. 3).
Figure 1. Genes with 24-hour rhythms of expression in the mouse eye.
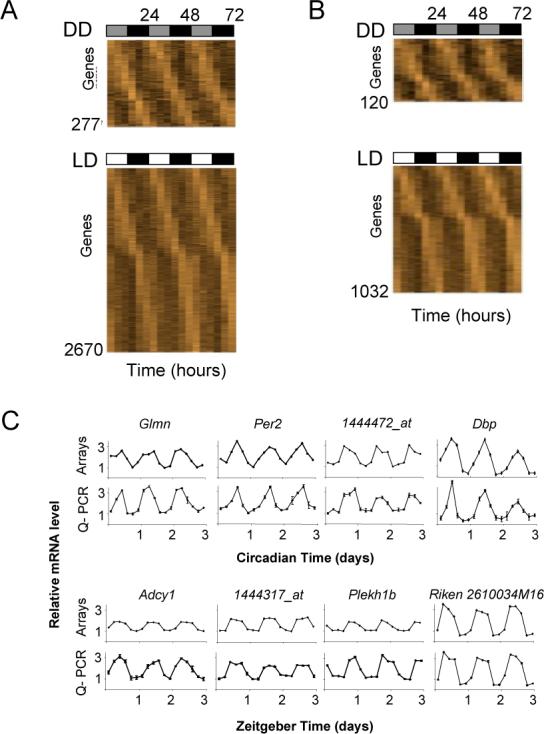
(A, B) Three-day expression profiles in which each column represents a time-point and each row a gene, with genes ordered by phase of peak expression. Light shades represent expression values above the mean for a gene; dark shades below the mean. Total time in constant darkness (DD) or in a light-dark cycle (LD) is indicated at top, and the bars represent subjective day and night in DD or light and dark conditions in LD. The number of genes is indicated at the lower left. (A) Threshold: 15% false discovery rate. (B) Threshold: 5% false discovery rate. (C) Validation of microarray profiles by quantitative reverse-transcriptase PCR (Q-PCR). Top, DD. Bottom, LD. Shown are comparisons of individual profiles from microarrays (arrays) and Q-PCR from the same RNA samples. For Q-PCR, the mean (N = 3) and SEM are shown (some error bars cannot be seen at this scale). Relative expression levels are plotted in arbitrary linear units. Glmn, glomulin; Per2, Period 2, Dbp, D-site albumin promoter binding protein; Adcy 1, adenylate cyclase 1; Plekh1b, plekstrin-homology 1b.
In LD, at the same statistical threshold, we identified 2670 genes with rhythmic expression (Figure 1A, bottom, LD). Included were 80% of the genes with rhythmic expression in DD and many genes with known expression in the retina (Supplemental Table 2). In addition to the ∼9-fold greater number of genes showing rhythmic expression in LD than DD, the distribution of phases in LD differed markedly from that in DD, with somewhat more than half of the genes showing a peak of expression during the night and the rest showing a roughly equal phase distribution (Figure 1A, bottom, LD). Both of these differences between the two conditions were observed at more stringent thresholds (Figure 1B), indicating that they do not arise from chance inclusion of noise. These results suggest that LD cycles drive expression of a large number of genes in the retina and, in particular, a large cluster of genes with a night-time peak of expression. Additional analyses indicated that any loss of circadian synchrony among mice or cells during the three-day period in DD did not contribute substantially to the differences between the DD and LD datasets (Supplemental Note 3; Supplemental Figure 5).
For validation, we selected 26 genes with rhythmic expression from DD and 28 from LD (identified in Supplemental Tables 1 and 2), as well as genes from each classified as non-rhythmic, for assessment by quantitative reverse transcriptase PCR (Q-PCR). In all cases but one, Q-PCR confirmed the rhythmic profiles (examples, Figure 1C). Microarray analysis thus accurately identified genes with rhythmic expression.
Importance of Bmal1 for retinal gene expression rhythms in a light-dark cycle
Given that the retina is a dedicated photosensory organ, the ∼9-fold excess of genes exhibiting rhythmic expression in LD compared to DD could simply result from regulation of many genes by light, independently of circadian clock function. To test this expectation, we compared temporal profiles of ocular gene expression in LD in wildtype mice and Bmal1−/− (Mop3−/−) (Bunger et al., 2000) littermates, which lack an essential component of the clock in all tissues and consequently are expected to lack all clock function. Genes regulated purely by LD cycles would be expected to retain full rhythms of expression in Bmal1−/− mice. In contrast, genes regulated by LD cycles in a manner that depends on clock function would be expected to exhibit altered regulation in LD, even if the genes are primarily driven by light and do not exhibit detectable rhythmic expression in DD. In initial Q-PCR studies of eyes from Bmal1−/− mice in DD, we observed loss of rhythms of expression of Per1, Per2, Dbp, and Rev-erbα, indicating that Bmal1 is required for circadian rhythms in the eye, as expected (Supplemental Figure 6A).
It is important to note that the null mutation of Bmal1 can affect clock-regulated processes in two ways (Supplemental Figure 5)---by eliminating the circadian feedback loop and thus abolishing circadian rhythms (Bunger et al., 2000) and by reducing expression of output genes driven directly by the CLOCK-BMAL1 transcription factor (Panda and Hogenesch, 2004). Therefore any phenotypes observed in Bmal1−/− mice could result from a loss of rhythmicity or from a reduction in one or more clock-driven transcriptional outputs, as well as by hypothetical non-circadian functions of Bmal1. For the purpose of discussion of Bmal1 mutants here and below, we define “circadian clock function” in this broad sense, including both intrinsic rhythmicity and transcriptional outputs of the clock mechanism.
First, to check for any confounding retinal developmental defects in Bmal1−/− mice, we studied retinas from adult Bmal1−/− mice and wildtype littermates by light and electron microscopy. Retinas from the two genotypes were indistinguishable in general architecture, cellular organization and density, the complement of organelles, and the distribution and structure of synaptic specializations of the principal retinal cell types (Figure 2). Although we cannot exclude the possibility of subtle quantitative differences, these results make it highly unlikely that a developmental or structural defect underlies any abnormalities of retinal gene expression or function in Bmal1−/− mice.
Figure 2. Normal retinal architecture, cellular organization, and ultrastructure in Bmal1−/− mutant mice.
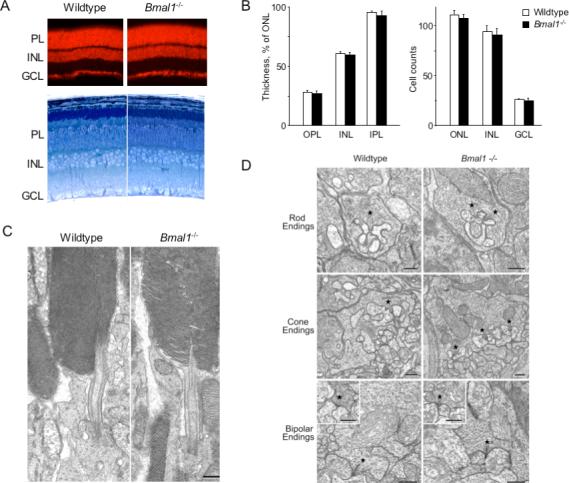
(A) Top, fluorescence images of retina sections from adult littermate wildtype or Bmal1−/− mutant mice stained with ethidium bromide to show cell nuclei. Organization of nuclear layers was indistinguishable in the two genotypes. Bottom, thick plastic sections of retinas from adult littermate wildtype or Bmal1−/− mice stained with toluidine blue to show cell morphology and structure. No difference between genotypes was observed. PL, photoreceptor layer; INL, inner nuclear layer; GCL, ganglion cell layer. (B) Quantitative comparison of thickness (left) and cell densities (right) (mean and SEM; N = 3) of retinal layers (at mid-periphery) of adult wildtype and Bmal1−/− littermate mice (Experimental Procedures). Results for central retina were similar (data not shown). There were no significant differences between genotypes. OPL, outer plexiform layer; IPL, inner plexiform layer. Cell counts: for ONL, INL--per 5000-μm2 area; GCL--per 200-μm segment of retina. (C) Representative electron micrographs of rod photoreceptors from adult littermate wildtype or Bmal1−/− mice. No difference between genotypes was observed in the fine structure of outer and inner segments. (D) Representative electron micrographs from adult littermate wildtype or Bmal1−/− mice. No differences between genotypes were evident in the morphology of synaptic endings of rods, cones, rod bipolars, or cone bipolars (insets) or in the complement of synaptic vesicles or structure of ribbon synapses (asterisks). (C,D) Scale bars, 500 nm.
Next we performed microarray analysis to compare ocular gene expression profiles of wildtype mice and Bmal1−/− littermates (on a C57BL/6 background) in LD at four-hour intervals over one daily cycle. To interpret these one-day datasets, we used information from the three-day experiment shown in Figure 1 (CBA/CaJ mice). For each of the genes showing rhythmic expression in LD in the three-day dataset, we examined the one-day profile in wildtype or Bmal1−/− littermates in LD. A one-day expression profile was classified as rhythmic if it showed a phase and waveform like that in the three-day experiment, defined as a significantly closer match to the mean of its corresponding three-day profile than to independent Gaussian noise (i.e., it behaved statistically like a “fourth day” of the rhythm; Supplemental Note 4).
In wildtype C57BL/6 mice (littermates of Bmal1−/− mice), many genes showed rhythmic regulation matching that in the three-day experiment (Figure 3A; the threshold for a match was set at a stringent 5% false discovery rate). In contrast, at the same threshold very few genes qualified as rhythmic in LD in Bmal1−/− littermates. A comparison of one-day expression profiles classified as rhythmic in wildtype mice with profiles of the same genes from the mutants revealed substantial disorganization in the mutants, although weak features similar to the wildtype pattern were apparent (Figure 3A). This result held true across different statistical thresholds (data not shown). Inspection of many individual gene profiles and validation of 20 by Q-PCR (identified in Supplemental Table 2) indicated that about 60% showed flat or noisy profiles, about 30% showed apparent regulation by light but with reduced amplitude and altered waveform, and about 10% showed fully-preserved rhythms (Figure 3B). In contrast, the Bmal1 mutation had no detectable effect on the expression of >3000 genes that are expressed constitutively in the eye in LD (Figure 3C).
Figure 3. Deficient rhythmic gene expression in LD in the eyes of Bmal1−/− mice.
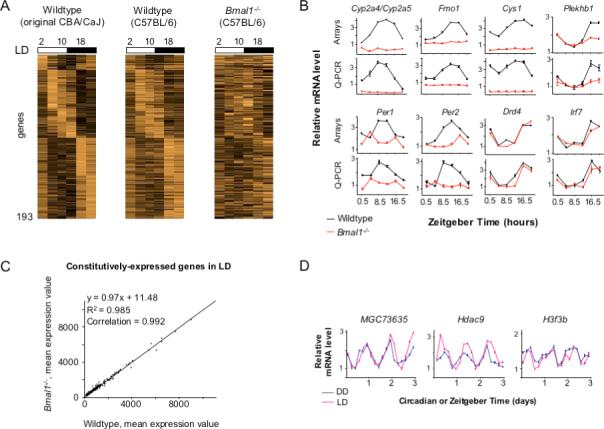
(A) One-day temporal profiles comparing ocular gene expression in LD for wildtype CBA/CAJ mice (the mean of three days—Figure1) and wildtype and littermate Bmal1−/− mice (C57BL/6); labels as in Figure 1. The same genes are shown in all three profiles, genes are ordered by the phase of peak expression in wildtype CBA/CaJ mice (left), and the brightness scale for expression is the same in all profiles. (B) Validation of microarray profiles by Q-PCR. Shown are examples of genes in Bmal1−/− mice with flat, altered, or normal rhythmic expression in comparison with wildtype littermates (C57BL/6). Cyp2a4/5, cytochrome P450−2a4/5; Fmo1, flavin-containing monooxygenase-1; Cys1, cystin 1; Plekh1b, plekstrin-homology 1b; Per1, Period 1; Drd4, D4 dopamine receptor; Irf7, interferon regulatory factor 7. (C) No effect of Bmal1 deletion on mean expression of 3,047 constitutively-expressed genes in the eye in LD (see Experimental Procedures). Genes are plotted in order of increasing mean expression from the wildtype dataset. (D) Daily rhythms of ocular expression of chromatin remodeling genes detected by microarray (Figure 1). MGC73635, similar to histone 2a; Hdac9, histone deacetylase 9; H3f3b, H3 histone, family 3B. For (B-D), relative expression values are plotted in arbitrary linear units.
Bmal1 thus plays a significant role in the light-dependent regulation of genes in the eye, most of which were not detectably rhythmic in DD. These results raise the possibility that a circadian clock broadly regulates transcriptional responses to light in the retina. Among genes with robust rhythms in both DD and LD were multiple histones and at least one histone deacetylase, genes with known actions in chromatin remodeling (Figure 3D; Supplemental Note 1 and Supplemental Tables 1 and 2). Thus it seems plausible that loss of circadian regulation of chromatin might underlie the aberrant transcriptional responses to LD cycles in Bmal1−/− mice.
Importance of Bmal1 for retinal electrical activity in response to light
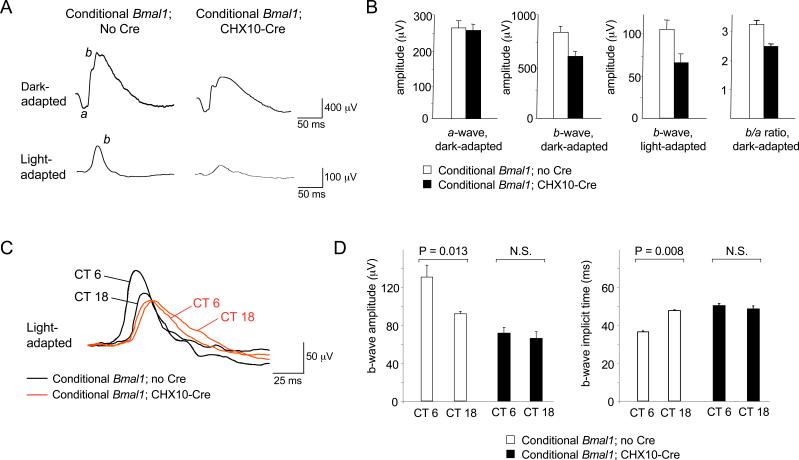
Given the importance of Bmal1 for retinal gene expression in LD, it seemed plausible that retinal visual physiology might be abnormal in Bmal1−/− mice. We therefore carried out ERG studies (Lu et al., 2001) to compare retinal electrical activity in response to light in wildtype and Bmal1−/− littermate mice (C57BL/6). ERG responses were recorded during mid-day hours, when the mouse light-adapted ERG b-wave amplitude is at or near the maximum in its daily rhythm (Barnard et al., 2006). Mice of the two genotypes were studied in alternating order to minimize possible effects of circadian time. Under dark-adapted conditions (rod pathway ERG), the amplitude of the ERG a-wave, the summed electrical response of photoreceptors, showed no significant difference between genotypes (Figure 4A,B). However, the b-wave, the post-receptor electrical response, was distinctly abnormal in Bmal1−/− mice, reduced in amplitude by 30% (P < 0.03; two-tailed t-test; N = 6 for each genotype). The selective reduction of b-wave amplitude is underscored by the highly significant decrease of the ratio of b-wave to a-wave amplitude in Bmal1−/− mice (P < 0.0005) (Figure 4B). Under light-adapted conditions (cone pathway ERG), the reduction of b-wave amplitude in Bmal1−/− mice was even greater, 60% (P < 0.015) (Figure 4A,B). Thus in the mutants the daytime ERG b-wave response is diminished such that it resembles the lower night-time amplitude of wildtype mice (Barnard et al., 2006; see below). These results indicate that Bmal1 is important for inner retinal processing of visual stimuli, but not for rod photoreceptor electrical responses.
Figure 4. Defective retinal electrical activity in response to light in Bmal1−/− mice but not SCN-lesioned wildtype mice.
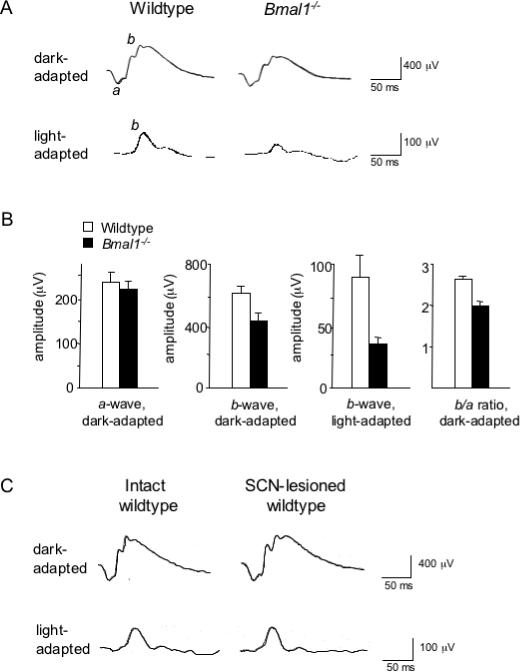
(A) Electroretinogram (ERG) traces in response to a flash of light for adult wildtype and littermate Bmal1−/− mice under dark- and light-adapted conditions; a- and b-waves labeled on wildtype traces. (B) Quantification of ERG responses in wildtype and Bmal1−/− littermates. Shown are mean and SEM; N = 6 for each genotype. (C) Representative ERG responses of intact wildtype and SCN-lesioned wildtype mice (each N = 5). No significant differences between groups were found for a- or b-wave amplitudes. Mice were C57BL/6, and ERGs were performed between Zeitgeber Time 4 and 9; wildtype and mutant (or intact and SCN-lesioned) mice were studied in alternating order.
To determine whether the SCN clock contributes to retinal performance, we made SCN lesions in wildtype mice and compared ERG responses in the behaviorally arrhythmic SCN-lesioned mice and intact controls (C57BL/6). No significant differences were found between groups for a- or b-wave amplitudes in either dark- or light-adapted conditions (Figure 4C; representative of N = 5 for each group). Thus the SCN circadian clock is not required for normal daytime retinal electrical activity in response to light. Given the ERG abnormality of Bmal1−/− mice (Figure 4A,B), any circadian clock important for retinal physiological function must therefore be outside the SCN.
Retina-specific deletion of Bmal1 and loss of retinal circadian clock function
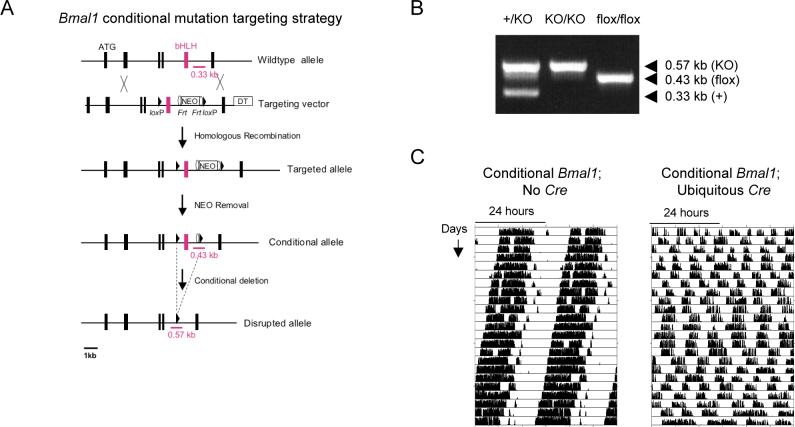
To examine the role of Bmal1 and circadian clock function specifically within the retina, we generated mice with a conditional allele of Bmal1. Upon the action of Cre recombinase, the exon encoding the BMAL1 basic helix-loop-helix (bHLH) domain is deleted (Figure 5A,B), resulting in a mutation nearly identical to the original null mutation in Bmal1−/− mice (Bunger et al., 2000). In validation studies, mice homozygous for the conditional allele had circadian behavioral rhythms indistinguishable from wildtype littermates, whereas after crossing in a ubiquitously-acting Cre transgene (Schwenk et al., 1995), the mice showed a complete loss of circadian behavioral rhythms and emergence of ultradian behavior (Figure 5C), copying the Bmal1−/− phenotype (Bunger et al., 2000). Thus the Bmal1 conditional allele has wildtype activity unless acted upon by Cre recombinase, after which it acts as a Bmal1 null mutation.
Figure 5. Conditional Bmal1 allele.
(A) Targeting strategy and conditional disruption. Closed boxes, exons; ATG, translation start site; bHLH, exon encoding basic helix-loop-helix domain; NEO, neomycin resistance marker; DT, diphtheria toxin cassette; triangles, loxP sites; ovals, Frt sites; bars with kilobase (kb) markers, sites and sizes of PCR products diagnostic of genotypes. (B) PCR products amplified from mouse tail DNA demonstrating the indicated genotypes. +, wildtype allele; KO, disrupted allele (from ubiquitously-acting Cre); flox, conditional allele. (C) Functional validation of Bmal1 condition allele: conditional loss of circadian rhythms of locomotor activity. Shown are representative double-plotted records of running-wheel activity in DD of mice homozygous for the conditional Bmal1 allele with or without a ubiquitously-acting Cre. Tick mark heights correspond to the number of running-wheel revolutions in a 6-min bin.
To generate a retina-specific Bmal1 mutation, we obtained a mouse line carrying a CHX10-Cre transgene, previously shown to act throughout the neural retina but to have little or no activity in the brain or other tissues (Rowan and Cepko, 2004). Crossing CHX10-Cre to an indicator line (Soriano, 1999) demonstrated Cre activity throughout the retina and little or no activity in non-retinal ocular tissues or the SCN and other brain regions, as expected (Figure 6A).
Figure 6. Retina-specific loss of Bmal1 function.
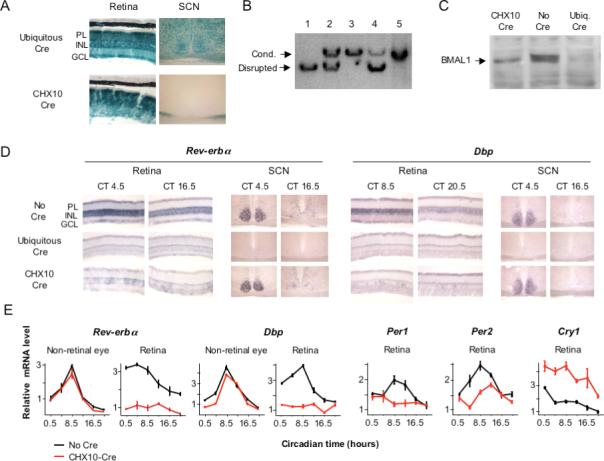
(A) Retina-specific Cre activity from CHX10-Cre transgene. Sections of retina and SCN from indicator mice showing blue precipitate reporting Cre recombinase activity. For CHX10-Cre, blue stain at bottom of the brain is from retinal ganglion cell projections. PL, photoreceptor layer; INL, inner nuclear layer; GCL, ganglion cell layer. (B) Retina-specific disruption of Bmal1 conditional allele. Genomic southern showing fragments diagnostic of the conditional or disrupted Bmal1 alleles. 1−4, Retina DNA. 1, Homozygous Bmal1 conditional, ubiquitous Cre; 2, Heterozygous for disrupted allele; 3, Homozygous Bmal1 conditional, no Cre; 4, Homozygous Bmal1 conditional, CHX10-Cre; 5, Hypothalamus DNA from same mouse as (4). (C) Loss of BMAL1 protein from the retina. Anti-BMAL1 western blot of protein extracts from retinas of homozygous conditional Bmal1 mice with the indicated Cre. (D) Loss of Bmal1 function in retina but not SCN. In situ hybridization to retina and SCN sections showing expression of Bmal1-dependent genes Rev-erbα and Dbp in homozygous conditional Bmal1 mice carrying the indicated Cre transgene. Circadian times (CT) correspond to the peak (left) or trough (right) of transcript rhythms. (E) Loss of molecular rhythms in retina. Temporal expression profiles (Q-PCR) of the indicated genes in mice homozygous for the conditional Bmal1 allele with or without CHX10-Cre. Relative expression levels are plotted in arbitrary linear units.
Next we generated mice homozygous for the Bmal1 conditional allele that carry a single copy of the CHX10-Cre transgene. As expected, these mice showed a retina-specific disruption of the Bmal1 conditional allele (Figure 6B). The residual non-recombined allele (Figure 6B, lane 4) is likely due to trace incomplete recombination in the neural retina and to DNA from retinal vasculature and contaminating retinal pigment epithelium, in which CHX10-Cre is not expected to act. In addition, the mice showed the expected substantial loss of BMAL1 protein from the retina (Figure 6C), with residual protein also likely derived from trace incomplete recombination in the neural retina and from retinal vascular and pigment epithelial cells. Interestingly, the residual BMAL1 appears to comprise only the faster-migrating form of the protein (Figure 6C), suggesting the possibility that BMAL1 has different post-translational modifications in neural and non-neural retinal cells.
To examine Bmal1 function, we monitored transcripts for Rev-erbα and Dbp, clock-regulated genes that depend on CLOCK-BMAL1 activity for expression (Ripperger et al., 2000). In conditional Bmal1 mice without Cre, in situ hybridization demonstrated the expected circadian rhythm of expression of Rev-erbα and Dbp in both the retina and the SCN (Figure 6D). With ubiquitously-acting Cre, expression of Rev-erbα and Dbp was lost in both the retina and the SCN (Figure 6D). With CHX10-Cre, Rev-erbα and Dbp showed loss of expression in the retina, except for variable small patches showing weak expression, whereas the SCN showed normal circadian rhythms of expression for both genes (Figure 6D). As expected, circadian rhythms of behavior were normal (data not shown). The patchy residual retinal expression of Rev-erbα and Dbp is consistent with the residual non-recombined allele in the retina (Figure 6B) and the trace retinal mosaicism previously reported for CHX10-Cre (Rowan and Cepko, 2004).
In conditional Bmal1 mice without Cre, Q-PCR demonstrated a circadian profile of Rev-erbα and Dbp expression in both the retina and non-retinal ocular tissues, as expected, whereas with CHX10-Cre, only non-retinal ocular tissues showed a circadian rhythm (Figure 6E). In addition, retinal rhythmic profiles of Per1 and Per2 expression were defective (Figure 6E), and Cry1, often weakly rhythmic or noisy in non-SCN sites (Storch et al., 2002), was abnormally high (Figure 6E), as expected in the absence of Bmal1 (Kondratov et al., 2006). Together these results indicate that conditional Bmal1 mice have wildtype Bmal1 function and robust retinal circadian rhythms, whereas in the presence of CHX10-Cre, the mice show a retina-specific loss of Bmal1 function that does not support demonstrable retinal circadian rhythms. It is possible, however, that some cells within patches of residual retinal Bmal1 function retain rhythms.
Importance of retinal Bmal1 for circadian rhythm of retinal responses to light
To test the importance of retinal Bmal1 for retinal physiology in vivo, we first compared daytime ERG responses of homozygous conditional Bmal1 mice (controls) and littermate homozygous conditional Bmal1 mice with a single copy of CHX10-Cre (Ret-Bmal1−/−; all mice were C57BL/6 × 129 hybrids). Under dark-adapted conditions, the amplitude of the a-wave showed no significant difference between genotypes (Figure 7A,B). In contrast, the amplitude of the b-wave was reduced by 27% in Ret-Bmal1−/− mice (Figure 7A,B) (P < 0.012; two-tailed t-test; N = 7 for each genotype), producing a highly significant reduction in the ratio of b-wave to a-wave amplitude (P < 0.0004) (Figure 7B). Under light-adapted conditions, the reduction in b-wave amplitude in Ret-Bmal1−/− mice was even greater, 44% (P < 0.018) (Figure 7A,B). Despite the difference in genetic background, the ERG phenotype of Ret-Bmal1−/− mice was essentially identical to that of Bmal1−/− mice (Figure 4A,B), indicating that retinal Bmal1 function fully accounts for the role of Bmal1 in retinal electrical responses to light.
Figure 7. Loss of circadian rhythm of inner retinal electrical activity in response to light in mice with a retina-specific deletion of Bmal1.
(A) Examples of daytime ERG traces in response to a flash of light for conditional Bmal1 littermates with or without the CHX10-Cre. (B) Quantification of ERG responses (mean and SEM; N = 7 for each genotype). (C) Examples of ERG traces in response to a flash of light for conditional Bmal1 littermates with or without CHX10-Cre at two circadian times (CT) in constant light (LL). (D) Quantification of ERG b-wave amplitudes and implicit times (mean and SEM; N = 8 for controls and N = 7 for ret-Bmal1). P-values, t-tests. N.S., not significant.
Recent work demonstrates that robust circadian regulation of the mouse light-adapted ERG b-wave amplitude is revealed under constant light (LL) conditions (Barnard et al., 2006). To determine the importance of retinal Bmal1 function for this rhythm, we compared light-adapted ERG responses of conditional Bmal1 controls and Ret-Bmal1−/− littermates in LL at times of high (CT 6) or low (CT 18) b-wave amplitude (it is not yet known if these circadian times represent the true peak and trough of the rhythm). Controls showed the expected circadian rhythm of b-wave responses (Figure 7C,D), with significantly higher amplitudes at CT 6 than CT 18 (Figure 7D). In contrast, Ret-Bmal1−/− mice had no detectable circadian rhythm of b-wave amplitude, with responses at both circadian times apparently fixed at a low amplitude, somewhat lower than the amplitude at CT 18 in controls (Fig. 7C,D). In addition, control mice exhibited a circadian rhythm of b-wave implicit time (the interval between the flash of light and the peak of the b-wave), a rhythm that has also been shown to be robust in DD (Barnard et al., 2006). In Ret-Bmal1−/− mice this rhythm was undetectable, and b-wave implicit times appeared to be fixed at a value similar to CT 18 in controls, much like b-wave amplitude (Figure 7C,D). These results indicate that Bmal1 function in the retina is required for circadian rhythms of inner retinal processing of visual stimuli. More specifically, retinal Bmal1 function is required to promote inner retinal visual processing during the subjective day, strongly suggesting that the rhythm in retinal electrophysiological function is driven by an intrinsic retinal circadian clock.
DISCUSSION
Our results indicate that in the mammalian eye hundreds of genes, many of which are expressed in the retina, are controlled by a circadian clock. We found that thousands more are regulated rhythmically in LD cycles, including a large cluster of >1000 genes with a night-time peak of expression. The rhythmically-expressed genes are associated with a wide range of functions, and the findings provide a rich context in which to investigate molecular processes underlying the impact of circadian clock function and light on retinal physiology and metabolism. Although less pronounced than that reported here, an excess of transcripts with daily cycles in LD compared to DD has been observed in chick retina (Bailey et al, 2004) and in Drosophila head (Lin et al., 2002; Ceriani et al., 2002; Wijnen et al., 2006), a structure predominantly composed of the eyes.
In Bmal1−/− mice, a large fraction of genes that normally show rhythmic ocular expression exclusively in LD exhibited non-rhythmic expression or reduced amplitude. In contrast, constitutively-expressed genes showed unaltered expression in Bmal1−/− mice. These results indicate that Bmal1 plays an important role in light-dependent but not global gene regulation in the retina. Although we cannot exclude a possible non-circadian role of Bmal1 in transcriptional responses to light, several lines of evidence suggest that defective circadian regulation likely underlies the phenotype. Circadian control of light-dependent gene induction in the mammalian retina has been documented (Masana et al., 1996; Humphries and Carter, 2004), regulation of chromatin by histone H3 has been implicated in light-dependent gene induction in the SCN (Crosio et al., 2000), and our finding that multiple histones (including histone H3 family members) and other chromatin remodeling genes exhibit circadian expression in the eye suggests that chromatin in the retina is under clock control. Together these observations suggest that regulators of chromatin state function in the retina as clock outputs in the control of transcriptional responses to light and that in the absence of Bmal1 one or more of these rhythmic regulators is deficient.
Our results indicate that Bmal1 function within the retina is required for circadian rhythms of inner retinal visual processing, specifically for the circadian rhythms of the light-adapted ERG b-wave amplitude and b-wave implicit time. In the absence of retinal Bmal1, both rhythms appear to be fixed at or near the low values typical for subjective night, indicating that retinal Bmal1 function is required for the facilitation of inner retinal visual processing during subjective day. This phenotype is strikingly similar to the unexpected ERG phenotype recently described for mice lacking melanopsin (Barnard et al., 2006), the photopigment of photoreceptive ganglion cells mediating circadian entrainment to LD cycles (Hatter et al., 2003). The essentially identical loss of circadian rhythms of retinal processing in two independent retina-specific mutations affecting the circadian system makes it very likely that circadian clock function within the retina directly drives rhythms of retinal visual physiology, and it provides support for the idea that melanopsin activity is required to synchronize a population of retinal clock cells controlling inner retinal electrical responses to light (Barnard et al., 2006).
A mild ERG b-wave amplitude defect has been reported for mice lacking the D4 dopamine receptor (Nir et al., 2002). Given the importance of dopamine in regulating inner retinal network properties (Gustincich et al., 1997) and evidence that retinal dopaminergic amacrine cells are circadian clock cells (Witkovsky et al., 2003; Gustincich et al., 2004; Ruan et al., 2006; Supplemental Figure 1A), it is possible that the loss of clock function within dopaminergic amacrine cells contributes to the ERG phenotype of Ret-Bmal1−/− mice.
Circadian clocks in mammals are widely distributed, but except for the SCN clock known to regulate behavior, their physiological functions in vivo are largely mysterious. The studies described here indicate that an intrinsic retinal circadian clock regulates retinal visual processing in vivo and that it does so autonomously, with no detectable contribution from the SCN or other clocks. Together with observations emerging from other tissue-specific manipulations of clock function (Durgan et al., 2006; McDearmon et al., 2006), our work provides evidence that circadian clocks outside the SCN contribute important physiological functions in vivo. Thus over evolutionary time different cell-types have likely recruited the circadian clock mechanism inherited from a single-celled ancestor for control of specialized tissue-specific processes.
EXPERIMENTAL PROCEDURES
Mice and tissue collection
For three-day microarray experiments, 108 adult male CBA/CaJ mice (The Jackson Laboratory) were entrained to a 12h:12h LD cycle for 3 weeks. During the dark phase, 54 mice were transferred to DD, the remainder kept in LD. At four-hour intervals, three mice from each group were euthanized (CO2), and eyes were collected, frozen in liquid nitrogen, and stored (−80°C). Total RNA was purified separately from the eyes of each mouse (Trizol, Invitrogen and RNAeasy, Qiagen), and equal aliquots of RNA from the three pairs of eyes collected at a single time-point were pooled. Studies were performed in accordance with the protocol approved by the Harvard Medical School Standing Committee on Animals.
Microroarray Analysis
Samples were hybridized to Affymetrix mouse 430.2 arrays representing the complete mouse genome (one array per time-point). Fluorescence images were normalized to median brightness (Li and Wong, 2001), and model-based expression values were computed using DNA-Chip Analyzer (Li and Wong, 2003). In each three-day, 18-array dataset, probe sets that were not classified as “expressed” (http://www.Affymetrix.com/products/algorithms_tech.htm) in at least three arrays were excluded, as were probe sets that had three or more missing expression values. The square-root transformation was applied to all expression values to make the error distribution more symmetric while limiting amplification of low-level noise. For a detailed description of procedures for identifying rhythmic expression, see Supplemental Note 4. To assess non-rhythmic transcripts, expression values determined by DNA-Chip Analyzer (Li and Wong, 2003) for the six arrays corresponding to the six time-points from wildtype mice in LD were used to select a “constitutively-expressed” set of genes. Genes with low mean expression value were excluded (<200 units; range: 0−10,900 units), and remaining genes with a near-constant level of expression across the six arrays were selected (standard deviation of expression < 5%, yielding 3,047 genes). Microarray data have been deposited in the ArrayExpress database (Accession # XXXXX).
Light and electron microscopy of retinas
For light microscopy, eyecups of three mice of each genotype were fixed (2% formaldehyde in 0.15M Sørensen phosphate buffer pH 7.4), cryoprotected (20% sucrose in PBS), and frozen in monochlorodifluomethane. Sections parallel to and including the horizontal meridian were obtained and stained with 300 nM 4′,6-diamidino-2-phenylindole (DAPI) in PBS. Fluorescence was detected with a Zeiss LSM 510 Meta confocal system and Zeiss Axioplan 2 microscope. Because photoreceptor activity was unaltered in Bmal1−/− mice, the thickness of the deep retinal layers was normalized to that of the ONL in each retina in order to avoid errors due to obliquity of sections. For EM, two mice of each genotype were perfused with 2% formaldehyde, 2.5% glutaraldehyde in 0.15M Sørenson phosphate buffer pH 7.4, followed by 1% OsO4, 1.5% potassium ferrocyanide, and stained en bloc with 1% uranyl acetate. Thick sections of the eyecup were stained with toluidine blue and examined with a Nikon Eclipse E600 microscope. Thin sections were stained with uranyl and lead, and micrographs were obtained with a JEOL X100 electron microscope.
Quantitative RT-PCR
Total RNA from whole eyes or dissected eye fractions (retinal, lens, and the rest of the eye) was transcribed into cDNA using random hexamers and Superscript reverse transcriptase (Invitrogen). cDNA derived from 25 ng of total RNA was PCR-amplified in a PTC-200 thermocycler with a Chromo4 module (MJ Research) using SYBR green (IQ SYBR Green Supermix, Bio-Rad) according to the manufacturer's instructions. Templates were amplified with the annealing temperature initially lowered from 70°C to 60°C in 2-cycle increments followed by 30 cycles of the following 3 steps: 20 sec at 94°C, 20 sec at 60°C, and 30 sec at 72°C. For quantitation, threshold cycle number difference was calculated based on the sample with the lowest expression, and the values obtained were 2n transformed. Input RNA concentration or the expression level of hypoxanthine guanine phosphoribosyl transferase-1 was used to normalize expression values. Sequences of primers are available upon request.
Electroretinography
ERG recordings were performed as described (Lu et al., 2001). Briefly, mice were dark-adapted overnight prior to recording. Mice were anesthetized, their pupils dilated, and responses were recorded with a chloride silver wire loop placed on the cornea. ERG responses were elicited with 10-μs flashes of white light (1.37 × 105 cd/m2) presented in a Ganzfeld dome at intervals of 1 min in darkness. Mice were then adapted to a background illumination (12 ft.-L., 10 min) and responses were elicited by 1-Hz flashes of white light (1.37 × 105 cd/m2). Circadian components of the light-adapted ERG were obtained from mice in constant light as described (Barnard et al, 2006), except the intensity of light was 300 Lux.
SCN lesions
Adult C57BL/6J mice were anesthetized with ketamine-xylazine (45 mg/kg-5 mg/kg), and an electrode (RNE-300X, Rhodes Medical Instruments) was lowered stereotaxically through the skull at the midsagittal sinus (anteroposterior −0.45 mm from Bregma) with the final tip position at 6.0 mm below the skull. Lesions were generated with constant current (2 mA, 10s; D.C. Constant Lesion Maker, Grass Instruments). From one week after surgery, locomotor activity in DD was monitored, and only mice showing arrhythmic behavior for >4 weeks were selected for ERG studies. Only mice with histologically-verified complete SCN lesions were included in the analysis (data not shown).
Generation of conditional Bmal1 mice
A cassette containing neomycin acetyltransferase (neo) flanked by two FRT sites and a single loxP site (EcoRI-BamHI fragment from vector PL452; Liu et al.; 2003) was inserted into a bacterial artificial chromosome encompassing the Bmal1 locus (RPCI23−331K23, BACPAC resources, Children's Hospital Oakland Research Institute) by homologous recombination in bacteria (Liu et al., 2003). The insertion site was immediately upstream of the PacI site in the intron 3' of the exon encoding the basic helix-loop-helix domain (exon 8). A 10.8-kb PmlI-fragment containing the Neo cassette with flanking genomic regions was subcloned into the HpaI-SmaI-digested targeting vector pKOII (Bardeesy et al., 2002), which contained a diphtheria toxin expression cassette. An oligonucleotide comprising a single loxP site preceded by a BamHI restriction site was cloned into the SanDI site of the intron preceding exon 8, resulting in the final targeting vector. Electroporation of ES cells, selection of neomycin-resistant clones, and injection of ES cells into blastocysts were performed using standard methods. The Neo cassette was removed by mating chimeras to mice carrying a Flpe knock-in at the Rosa26 locus (Farley et al., 2000). Genotyping was performed using multiplex PCR and primers L1, ACTGGAAGTAACTTTATCAAACTG; L2, AATCCGCCTGCCTACTGCCTCC (reverse primer); and R4, GGGTGGAGTATGATATGACC. For assessing the efficiency of loxP recombination, retinal genomic DNA was digested with AvrII, and Southern blot hybridization was performed using a 350-bp probe specific for sequences immediately following the 3' flanking region used for targeting.
In situ hybridization, histology, and western blots
For in situ hybridization, dissected retinas were fixed (cold 4% formaldehyde in PBS, 10 min), embedded (Tissue-Tek), cut frozen (20 μm), and processed as described (Kraves and Weitz, 2006). Sequences of riboprobes are available upon request. β-galactosidase activity in 20-μm brain or retina sections was detected with 5-bromo-4-chloro-3-indolyl β-D-galactopyranoside (X-gal). Western analysis was performed with rabbit anti-BMAL1 antiserum (1:500).
ACKNOWLEDGMENTS
We thank Susan Dymecki and Connie Cepko for data on CHX10-Cre; Christopher Bradfield for Bmal1−/− (Mop3−/−) mice; Ueli Schibler for anti-BMAL1 antiserum; the Gene Manipulation Core of the Developmental Disabilities Research Center of Children's Hospital, Boston for expert service, and Ming Liu and Xiao Ling Long for expert technical assistance. This work was supported by grants from the National Institutes of Health to C.J.W. (NS055831), E.R. (EY001344), and T.L. (EY10309).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary Material
SUPPLEMENTAL DATA
Supplemental Notes 1−4, Supplemental Figures 1−6, Supplemental Tables 1 and 2.
REFERENCES
- Abe M, Herzog ED, Yamazaki S, Straume M, Tei H, Sakaki Y, Menaker M, Block GD. Circadian rhythms in isolated brain regions. J. Neurosci. 2002;22:350–356. doi: 10.1523/JNEUROSCI.22-01-00350.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar RA, Reddy AB, Maywood ES, Clayton JD, King VM, Smith AG, Gant TW, Hastings MH, Kyriacou CP. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr. Biol. 2002;12:540–550. doi: 10.1016/s0960-9822(02)00759-5. [DOI] [PubMed] [Google Scholar]
- Bailey MJ, Beremand PD, Hammer R, Reidel E, Thomas TL, Cassone VM. Transcriptional profiling of circadian patterns of mRNA expression in the chick retina. J. Biol. Chem. 2004;279:52247–52254. doi: 10.1074/jbc.M405679200. [DOI] [PubMed] [Google Scholar]
- Barnard AR, Hattar S, Hankins MW, Lucas RJ. Melanopsin regulates visual processing in the mouse retina. Curr. Biol. 2006;16:389–395. doi: 10.1016/j.cub.2005.12.045. [DOI] [PubMed] [Google Scholar]
- Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in cultured Rat-1 fibroblasts. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- Bardeesy N, Sinha M, Hezel AF, Signoretti S, Hathaway NA, Sharpless NE, Loda M, Carrasco DR, DePinho RA. Loss of the Lkb1 tumour suppressor provokes intestinal polyposis but resistance to transformation. Nature. 2002;419:162–167. doi: 10.1038/nature01045. [DOI] [PubMed] [Google Scholar]
- Besharse JC, Iuvone PM. Circadian clock in Xenopus eye controlling retinal serotonin N-acetyltransferase. Nature. 1983;305:133–135. doi: 10.1038/305133a0. [DOI] [PubMed] [Google Scholar]
- Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, Bradfield CA. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103:1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill GM, Besharse JC. Circadian clock functions localized in xenopus retinal photoreceptors. Neuron. 1993;10:573–577. doi: 10.1016/0896-6273(93)90160-s. [DOI] [PubMed] [Google Scholar]
- Ceriani MF, Hogenesch JB, Yanovsky M, Panda S, Straume M, Kay SA. Genome-wide expression analysis in Drosophila reveals genes controlling circadian behavior. J. Neurosci. 2002;22:9305–9319. doi: 10.1523/JNEUROSCI.22-21-09305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosio C, Cermakian N, Allis CD, Sassone-Corsi P. Light induces chromatin modification in cells of the mammalian circadian clock. Nat. Neurosci. 2000;3:1241–1247. doi: 10.1038/81767. [DOI] [PubMed] [Google Scholar]
- Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle SE, Grace MS, McIvor W, Menaker M. Circadian rhythms of dopamine in mouse retina: the role of melatonin. Vis. Neurosci. 2002;19:593–601. doi: 10.1017/s0952523802195058. [DOI] [PubMed] [Google Scholar]
- Duffield GE, Best JD, Meurers BH, Bittner A, Loros JJ, Dunlap JC. Circadian programs of transcriptional activation, signaling, and protein turnover revealed by microarray analysis of mammalian cells. Curr. Biol. 2002;12:551–557. doi: 10.1016/s0960-9822(02)00765-0. [DOI] [PubMed] [Google Scholar]
- Durgan DJ, Trexler NA, Egbejimi O, McElfresh TA, Suk HY, Petterson LE, Shaw CA, Hardin PE, Bray MS, Chandler MP, et al. The circadian clock within the cardiomyocyte is essential for responsiveness of the heart to fatty acids. J. Biol. Chem. 2006;281:24254–24269. doi: 10.1074/jbc.M601704200. [DOI] [PubMed] [Google Scholar]
- Farley FW, Soriano P, Steffen LS, Dymecki SM. Widespread recombinase expression using FLPeR (flipper) mice. Genesis. 2000;28:106–110. [PubMed] [Google Scholar]
- Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- Goto M, Oshima I, Tomita T, Ebihara S. Melatonin content of the pineal gland in different mouse strains. J. Pineal Res. 1989;7:195–204. doi: 10.1111/j.1600-079x.1989.tb00667.x. [DOI] [PubMed] [Google Scholar]
- Gustincich S, Contini M, Gariboldi M, Puopolo M, Kadota K, Bono H, LeMieux J, Walsh P, Carninci P, Hayashizaki Y, et al. Gene discovery in genetically labeled single dopaminergic neurons of the retina. Proc. Natl. Acad. Sci. U S A. 2004;101:5069–5074. doi: 10.1073/pnas.0400913101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustincich S, Feigenspan A, Wu DK, Koopman LJ, Raviola E. Control of dopamine release in the retina: a transgenic approach to neural networks. Neuron. 1997;18:723–736. doi: 10.1016/s0896-6273(00)80313-x. [DOI] [PubMed] [Google Scholar]
- Hattar S, Lucas RJ, Mrosovsky N, Thompson S, Douglas RH, Hankins MW, Lem J, Biel M, Hofmann F, Foster RG, et al. Melanopsin and rod-cone photoreceptive systems account for all major accessory visual functions in mice. Nature. 2003;424:76–81. doi: 10.1038/nature01761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayasaka N, LaRue SI, Green CB. In vivo disruption of Xenopus CLOCK in the retinal photoreceptor cells abolishes circadian melatonin rhythmicity without affecting its production levels. J. Neurosci. 2002;22:1600–1607. doi: 10.1523/JNEUROSCI.22-05-01600.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries A, Carter DA. Circadian dependency of nocturnal immediate-early protein induction in rat retina. Biochem. Biophys. Res. Commun. 2004;320:551–556. doi: 10.1016/j.bbrc.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Ko CH, Takahashi JS. Molecular components of the mammalian circadian clock. Hum. Mol. Genet. 2006;15:R271–277. doi: 10.1093/hmg/ddl207. [DOI] [PubMed] [Google Scholar]
- Kondratov RV, Shamanna RK, Kondratova AA, Gorbacheva VY, Antoch MP. Dual role of the CLOCK/BMAL1 circadian complex in transcriptional regulation. FASEB J. 2006;20:530–532. doi: 10.1096/fj.05-5321fje. [DOI] [PubMed] [Google Scholar]
- Kraves S, Weitz CJ. A role for cardiotrophin-like cytokine in the circadian control of mammalian locomotor activity. Nat. Neurosci. 2006;9:212–219. doi: 10.1038/nn1633. [DOI] [PubMed] [Google Scholar]
- LaVail MM, Ward PA. Studies on the hormonal control of circadian outer segment disc shedding in the rat retina. Invest. Ophthalmol. Vis. Sci. 1978;17:1189–1183. [PubMed] [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: model validation, design issues and standard error application. Genome Biol. 2001;2:research0032.1–0032.11. doi: 10.1186/gb-2001-2-8-research0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wong WH. DNA-Chip Analyzer (dChip). In: Parmigiani G, Garrett ES, Irizarry R, Zeger SL, editors. The Analysis of Gene Expression Data: Methods and Software. Springer; New York, NY: 2003. pp. 120–141. [Google Scholar]
- Lin Y, Han M, Shimada B, Wang L, Gibler TM, Amarakone A, Awad TA, Stormo GD, Van Gelder RN, Taghert PH. Influence of the period-dependent circadian clock on diurnal, circadian, and aperiodic gene expression in Drosophila melanogaster. Proc. Natl. Acad. Sci. U S A. 2002;99:9562–9567. doi: 10.1073/pnas.132269699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Jenkins NA, Copeland NG. A highly efficient recombineering-based method for generating conditional knockout mutations. Genome Res. 2003;13:476–484. doi: 10.1101/gr.749203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Peng YW, Shang J, Pawlyk BS, Yu F, Li T. The mammalian retinal degeneration B2 gene is not required for photoreceptor function and survival. Neuroscience. 2001;107:35–41. doi: 10.1016/s0306-4522(01)00337-2. [DOI] [PubMed] [Google Scholar]
- Manglapus MK, Uchiyama H, Buelow NF, Barlow RB. Circadian rhythms of rod-cone dominance in the Japanese quail retina. J. Neurosci. 1998;18:4775–4784. doi: 10.1523/JNEUROSCI.18-12-04775.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masana MI, Benloucif S, Dubocovich ML. Light-induced c-fos mRNA expression in the suprachiasmatic nucleus and the retina of C3H/HeN mice. Brain Res. Mol. Brain. Res. 1996;42:193–201. doi: 10.1016/s0169-328x(96)00031-9. [DOI] [PubMed] [Google Scholar]
- McDearmon EL, Patel KN, Ko CH, Walisser JA, Schook AC, Chong JL, Wilsbacher LD, Song EJ, Hong HK, Bradfield CA, et al. Dissecting the functions of the mammalian clock protein BMAL1 by tissue-specific rescue in mice. Science. 2006;314:1304–1308. doi: 10.1126/science.1132430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGoogan JM, Cassone VM. Circadian regulation of chick electroretinogram: effects of pinealectomy and exogenous melatonin. Am. J. Physiol. 1999;277:R1418–1427. doi: 10.1152/ajpregu.1999.277.5.R1418. [DOI] [PubMed] [Google Scholar]
- Miranda-Anaya M, Bartell PA, Menaker M. Circadian rhythm of iguana electroretinogram: the role of dopamine and melatonin. J. Biol. Rhythms. 2002;17:526–538. doi: 10.1177/0748730402238235. [DOI] [PubMed] [Google Scholar]
- Nir I, Harrison JM, Haque R, Low MJ, Grandy DK, Rubinstein M, Iuvone PM. Dysfunctional light-evoked regulation of cAMP in photoreceptors and abnormal retinal adaptation in mice lacking dopamine D4 receptors. J. Neurosci. 2002;22:2063–2073. doi: 10.1523/JNEUROSCI.22-06-02063.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang Y, Andersson CR, Kondo T, Golden SS, Johnson CH. Resonating circadian clocks enhance fitness in cyanobacteria. Proc. Natl. Acad. Sci. USA. 1998;95:8660–8664. doi: 10.1073/pnas.95.15.8660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- Panda S, Hogenesch JB. It's all in the timing: many clock, many outputs. J. Biol. Rhythms. 2004;19:374–387. doi: 10.1177/0748730404269008. [DOI] [PubMed] [Google Scholar]
- Pierce ME, Sheshberadaran H, Zhang Z, Fox LE, Applebury ML, Takahashi JS. Circadian regulation of iodopsin gene expression in embryonic photoreceptors in retinal cell culture. Neuron. 1993;10:579–584. doi: 10.1016/0896-6273(93)90161-j. [DOI] [PubMed] [Google Scholar]
- Ripperger JA, Shearman LP, Reppert SM, Schibler U. CLOCK, an essential pacemaker component, controls expression of the circadian transcription factor DBP. Genes Dev. 2000;14:679–689. [PMC free article] [PubMed] [Google Scholar]
- Rowan S, Cepko CL. Genetic analysis of the homeodomain transcription factor Chx10 in the retina using a novel multifunctional BAC transgenic mouse reporter. Dev. Biol. 2004;271:388–402. doi: 10.1016/j.ydbio.2004.03.039. [DOI] [PubMed] [Google Scholar]
- Ruan GX, Zhang DQ, Zhou T, Yamazaki S, McMahon DG. Circadian organization of the mammalian retina. Proc. Natl. Acad. Sci. U S A. 2006;103:9703–9708. doi: 10.1073/pnas.0601940103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenk F, Baron U, Rajewsky K. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 1995;23:5080–5081. doi: 10.1093/nar/23.24.5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat. Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- Storey JD, Taylor JE, Siegmund D. Strong control, conservative point estimation and simultaneous conservative consistency of false discovery rates: a unified approach. J. ROY. STAT. SOC. B. 2004;66:187–205. [Google Scholar]
- Tanoue S, Krishnan P, Krishnan B, Dryer SE, Hardin PE. Circadian clocks in antennal neurons are necessary and sufficient for olfaction rhythms in Drosophila. Curr. Biol. 2004;14:638–649. doi: 10.1016/j.cub.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Tosini G, Menaker M. Circadian rhythms in cultured mammalian retina. Science. 1996;272:419–421. doi: 10.1126/science.272.5260.419. [DOI] [PubMed] [Google Scholar]
- Wijnen H, Naef F, Boothroyd C, Claridge-Chang A, Young MW. Control of daily transcript oscillations in Drosophila by light and circadian clock. PLoS Genet. 2006;2:e39. doi: 10.1371/journal.pgen.0020039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witkovsky P, Veisenberger E, LeSauter J, Yan L, Johnson M, Zhang DQ, McMahon D, Silver R. Cellular location and circadian rhythm of expression of the biological clock gene Period 1 in the mouse retina. J. Neurosci. 2003;23:7670–7676. doi: 10.1523/JNEUROSCI.23-20-07670.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki S, Numano R, Abe M, Hida A, Takahashi R, Ueda M, Block GD, Sakaki Y, Menaker M, Tei H. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288:682–685. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPLEMENTAL DATA
Supplemental Notes 1−4, Supplemental Figures 1−6, Supplemental Tables 1 and 2.