Abstract
Extracellular matrix (ECM) regulates cell morphology and gene expression in vivo; these relationships are maintained in three-dimensional (3D) cultures of mammary epithelial cells. In the presence of laminin-rich ECM (lrECM), mammary epithelial cells round up and undergo global histone deacetylation, a process critical for their functional differentiation. However, it remains unclear whether lrECM-dependent cell rounding and global histone deacetylation are indeed part of a common physical-biochemical pathway. Using 3D cultures as well as nonadhesive and micropatterned substrata, here we showed that the cell ‘rounding’ caused by lrECM was sufficient to induce deacetylation of histones H3 and H4 in the absence of biochemical cues. Microarray and confocal analysis demonstrated that this deacetylation in 3D culture is associated with a global increase in chromatin condensation and a reduction in gene expression. Whereas cells cultured on plastic substrata formed prominent stress fibers, cells grown in 3D lrECM or on micropatterns lacked these structures. Disruption of the actin cytoskeleton with cytochalasin D phenocopied the lrECM-induced cell rounding and histone deacetylation. These results reveal a novel link between ECM-controlled cell shape and chromatin structure, and suggest that this link is mediated by changes in the actin cytoskeleton.
Keywords: cell morphology, chromatin structure, differentiation, histone acetylation
Introduction
Cell structure and function were postulated to be intimately connected in maintenance of tissue homeostasis [1]. Manipulating cellular and nuclear morphology in culture can induce a variety of functional changes, including glucose uptake and metabolism [2], proliferation [3–5], apoptosis [4], differentiation and gene expression [6–10]. These morphologically driven processes are controlled by the local microenvironment as exemplified in the mammary gland and mammary epithelial cells (for review see [11]). Isolated mouse mammary epithelial cells can be induced to undergo structural and functional differentiation in three-dimensional (3D) cultures. The acquisition of the functionally differentiated phenotype, i.e. expression of milk proteins, requires lactogenic hormones and the basement membrane protein laminin-111 (laminin-1) [12, 13]. The latter induces dramatic morphological changes in that cells become rounded and actin becomes cortical. These morphological changes are necessary for expression of some milk proteins including β-casein [9]. Artificially pre-rounding the cells by plating on the nonadhesive substratum poly(2-hydroxyethyl methacrylate) (polyHEMA) poises them for responsiveness to prolactin and laminin-rich ECM (lrECM)[9], whereas preventing cell rounding by treatment with the phorbol ester 12-o-tetradecanoylphorbol 13-acetate (TPA) inhibits milk protein synthesis [14]. These previous findings strongly suggested that cell rounding, mediated by reorganization of actin filaments and other cytoskeletal components, may be an important physical signal conveyed by the ECM leading to changes in gene expression.
Cell fate and differentiated function are controlled by patterns of gene expression, which in turn are dictated by chromatin organization. The structure of chromatin is regulated by a number of post-translational modifications at the amino-terminal tails of nucleosomal histones, including acetylation/deacetylation, phosphorylation, methylation, and ADP ribosylation [15]. The acetylation status of chromatin is dynamic, balanced by the activities of histone acetyltransferases (HATs) and histone deacetylases (HDACs) [16]. Gene expression generally correlates with histone hyperacetylation at promoter regions and other regulatory cis-elements [17–19] whereas histone deacetylation represses transcription by promoting chromatin condensation into heterochromatin [20, 21]. Histone deacetylation was shown to correlate with changes in gene expression in the developing brain in rats [22], and abrogating the deacetylation by treatment with an HDAC inhibitor resulted in impaired brain development and delayed expression of differentiation markers. Functional differentiation of many cell types is associated with activation of specific subsets of genes, silencing of other genes, and extensive formation of heterochromatin [23].
Acinar morphogenesis of a non-malignant human mammary epithelial cell line HMT3522-S1, is accompanied by rearrangements in chromatin structure and nuclear architecture [24–26]. These reorganizations appear to be important for establishing and maintaining the normal phenotype of cells in 3D [24, 25, 27]. We previously found that the global level of histone acetylation is critical for functional differentiation of mammary epithelial cells [24, 28]. Altering chromatin structure by increasing histone acetylation with trichostatin A induces human mammary epithelial cells to alter their morphology, re-enter the cell cycle, and become disorganized [24]; overexpressing a HAT in mouse mammary epithelial cells inhibits production of endogenous β-casein in response to lrECM [28]. Conversely, treatment with lrECM induces global histone deacetylation in human and mouse mammary epithelial cells [25, 28].
These previous studies showed that ECM has profound effects on cell shape and chromatin structure, particularly histone deacetylation, but it was not understood whether these effects were linked. In this report, we used several strategies to change cell shape, including 3D culture on different substrata with different adhesiveness and micropatterned substrata. We find that cell rounding per se induces global histone deacetylation and an increase in chromatin condensation, and that these processes are associated with a global reduction of gene expression. These results reveal a process by which ECM integrates structure and function in mammary epithelial cells through a shape-dependent global histone deacetylation.
Materials and Methods
Cell culture
HMT-3522-S1 and -T4-2 human mammary epithelial cells [29, 30] were grown as two-dimensional (2D) monolayers on plastic or within 3D lrECM (Matrigel, Collaborative Research) and maintained as previously described [31, 32]. Cells were grown in 2D and 3D for 10 days before harvests. For culture on nonadhesive substrata, ~4000 cells/cm2 were seeded on polyHEMA (Sigma)-coated plates prepared as previously described for mouse mammary epithelial cells [9].
Micropatterning
Micropatterned substrata consisting of collagen-coated islands were created as described [33]. Briefly, elastomeric stamps containing a relief of the desired pattern were coated with type I collagen (50 μg/mL in water; Vitrogen 100, Cohesion Technologies, Palo Alto, CA) for 2 hours, washed with water, and dried under a stream of nitrogen. Flat poly(dimethylsiloxane) (PDMS; Sylgard 184, Ellsworth Adhesives, Germantown, WI) elastomer-coated substrata were UV-oxidized for 7 minutes (UVO Cleaner, Jelight Co., Irvine, CA), stamped with collagen, blocked with 1% pluronic F108 (BASF Corp., Florham Park, NJ) in water for 1 hour, and rinsed in PBS before seeding T4-2 cells. Cells were allowed to attach to the patterned islands for approximately 30 minutes before washing away the remaining floating cells. A flat block of PDMS was coated with collagen and stamped for the unpatterned substratum control. For measurements of projected cell area, phase contrast images of individual cells were outlined and processed with Scion Image software.
Global gene expression analysis
cDNA microarrays with ~8000 known genes spotted on poly-L-lysine-coated chips (custom arrayed at Lawrence Berkeley National Laboratory using Research Genetics 8k human clones) were used. mRNA samples of interest were directly compared to each other by co-hybridization to the same slide using dendrimer technology to label with red-Cy5 and green-Cy3 (Genisphere). Total RNA (1μg) isolated with Qiagen RNEasy reagents was used for each sample hybridized. Cells in 3D lrECM were extracted using 5mM EDTA in cold PBS to dissolve the Matrigel. For each comparison, 3 independent sets of cells cultured for 10 days were processed, and 4 slides were hybridized. This corresponded to 3 sets of RNA from independent culture sets plus a dye-swap experiment in which the red and green label was switched for the two samples in question to account for dye-specific effects. Arrays were scanned using a Genepix scanner (Axon). Raw data for each channel (red and green) was loaded onto Genespring (Silicon Genetics) for normalization and analysis. For each chip, per-spot and per-chip intensity-dependent Lowess normalization was performed using 20% of data for smoothing at a cutoff value for signal of 10. This was followed by normalization of each chip to the 50th percentile of the measurements taken from that chip, without extra background correction. Differential gene expression was determined at the t-test p-value of 0.01 or lower.
Immunofluorescence analysis
Samples were fixed in 2% paraformaldehyde in PBS, rinsed in 50 mM glycine in PBS, blocked in 10% goat serum, and stained with a 1:500 dilution of FITC-phalloidin (Molecular Probes) or with a 1:100 dilution of AcH4 antibody in PBS. Stained samples were imaged using a Spot RT camera attached to a Zeiss upright epifluorescence microscope or a Stanford Photonics XR/Mega-10 ICCD camera attached to a Zeiss spinning disk confocal microscope.
DNA was stained with DAPI and the corresponding fluorescence was measured by acquiring confocal sections separated by 0.16 μm using a spinning disk confocal microscope. Confocal images were corrected for background. To assess the nuclear volume, a common intensity threshold was established to define the edge of the nuclei in each confocal section and calculate the enclosed area. The total fluorescence intensity was divided by the nuclear volume to obtain the average dye spatial density, which correlates with the average chromatin packing ratio (Mascetti et al 2001). All image processing was performed using Image J software.
Western blotting
Total protein from S1 or T4-2 cells was extracted with lysis buffer [50 mM Tris (pH 7.4), 30 mM NaCl, 2 % (w/v) SDS, and protease inhibitor cocktail]. After sonication, an equal amount of protein from each sample was subjected to SDS gel electrophoresis and then transferred to nitrocellulose membrane (Schleicher & Schuell). The membrane was subsequently blocked in TBST buffer [50 mM Tris (pH 8.0), 150 mM NaCl, 0.1% (v/v) Tween 20] containing 5% nonfat dried milk, then incubated in blocking buffer containing primary antibody (AcH3 and AcH4 from Upstate; total H3 from Santa Cruz). All blots were further incubated in blocking buffer containing horseradish peroxidase-conjugated secondary antibodies and subjected to enhanced chemiluminescence (ECL) using the SuperSignal chemiluminescent substrate (Pierce, Rockford, IL). The results were quantified with AlphaEasyFC software, and student t test were performed using SigmaPlot.
Quantitative real-time PCR analysis
Total RNA was extracted from cells with Trizol reagent (Invitrogen). cDNA was synthesized using Superscript first strand synthesis kit (Invitrogen) from equal amounts of RNA. Quantitative real-time PCR analysis was performed with the Lightcycler System (Roche) using the Lightcycler FastStart DAN Master SYBR Green I kit (Roche). The following primers were used to amplify p21 and 18S sequences: forward primer of the p21 gene 5′-CTG GGG ATG TCC GTC AGA AC-3′ and reverse primer 5′- AGC GAG GCA CAA GGG TAC AA-3′; forward primer of the 18S gene: 5′- ACG GAC CAG AGC GAA AGC AT -3′ and reverse primer 5′- GGA CAT CTA AGG GCA TCA CAG AC -3′. The following Lightcycler PCR amplification protocol was used: 95°C for 10 minutes, and 45 amplification cycles (95°C for 5 seconds, 60°C for 10 seconds, 72°C for 5 seconds). Amplification was followed by melting curve analysis to verify the presence of a single PCR product [34]; 18S was amplified as a reference gene using the same protocol.
Chromatin immunoprecipitation (ChIP) assay
ChIP assays were performed using a commercially available kit (ChIP kit; Upstate Biotechnology, Lake Placid, NY) per manufacturer instructions. For 2D conditions, 1 × 107 S1 cells grown in a 100-mm dish were cross-linked with 1% formaldehyde at room temperature for 10 minutes. For 3D conditions, S1 cells were isolated from lrECM using PBS/EDTA and cross-linked as above. Cells were washed with PBS, resuspended and lysed in ChIP lysis buffer (1% SDS, 10mM EDTA, 50 mM Tris-HCl pH8.0). Sonicated lysates were diluted with ChIP dilution buffer and bound to protein A-agarose beads. The precleared lysates were then immunoprecipitated with AcH3 or AcH4 antibodies (Upstate Biotechnology), collected with protein A-agarose beads, and washed sequentially with each of the following buffers: low salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl pH8.0, 150 mM NaCl); high salt wash buffer (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl pH8.0, 500 mM NaCl); LiCl buffer (0.25 M LiCl, 1% NP-40, 1% SDC, 1 mM EDTA, 10 mM Tris-HCl pH8.0); TE buffer (20 mM Tris-HCl pH8.0, 1 mM EDTA pH8.0). The remaining bound p21 promoter DNA was PCR-amplified using the following primers: forward primer 5′-GGT GTC TAG GTG CTC CAG GT-3′ and reverse primer 5′-GCA CTC TCC AGG AGG ACA CA-3′.
Results
Culturing cells in 3D lrECM induces alterations in cellular morphology and global histone deacetylation
Culturing non-malignant breast epithelial S1 cells in 3D within lrECM allows the cells to form spherical polarized structures that resemble mammary acini in vivo (Fig. 1A), whereas the cells form monolayers when they are cultured in 2D. Immunofluorescence and western blot analysis showed that levels of both acetylated histones H3 and H4 were reduced in 3D cultures. (Fig. 1A, B, C).
Figure 1.
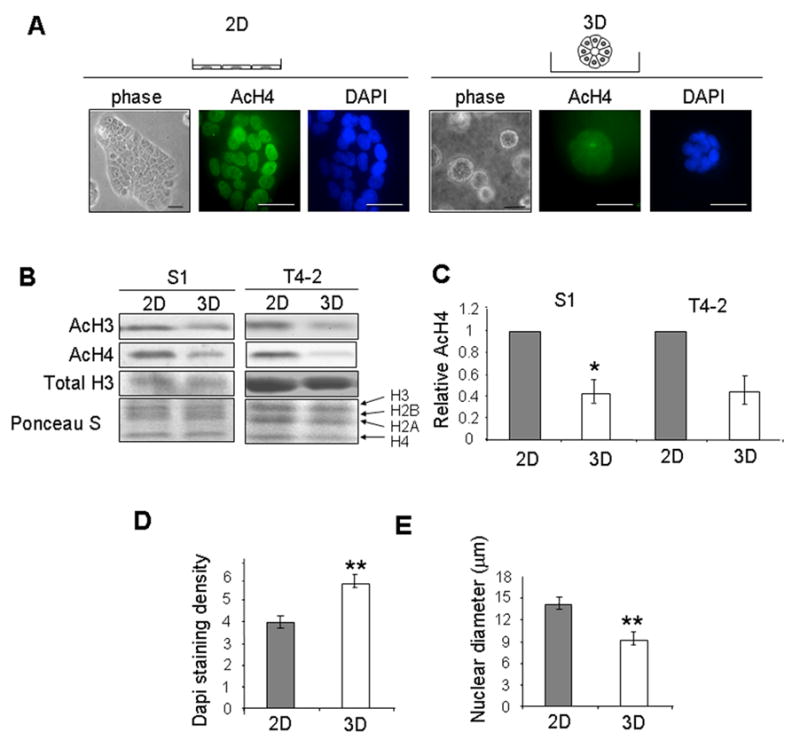
Culturing mammary epithelial cells in 3D lrECM induces alterations in cellular morphology and global histone deacetylation. (A) Phase contrast and immunofluorescence images of AcH4 and DAPI staining in S1 cells on tissue culture plastic (2D) or within lrECM (3D). Scale bars, 50 μm; (B) Western blot analysis of AcH3 and AcH4 in S1 and T4-2 cells cultured in 2D and 3D; (C) Bar graphs quantifying the relative AcH4 levels in S1 (n=4) and T4-2 (n=2) cells cultured in 2D and 3D; error bars indicate s.e.m. (*) p<0.05; (D) Quantification of nuclear DAPI staining for S1 cells cultured in 2D and 3D. (**) p<0.01; n=20; (E) Quantification of nuclear diameter for S1 cells cultured in 2D and 3D; error bars indicate s.e.m. (**) p<0.01; n=20.
Histone deacetylation is associated with chromatin condensation, and hypoacetylation of histones is commonly used as a marker of heterochromatin [35]. Here we used the average 4′,6-diamidino-2-phenylindole (DAPI) spatial density to assess global changes in chromatin condensation because the density of DAPI staining in nuclei increases with the chromatin packing ratio [36, 37]. We found that S1 cells in 3D lrECM had higher DAPI density than cells cultured in 2D (Fig. 1D). These data indicate that the histone deacetylation in 3D cultures is associated with increased chromatin compaction. In addition, the tissue-like morphogenesis observed in 3D was accompanied by a significant decrease in nuclear diameter (Fig. 1E).
T4-2, the tumorigenic derivative of S1 cells, form large non-polarized and disorganized colonies in 3D lrECM reminiscent of tumors in vivo, but they still respond to lrECM by changing cell shape (Supplemental Fig A). To determine whether global histone deacetylation was the result of acinar morphogenesis, we measured the levels of acetylated histones H3 and H4 in T4-2 cells which form disorganized structure in lrECM. Histone acetylation was decreased in 3D culture, whether or not the cells were polarized (Fig 1B, C), indicating that the reduction in histone deacetylation is a response either to ECM-induced cell rounding or to ECM-induced signaling.
Cell rounding induces global histone deacetylation
To test whether changes in cell morphology could alter histone acetylation, we compared cells cultured on plastic with those cultured on the nonadhesive substratum polyHEMA, which prevents cell attachment and spreading and allows examination of the role of cellular shape independently of ECM signaling [3, 9]. Both S1 (Fig. 2A) and T4-2 (data not shown) cells adopted a rounded morphology when cultured on polyHEMA. Culturing cells on polyHEMA for 4 days also caused them to cluster together (Fig. 2A). Immunofluorescence and western blot analysis of acetylated histones H3 and H4 demonstrated that cultivation on polyHEMA leads to histone deacetylation in both S1 and T4-2 cells (Fig. 2A, B, C). Additionally, histone deacetylation in cells on polyHEMA is associated with chromatin condensation and a decrease in nuclear size (Fig. 2D, E). Adding lrECM to cells cultured on polyHEMA did not decrease histone acetylation levels further (data not shown), indicating that histone deacetylation induced by lrECM is due specifically to an effect of cell rounding or cell clustering, and that ECM ligand-induced signaling does not potentiate this effect.
Figure 2.
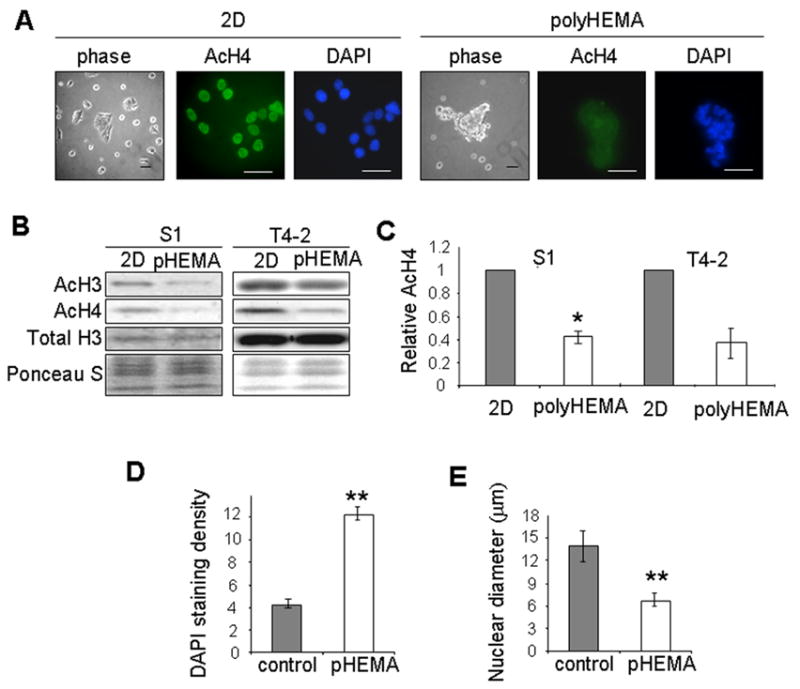
Cell rounding on the nonadhesive substratum polyHEMA induces global histone deacetylation. (A) Phase contrast images of S1 cells cultured in 2D on tissue culture plastic or rounded on polyHEMA. Immunofluorescence images of AcH4 and DAPI staining in S1 cells cultured in 2D and on polyHEMA; (B) Western blot analysis of AcH3 and AcH4 in S1 and T4-2 cells cultured in 2D or on polyHEMA; (C) Bar graphs quantifying the relative AcH4 levels in S1 (n=4) and T4-2 (n=2) cells cultured in 2D and on polyHEMA. (**) p<0.01; (D) Quantification of nuclear DAPI staining in S1 cells cultured in 2D and on polyHEMA. (**) p<0.01; n=20; (E) Quantification of nuclear diameter for S1 cells cultured in 2D and on polyHEMA. (**) p<0.01; n=20.
To delineate the effects of cell rounding from those of cell clustering, we used a micropatterning approach to maintain single attached cells in a rounded morphology. Cells were cultured on micropatterned substrata that contained micrometer-sized islands of collagen surrounded by nonadhesive regions. Cells plated on these substrata could only attach to the collagen-coated islands; individual cells cultured on small islands were prevented from spreading. We compared unpatterned and freely spread T4-2 cells with those patterned on 25-μm square islands (Fig. 3A). T4-2 cells plated on micropatterned substrata remained rounded compared to those plated on unpatterned collagen-coated substrata (Fig. 3A). Whereas control cells formed stress fibers, patterned cells exhibited mainly cortical actin (Fig. 3A). The mean projected area of individual cells plated on the micropatterned substrata was significantly reduced when compared to cells plated on tissue culture plastic (Supplemental Fig. B). Western blot analysis showed that levels of acetylated histones H3 and H4 were reduced in patterned cells compared to unpatterned cells (Fig. 3B, C). Quantification of DAPI staining demonstrated that patterned cells had more compacted chromatin than unpatterned cells (Fig. 3D), accompanied by a reduction in nuclear size in the patterned cells (Fig. 3E). These data show that ECM-induced cell rounding directly controls histone deacetylation and chromatin condensation in mammary epithelial cells. It further indicates that even malignant T4-2 cells remain sensitive to shape-induced changes in their microenvironment.
Figure 3.
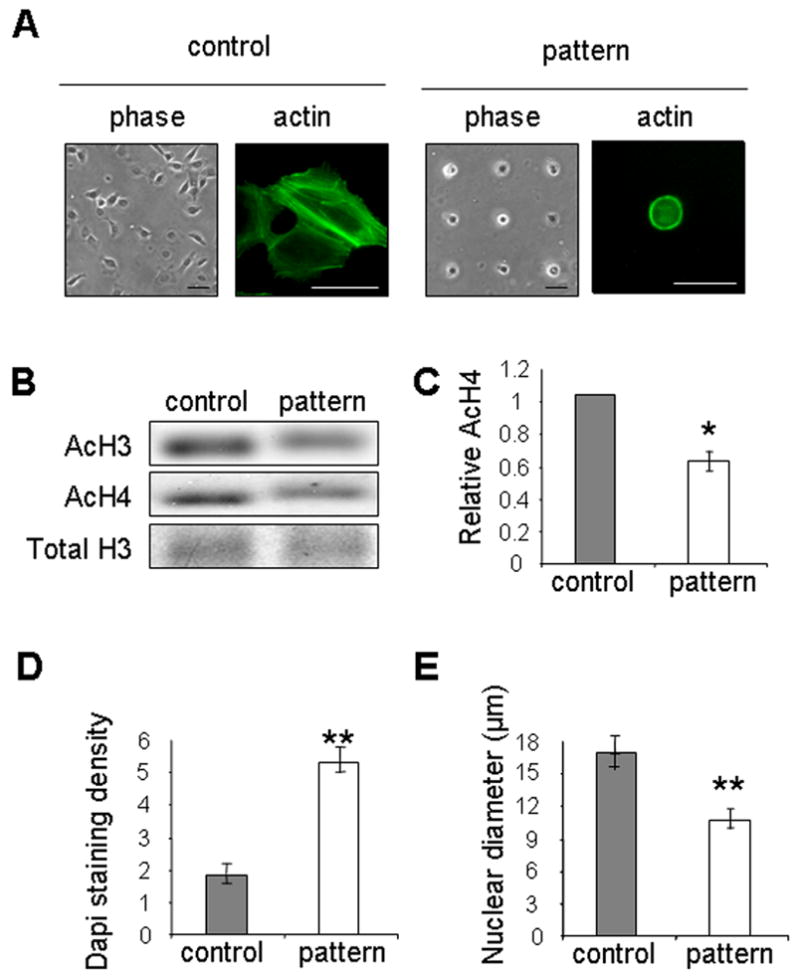
Cell rounding on micropatterned substrata induces histone deacetylation. (A) Phase contrast and immunofluorescence images of phalloidin-stained T4-2 cells cultured on unpatterned substratum (control) or substratum patterned with 25-μm square islands (pattern) for 24 hours. Scale bars, 50 μm; (B) Western blot analysis of AcH3 and AcH4 in T4-2 cells cultured on unpatterned and patterned substrata; (C) Bar graphs quantifying the relative AcH4 levels in T4-2 cells. (*) p<0.05, n=4; (D) Quantification of nuclear DAPI staining for T4-2 cells cultured on unpatterned and patterned substrata. (**) p<0.01; n=20; (E) Quantification of nuclear diameter for T4-2 cells cultured on unpatterned and patterned substrata. (**) p<0.01; n=20.
Cell shape is largely governed by the actin cytoskeleton [38]. Shape dependence therefore implies a requirement for a particular actin organization. Indeed, one of the striking differences we noticed between cells spread on plastic and those rounded in 3D lrECM or on micropatterned substrata was the organization of the actin cytoskeleton. Both S1 and T4-2 cells cultured in 2D exhibited prominent stress fibers as well as cortical actin when stained with fluorescently-tagged phalloidin (Fig. 4A). In contrast, even though the gross architectures of the colonies formed by these cells differed substantially, individual S1 and T4-2 cells appeared similarly rounded in 3D (Fig. 1A and Supplemental Fig. A) with mainly cortical actin (Fig. 4A). Cells rounded by culture on micropatterned substrata also showed prominent cortical actin (Fig 4B). To address the involvement of shape and actin organization in regulation of histone acetylation, S1 cells grown on tissue culture plastic were treated with the actin polymerization inhibitor cytochalasin D. Treatment with this drug disrupted actin microfilaments, induced cell rounding (Fig. 4B), and reduced the levels of acetylated histones H3 and H4 (Fig. 4C, D). Importantly, removal of the drug reversed cell morphology (data not shown) and induced an increase in histone acetylation (Fig. 4D). Therefore, cytoskeletal changes associated with cell rounding induce global histone deacetylation.
Figure 4.
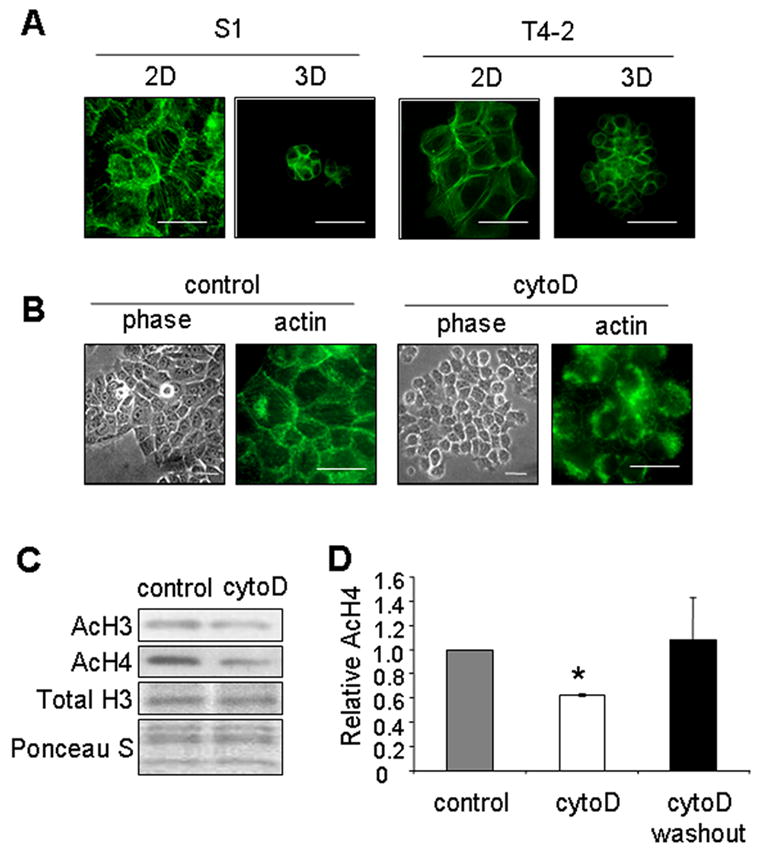
Actin cytoskeleton regulates cell rounding and histone deacetylation. (A) Immunofluorescence images of phalloidin-stained S1 and T4-2 cells cultured in 2D or 3D; (B) Phase contrast and immunofluorescence images of phalloidin-stained S1 cells in the presence or absence of cytochalasin D (cytoD). Scale bars, 50 μm; (C) Western blot for AcH3 and AcH4 in S1 cells in the presence or absence of cytochalasin D. (D) Bar graphs of relative AcH4 levels in the control S1 cells (control), cells treated with cytochalasin D (cytoD), and cells treated with cytochalasin D followed by removal of the drug (cytoD washout). (*) p<0.05, n=4.
Culture in 3D lrECM induces a global reduction in gene expression
One question that arises from these observations is whether the histone deacetylation in 3D culture regulates gene expression. Indeed, broad acetylation of histones H3 and H4 was shown to lead to chromatin decondensation and a structure permissive for transcription (reviewed in [39]), whereas histone deacetylation correlated with the repressed state of chromatin and alterations in patterns of gene expression [40]. To determine whether culture in 3D lrECM induced a reduction in gene expression in addition to global histone deacetylation, we performed microarray analysis. mRNA samples from S1 cells cultured on plastic or in 3D lrECM were fluorescently tagged and co-hybridized to cDNA microarrays, and the intensity of hybridizations were compared. Genes expressed differentially between the two conditions were selected for analysis. We found that cells cultured in 3D lrECM showed an appreciable reduction in gene expression as compared to cells cultured on plastic (Fig. 5A; Supplemental Table.1). Of the differentially expressed genes, 162 genes were expressed at lower levels in 3D, compared to 91 expressed at higher levels. This decrease in gene expression is consistent with the global histone deacetylation observed in 3D cultures.
Figure 5.
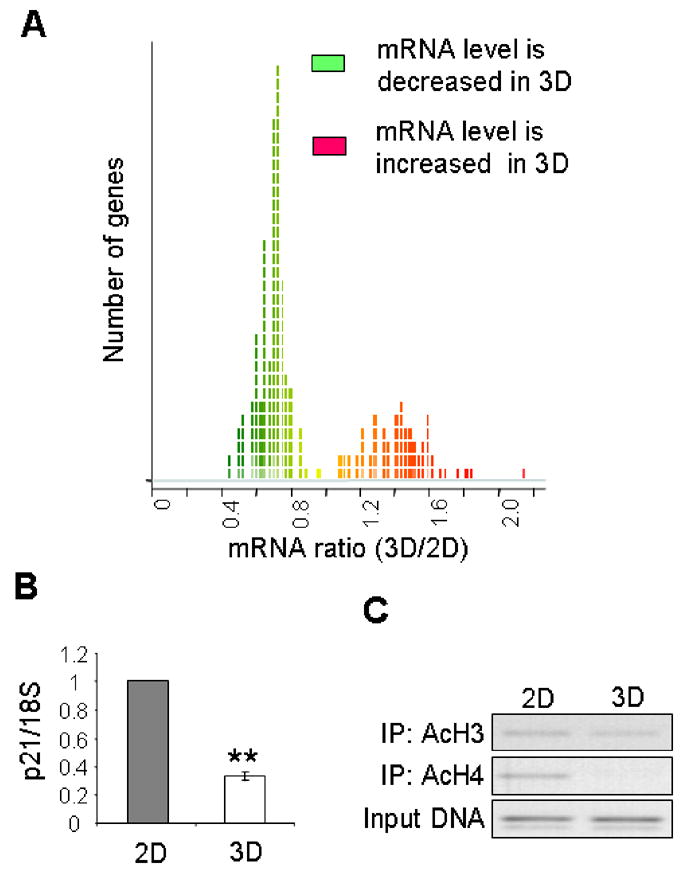
Culturing mammary epithelial cells in 3D lrECM induces a global reduction in gene expression. (A) Ratio of global mRNA levels for S1 cells cultured in 2D and 3D lrECM. The x-axis shows mean ratio of 3D/2D for four experiments; all genes with p< 0.01 are displayed: 91 genes have higher mRNA levels in 3D (red); 162 genes have lower levels in 3D (green). (B) Quantitative RT/PCR analysis for p21 normalized to levels of 18S in the same samples. (**) p<0.01; (C) ChIP assay measuring levels of AcH3 and AcH4 in the p21 promoter for S1 cells cultured in 2D and 3D.
Acetylation of histones in the promoter regions and other regulatory cis-elements clearly affects transcription of a large number of genes [17, 18, 41]. To address the question of whether the lrECM-dependent global deacetylation could specifically affect the promoter of a functionally relevant gene, we focused our attention on the cell cycle inhibitor p21. It was previously shown that the expression of the p21 gene is controlled by the acetylation status of histones associated with its promoter [42, 43]. Indeed, from our array data, the transcript level of p21 in S1 cells in 3D was reduced to 76% of that in cells in 2D after 10 days culture (Supplemental Table 1). Quantitative real-time RT/PCR confirmed that p21 mRNA levels were reduced in 3D compared to 2D (Fig. 5B). To determine whether this reduction in expression is correlated with reduced acetylation at the promoter region of the p21 gene, we performed chromatin immunoprecipitation (ChIP) experiments. Lysates from S1 cells cultured in 2D or 3D were immunoprecipitated for acetylated histones H3 or H4, and the remaining associated DNA was PCR-amplified using primers specific for the p21 promoter. Culturing the cells in 3D lrECM induced a dramatic reduction of histone acetylation in the p21 promoter (Fig. 5C). This result is consistent with the reduced expression of p21 observed by microarray analysis and RT/PCR. These data indicate that lrECM-induced histone deacetylation is associated with a functional reduction of gene expression.
Discussion
The interactions between a cell and its local microenvironment have profound effects on cell shape, cytoskeletal and nuclear matrix organization, chromatin structure, and gene expression [24, 44–46]. In this study, we found that culture of human mammary epithelial cells within 3D lrECM induced cell rounding and global histone deacetylation. Histone deacetylation correlates with chromatin condensation and reduced gene expression. These data are corroborated by previous studies demonstrating that lrECM-induced morphogenesis correlates with reduced histone acetylation and increased histone methylation [24, 25, 28, 44]. Cells receive at least two types of signals from laminin-rich ECM gels: biochemical signals from the engagement of cell surface receptors, and biophysical signals from the change in their morphology [9]. Altering cellular shape has been shown to regulate nuclear morphology and chromatin structure [37]. Using non-adhesive and micropatterned substrata as well as inhibitors of actin polymerization, we determined that cell rounding is a sufficient stimulus to induce histone deacetylation and chromatin condensation in the absence of biochemical signals transduced from the ECM; notably, adding ECM to pre-rounded cells did not potentiate the shape-induced histone deacetylation.
There is increasing evidence that cell rounding inhibits cell proliferation [3, 4], and that histone acetylation levels correlate with cell cycle status [47]. We found that culturing cells on plastic and in 3D lrECM led to a similar percentage of growth-arrested cells (Supplemental Fig. C). Furthermore, global histone deacetylation was detected in T4-2 cells cultured in 3D, although these cells failed to growth arrest (data not shown). Therefore, the ECM-induced reduction in histone acetylation does not correlate with decreased proliferation in this system. S1 cells undergo growth arrest after 10 days culture in both 2D and 3D. However, transcription levels of the p21 gene were lower in 3D than in 2D culture, suggesting that the growth arrest in 3D may be regulated by other proteins or signaling pathways. Consistent with this possibility, we previously reported that cell cycle arrest in 3D culture was accompanied by progressive hypophosphorylation of the retinoblastoma (Rb) gene and induction of the cyclin-dependent kinase (CDK) inhibitor p27kip1[48].
Results over the past two decades appear to support the hypothesis that the ECM is dynamically coupled to the nucleus through the cytoskeleton and the nuclear matrix [1, 49–51]. One attractive potential mechanism to regulate chromatin structure involves signaling from the cytoskeleton. We found that cell rounding (either in 3D lrECM or on micropatterned substrata) altered the actin cytoskeleton and prevented the formation of stress fibers, and that blocking actin polymerization/depolymerization with cytochalasin D led to histone deacetylation. Disrupting microtubules also causes cell rounding [52] and has been shown to induce histone deacetylation [53]. These results suggest a role for low intracellular tension generated by cytoskeletal organization in the control of histone acetylation. It has been shown that disrupting actomyosin tension generation by inhibiting myosin light chain kinase, RhoA, or Rho-kinase leads to global histone deacetylation, and conversely, increasing actomyosin contractility enhances histone acetylation levels in gastric carcinoma cells [41]. Increasing tension by application of shear stress leads to histone acetylation, chromatin remodeling, and changes in gene expression in endothelial and embryonic stem cells [54, 55]. In agreement, experimental and theoretical evidence supports a specific role for the actin cytoskeleton in physically connecting sites of cell adhesion to ECM with the nucleus [51] as well as integrating changes in cell and nuclear shape [56, 57]. In this scenario it is thought that tension developed by actin bundles in spread cells deforms the nucleus to increase its projected area, while a decrease in tension (as occurs in round cells) would have the opposite effects. In support of this model we observed a systematic reduction in nuclear diameter in round cells compared to control cells. Based on the collective data described, it is tempting to speculate that cell shape regulates histone acetylation through mechanical transmission of actin/cytoskeletal tension to the nucleus.
Histone acetylation and deacetylation are catalyzed by HATs and HDACs; therefore cell rounding could regulate histone acetylation by altering the total cellular levels or activities of these enzymes, changing their levels in the nucleus, or their association with the nuclear matrix. Cell rounding reduces the calibre of nuclear pore complexes and reduces the rate of nucleocytoplasmic transport [58, 59], which could directly affect the shuttling of HATs and HDACs and their relative levels within the nucleus. Biochemically, HAT and HDAC shuttling is regulated by phosphorylation which changes their binding affinity to chaperone proteins that usher them into or out of the nucleus [60]. Indeed, during skeletal muscle differentiation Ca2+/calmodulin-dependent protein kinase (CaMK) activity regulates class II HDAC phosphorylation, binding to 14-3-3 protein chaperone, and subsequent nuclear shuttling [61]. Additionally, some HDACs have been found to bind to actin filaments and microtubules in the cytoplasm, potentially sequestering them away from the nuclear compartment [62]. Depolymerizing portions of the cytoskeleton may release HDACs and allow them to shuttle from the cytoplasm into the nucleus, tipping the balance in favour of histone deacetylation. In general, regulation of HDAC and HAT activities is a large field of study which is opening an ever greater array of possible mechanisms by which cell shape might regulate histone acetylation.
Despite the fact that ECM-induced cell rounding leads to global histone deacetylation, and that the deacetylation is associated with a reduction in overall gene expression in 3D cultures, histones associated with genes specifically upregulated during differentiation are most probably acetylated locally. For example, histone H4 acetylation is increased in the promoter region of the α-casein milk protein gene in mammary epithelial cells in the presence of collagen [63], and histone H3 and H4 acetylation is increased in the promoter region of the β-casein gene during differentiation [64, 65]. It will be interesting to determine how cell shape affects the acetylation status of these and other differentiation-associated genes. Although global histone deacetylation is observed during functional differentiation of mammary epithelial cells [24, 28] and is regulated by cell rounding, the latter per se may not explain all the effects of ECM on functional differentiation; indeed, the assembly of a basement membrane is required in addition to cell rounding for the reorganization of the nuclear protein NuMA and tissue-specific functions [24], and biochemical signals transmitted through integrin and non-integrin laminin receptors are required for functional differentiation and β-casein expression [9, 12, 66, 67].
We have provided strong evidence for a link between ECM-regulated cell shape, chromatin structure and gene expression. Since ECM influences morphology and gene expression in a wide variety of tissues [reviewed in [68–72]], it is most likely that cell shape-induced changes in global histone acetylation and chromatin structure are a common mechanism in regulation of tissue- and context-specific gene expression in many organs.
Supplementary Material
Supplemental Table 1. List of genes differentially expressed between S1 cells grown in 2D and 3D lrECM culture. “Common” refers to the gene name in use, “Genbank” column contains unique gene identifiers, “Normalized” values refer to the mean value of the ratio of 3D/2D across four microarray experiments in which Lowess normalization was performed, and t-test p-value is across four independent array experiments.
Supplemental Figure A. Phase contrast images of T4-2 cells on tissue culture plastic (2D) or within lrECM (3D). Scale bars, 50 μm.
Supplemental Figure B. Bar graph of mean projected cell area for T4-2 cells cultured on unpatterned or patterned substrata.
Supplemental Figure C. Table showing flow cytometric analysis of cell cycle distribution of S1 cells in 2D or 3D lrECM.
Acknowledgments
We thank Virginia Spencer and other members of the Bissell laboratory for helpful discussions. This work was supported by grants from the Office of Biological and Environmental Research of the Department of Energy (DE-AC03-76SF00098 and a Distinguish Fellow Award to MJB), the National Cancer Institute (CA64786 to M.J.B. and Ole William Peterson, and CA57621 to M.J.B. and Zena Werb), and the Breast Cancer Research Program (BCRP) of the Department of Defense (DOD) (an Innovator Award to MJB; postdoctoral fellowships DAMD17-02-1-0441 and W81XWH-04-1-0582 to RX and CMN). CMN holds a Career Award at the Scientific Interface from the Burroughs Wellcome Fund.
Abbreviations
- ECM
extracellular matrix
- lrECM
laminin-rich reconstituted ECM
- HATs
histone acetyltransferases
- HDACs
histone deacetylases
- polyHEMA
poly(2-hydroxyethyl methacrylate)
- TPA
12-o-tetradecanoylphorbol 13-acetate
- 2D
two-dimension(al)
- 3D
three-dimension(al)
- AcH3 and AcH4
acetylated histone H3 and H4
- ChIP
chromatin immunoprecipitation
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bissell MJ, Hall HG, Parry G. How does the extracellular matrix direct gene expression? J Theor Biol. 1982;99:31–68. doi: 10.1016/0022-5193(82)90388-5. [DOI] [PubMed] [Google Scholar]
- 2.Bissell MJ, Farson D, Tung AS. Cell shape and hexose transport in normal and virus-transformed cells in culture. J Supramol Struct. 1977;6:1–12. doi: 10.1002/jss.400060102. [DOI] [PubMed] [Google Scholar]
- 3.Folkman J, Moscona A. Role of cell shape in growth control. Nature. 1978;273:345–9. doi: 10.1038/273345a0. [DOI] [PubMed] [Google Scholar]
- 4.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Geometric control of cell life and death. Science. 1997;276:1425–8. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 5.Singhvi R, Kumar A, Lopez GP, Stephanopoulos GN, Wang DI, Whitesides GM, Ingber DE. Engineering cell shape and function. Science. 1994;264:696–8. doi: 10.1126/science.8171320. [DOI] [PubMed] [Google Scholar]
- 6.Farmer SR, Ben-Ze’av A, Benecke BJ, Penman S. Altered translatability of messenger RNA from suspended anchorage-dependent fibroblasts: reversal upon cell attachment to a surface. Cell. 1978;15:627–37. doi: 10.1016/0092-8674(78)90031-4. [DOI] [PubMed] [Google Scholar]
- 7.Thomas CH, Collier JH, Sfeir CS, Healy KE. Engineering gene expression and protein synthesis by modulation of nuclear shape. Proc Natl Acad Sci U S A. 2002;99:1972–7. doi: 10.1073/pnas.032668799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ben-Ze’ev A, Farmer SR, Penman S. Protein synthesis requires cell-surface contact while nuclear events respond to cell shape in anchorage-dependent fibroblasts. Cell. 1980;21:365–72. doi: 10.1016/0092-8674(80)90473-0. [DOI] [PubMed] [Google Scholar]
- 9.Roskelley CD, Desprez PY, Bissell MJ. Extracellular matrix-dependent tissue-specific gene expression in mammary epithelial cells requires both physical and biochemical signal transduction. Proc Natl Acad Sci U S A. 1994;91:12378–82. doi: 10.1073/pnas.91.26.12378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emerman JT, Burwen SJ, Pitelka DR. Substrate properties influencing ultrastructural differentiation of mammary epithelial cells in culture. Tissue Cell. 1979;11:109–19. doi: 10.1016/0040-8166(79)90011-9. [DOI] [PubMed] [Google Scholar]
- 11.Bissell MJ, Rizki A, Mian IS. Tissue architecture: the ultimate regulator of breast epithelial function. Curr Opin Cell Biol. 2003;15:753–62. doi: 10.1016/j.ceb.2003.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Streuli CH, Bailey N, Bissell MJ. Control of mammary epithelial differentiation: basement membrane induces tissue-specific gene expression in the absence of cell-cell interaction and morphological polarity. J Cell Biol. 1991;115:1383–95. doi: 10.1083/jcb.115.5.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Streuli CH, Schmidhauser C, Bailey N, Yurchenco P, Skubitz AP, Roskelley C, Bissell MJ. Laminin mediates tissue-specific gene expression in mammary epithelia. J Cell Biol. 1995;129:591–603. doi: 10.1083/jcb.129.3.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taketani Y, Oka T. Tumor promoter 12-O-tetradecanoylphorbol 13-acetate, like epidermal growth factor, stimulates cell proliferation and inhibits differentiation of mouse mammary epithelial cells in culture. Proc Natl Acad Sci U S A. 1983;80:1646–9. doi: 10.1073/pnas.80.6.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grant PA. A tale of histone modifications. Genome Biol. 2001;2:1–6. doi: 10.1186/gb-2001-2-4-reviews0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davie JR. Histone modifications, chromatin structure, and the nuclear matrix. J Cell Biochem. 1996;62:149–57. doi: 10.1002/(sici)1097-4644(199608)62:2<149::aid-jcb2>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 17.Roh TY, Cuddapah S, Zhao K. Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes Dev. 2005;19:542–52. doi: 10.1101/gad.1272505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernstein BE, Humphrey EL, Erlich RL, Schneider R, Bouman P, Liu JS, Kouzarides T, Schreiber SL. Methylation of histone H3 Lys 4 in coding regions of active genes. Proc Natl Acad Sci U S A. 2002;99:8695–700. doi: 10.1073/pnas.082249499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B. A high-resolution map of active promoters in the human genome. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 21.Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, Bonaldi T, Haydon C, Ropero S, Petrie K, Iyer NG, Perez-Rosado A, Calvo E, Lopez JA, Cano A, Calasanz MJ, Colomer D, Piris MA, Ahn N, Imhof A, Caldas C, Jenuwein T, Esteller M. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet. 2005;37:391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- 22.Shen S, Li J, Casaccia-Bonnefil P. Histone modifications affect timing of oligodendrocyte progenitor differentiation in the developing rat brain. J Cell Biol. 2005;169:577–89. doi: 10.1083/jcb.200412101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Francastel C, Schubeler D, Martin DI, Groudine M. Nuclear compartmentalization and gene activity. Nat Rev Mol Cell Biol. 2000;1:137–43. doi: 10.1038/35040083. [DOI] [PubMed] [Google Scholar]
- 24.Lelievre SA, Weaver VM, Nickerson JA, Larabell CA, Bhaumik A, Petersen OW, Bissell MJ. Tissue phenotype depends on reciprocal interactions between the extracellular matrix and the structural organization of the nucleus. Proc Natl Acad Sci U S A. 1998;95:14711–6. doi: 10.1073/pnas.95.25.14711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Plachot C, Lelievre SA. DNA methylation control of tissue polarity and cellular differentiation in the mammary epithelium. Exp Cell Res. 2004;298:122–32. doi: 10.1016/j.yexcr.2004.04.024. [DOI] [PubMed] [Google Scholar]
- 26.Knowles DW, Sudar D, Bator-Kelly C, Bissell MJ, Lelievre SA. Automated local bright feature image analysis of nuclear protein distribution identifies changes in tissue phenotype. Proc Natl Acad Sci U S A. 2006;103:4445–50. doi: 10.1073/pnas.0509944102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaminker P, Plachot C, Kim SH, Chung P, Crippen D, Petersen OW, Bissell MJ, Campisi J, Lelievre SA. Higher-order nuclear organization in growth arrest of human mammary epithelial cells: a novel role for telomere-associated protein TIN2. J Cell Sci. 2005;118:1321–30. doi: 10.1242/jcs.01709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pujuguet P, Radisky D, Levy D, Lacza C, Bissell MJ. Trichostatin A inhibits beta-casein expression in mammary epithelial cells. J Cell Biochem. 2001;83:660–70. doi: 10.1002/jcb.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Briand P, Petersen OW, Van Deurs B. A new diploid nontumorigenic human breast epithelial cell line isolated and propagated in chemically defined medium. In Vitro Cell Dev Biol. 1987;23:181–8. doi: 10.1007/BF02623578. [DOI] [PubMed] [Google Scholar]
- 30.Briand P, Nielsen KV, Madsen MW, Petersen OW. Trisomy 7p and malignant transformation of human breast epithelial cells following epidermal growth factor withdrawal. Cancer Res. 1996;56:2039–44. [PubMed] [Google Scholar]
- 31.Petersen OW, Ronnov-Jessen L, Howlett AR, Bissell MJ. Interaction with basement membrane serves to rapidly distinguish growth and differentiation pattern of normal and malignant human breast epithelial cells. Proc Natl Acad Sci U S A. 1992;89:9064–8. doi: 10.1073/pnas.89.19.9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weaver VM, Petersen OW, Wang F, Larabell CA, Briand P, Damsky C, Bissell MJ. Reversion of the malignant phenotype of human breast cells in three-dimensional culture and in vivo by integrin blocking antibodies. J Cell Biol. 1997;137:231–45. doi: 10.1083/jcb.137.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tan JL, Liu W, Nelson CM, Raghavan S, Chen CS. Simple approach to micropattern cells on common culture substrates by tuning substrate wettability. Tissue Eng. 2004;10:865–72. doi: 10.1089/1076327041348365. [DOI] [PubMed] [Google Scholar]
- 34.Sugimoto S, Mitaka T, Ikeda S, Harada K, Ikai I, Yamaoka Y, Mochizuki Y. Morphological changes induced by extracellular matrix are correlated with maturation of rat small hepatocytes. J Cell Biochem. 2002;87:16–28. doi: 10.1002/jcb.10274. [DOI] [PubMed] [Google Scholar]
- 35.Grunstein M. Yeast heterochromatin: regulation of its assembly and inheritance by histones. Cell. 1998;93:325–8. doi: 10.1016/s0092-8674(00)81160-5. [DOI] [PubMed] [Google Scholar]
- 36.Mascetti G, Carrara S, Vergani L. Relationship between chromatin compactness and dye uptake for in situ chromatin stained with DAPI. Cytometry. 2001;44:113–9. doi: 10.1002/1097-0320(20010601)44:2<113::aid-cyto1089>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 37.Vergani L, Grattarola M, Nicolini C. Modifications of chromatin structure and gene expression following induced alterations of cellular shape. Int J Biochem Cell Biol. 2004;36:1447–61. doi: 10.1016/j.biocel.2003.11.015. [DOI] [PubMed] [Google Scholar]
- 38.Sims JR, Karp S, Ingber DE. Altering the cellular mechanical force balance results in integrated changes in cell, cytoskeletal and nuclear shape. J Cell Sci. 1992;103 ( Pt 4):1215–22. doi: 10.1242/jcs.103.4.1215. [DOI] [PubMed] [Google Scholar]
- 39.Eberharter A, Becker PB. Histone acetylation: a switch between repressive and permissive chromatin. Second in review series on chromatin dynamics. EMBO Rep. 2002;3:224–9. doi: 10.1093/embo-reports/kvf053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kurdistani SK, Tavazoie S, Grunstein M. Mapping global histone acetylation patterns to gene expression. Cell. 2004;117:721–33. doi: 10.1016/j.cell.2004.05.023. [DOI] [PubMed] [Google Scholar]
- 41.Kim YB, Yu J, Lee SY, Lee MS, Ko SG, Ye SK, Jong HS, Kim TY, Bang YJ, Lee JW. Cell adhesion status-dependent histone acetylation is regulated through intracellular contractility-related signaling activities. J Biol Chem. 2005 doi: 10.1074/jbc.M412608200. [DOI] [PubMed] [Google Scholar]
- 42.Davis T, Kennedy C, Chiew YE, Clarke CL, deFazio A. Histone deacetylase inhibitors decrease proliferation and modulate cell cycle gene expression in normal mammary epithelial cells. Clin Cancer Res. 2000;6:4334–42. [PubMed] [Google Scholar]
- 43.Huang L, Sowa Y, Sakai T, Pardee AB. Activation of the p21WAF1/CIP1 promoter independent of p53 by the histone deacetylase inhibitor suberoylanilide hydroxamic acid (SAHA) through the Sp1 sites. Oncogene. 2000;19:5712–9. doi: 10.1038/sj.onc.1203963. [DOI] [PubMed] [Google Scholar]
- 44.Myers CA, Schmidhauser C, Mellentin-Michelotti J, Fragoso G, Roskelley CD, Casperson G, Mossi R, Pujuguet P, Hager G, Bissell MJ. Characterization of BCE-1, a transcriptional enhancer regulated by prolactin and extracellular matrix and modulated by the state of histone acetylation. Mol Cell Biol. 1998;18:2184–95. doi: 10.1128/mcb.18.4.2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beningo KA, Dembo M, Wang YL. Responses of fibroblasts to anchorage of dorsal extracellular matrix receptors. Proc Natl Acad Sci U S A. 2004;101:18024–9. doi: 10.1073/pnas.0405747102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sidhu JS, Liu F, Omiecinski CJ. Phenobarbital responsiveness as a uniquely sensitive indicator of hepatocyte differentiation status: requirement of dexamethasone and extracellular matrix in establishing the functional integrity of cultured primary rat hepatocytes. Exp Cell Res. 2004;292:252–64. doi: 10.1016/j.yexcr.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 47.Ramaswamy V, Williams JS, Robinson KM, Sopko RL, Schultz MC. Global control of histone modification by the anaphase-promoting complex. Mol Cell Biol. 2003;23:9136–49. doi: 10.1128/MCB.23.24.9136-9149.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fournier MV, Martin KJ, Kenny PA, Xhaja K, Bosch I, Yaswen P, Bissell MJ. Gene expression signature in organized and growth-arrested mammary acini predicts good outcome in breast cancer. Cancer Res. 2006;66:7095–102. doi: 10.1158/0008-5472.CAN-06-0515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pienta KJ, Coffey DS. Nuclear-cytoskeletal interactions: evidence for physical connections between the nucleus and cell periphery and their alteration by transformation. J Cell Biochem. 1992;49:357–65. doi: 10.1002/jcb.240490406. [DOI] [PubMed] [Google Scholar]
- 50.Fey EG, Wan KM, Penman S. Epithelial cytoskeletal framework and nuclear matrix-intermediate filament scaffold: three-dimensional organization and protein composition. J Cell Biol. 1984;98:1973–84. doi: 10.1083/jcb.98.6.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maniotis AJ, Chen CS, Ingber DE. Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc Natl Acad Sci U S A. 1997;94:849–54. doi: 10.1073/pnas.94.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Flusberg DA, Numaguchi Y, Ingber DE. Cooperative control of Akt phosphorylation, bcl-2 expression, and apoptosis by cytoskeletal microfilaments and microtubules in capillary endothelial cells. Mol Biol Cell. 2001;12:3087–94. doi: 10.1091/mbc.12.10.3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nishiyama A, Dey A, Miyazaki J, Ozato K. Brd4 is required for recovery from antimicrotubule drug-induced mitotic arrest: preservation of acetylated chromatin. Mol Biol Cell. 2006;17:814–23. doi: 10.1091/mbc.E05-08-0729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Illi B, Nanni S, Scopece A, Farsetti A, Biglioli P, Capogrossi MC, Gaetano C. Shear stress-mediated chromatin remodeling provides molecular basis for flow-dependent regulation of gene expression. Circ Res. 2003;93:155–61. doi: 10.1161/01.RES.0000080933.82105.29. [DOI] [PubMed] [Google Scholar]
- 55.Illi B, Scopece A, Nanni S, Farsetti A, Morgante L, Biglioli P, Capogrossi MC, Gaetano C. Epigenetic histone modification and cardiovascular lineage programming in mouse embryonic stem cells exposed to laminar shear stress. Circ Res. 2005;96:501–8. doi: 10.1161/01.RES.0000159181.06379.63. [DOI] [PubMed] [Google Scholar]
- 56.Jean RP, Gray DS, Spector AA, Chen CS. Characterization of the nuclear deformation caused by changes in endothelial cell shape. J Biomech Eng. 2004;126:552–8. doi: 10.1115/1.1800559. [DOI] [PubMed] [Google Scholar]
- 57.Guilak F. Compression-induced changes in the shape and volume of the chondrocyte nucleus. J Biomech. 1995;28:1529–41. doi: 10.1016/0021-9290(95)00100-x. [DOI] [PubMed] [Google Scholar]
- 58.Hansen LK, Ingber DE. Regulation of nucleocytoplasmic transport by mechanical forces transmitted through the cytoskeleton. In: Feldherr CM, editor. Nuclear Trafficking. Academic Press; Orlando, FL: 1992. [Google Scholar]
- 59.Feldherr CM, Akin D. The permeability of the nuclear envelope in dividing and nondividing cell cultures. J Cell Biol. 1990;111:1–8. doi: 10.1083/jcb.111.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Legube G, Trouche D. Regulating histone acetyltransferases and deacetylases. EMBO Rep. 2003;4:944–7. doi: 10.1038/sj.embor.embor941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McKinsey TA, Zhang CL, Lu J, Olson EN. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature. 2000;408:106–11. doi: 10.1038/35040593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Waltregny D, Glenisson W, Tran SL, North BJ, Verdin E, Colige A, Castronovo V. Histone deacetylase HDAC8 associates with smooth muscle alpha-actin and is essential for smooth muscle cell contractility. Faseb J. 2005;19:966–8. doi: 10.1096/fj.04-2303fje. [DOI] [PubMed] [Google Scholar]
- 63.Jolivet G, Pantano T, Houdebine LM. Regulation by the extracellular matrix (ECM) of prolactin-induced alpha s1-casein gene expression in rabbit primary mammary cells: role of STAT5, C/EBP, and chromatin structure. J Cell Biochem. 2005;95:313–27. doi: 10.1002/jcb.20397. [DOI] [PubMed] [Google Scholar]
- 64.Kabotyanski EB, Huetter M, Xian W, Rijnkels M, Rosen JM. Integration of prolactin and glucocorticoid signaling at the {beta}-casein promoter and enhancer by ordered recruitment of specific transcription factors and chromatin modifiers. Mol Endocrinol. 2006 doi: 10.1210/me.2006-0160. [DOI] [PubMed] [Google Scholar]
- 65.Xu R, Spencer VA, Bissell MJ. Extracellular matrix-regulated gene expression requires cooperation of SWI/SNF and transcription factors. J Biol Chem. 2007 doi: 10.1074/jbc.M610316200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Muschler J, Lochter A, Roskelley CD, Yurchenco P, Bissell MJ. Division of labor among the alpha6beta4 integrin, beta1 integrins, and an E3 laminin receptor to signal morphogenesis and beta-casein expression in mammary epithelial cells. Mol Biol Cell. 1999;10:2817–28. doi: 10.1091/mbc.10.9.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weir ML, Oppizzi ML, Henry MD, Onishi A, Campbell KP, Bissell MJ, Muschler JL. Dystroglycan loss disrupts polarity and {beta}-casein induction in mammary epithelial cells by perturbing laminin anchoring. J Cell Sci. 2006;119:4047–58. doi: 10.1242/jcs.03103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Adams JC, Watt FM. Regulation of development and differentiation by the extracellular matrix. Development. 1993;117:1183–98. doi: 10.1242/dev.117.4.1183. [DOI] [PubMed] [Google Scholar]
- 69.Boudreau N, Myers C, Bissell MJ. From laminin to lamin: regulation of tissue-specific gene expression by the ECM. Trends Cell Biol. 1995;5:1–4. doi: 10.1016/s0962-8924(00)88924-2. [DOI] [PubMed] [Google Scholar]
- 70.Ashkenas J, Muschler J, Bissell MJ. The extracellular matrix in epithelial biology: shared molecules and common themes in distant phyla. Dev Biol. 1996;180:433–44. doi: 10.1006/dbio.1996.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Teller IC, Beaulieu JF. Interactions between laminin and epithelial cells in intestinal health and disease. Expert Rev Mol Med. 2001;2001:1–18. doi: 10.1017/S1462399401003623. [DOI] [PubMed] [Google Scholar]
- 72.Ingber DE. Mechanical signaling and the cellular response to extracellular matrix in angiogenesis and cardiovascular physiology. Circ Res. 2002;91:877–87. doi: 10.1161/01.res.0000039537.73816.e5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1. List of genes differentially expressed between S1 cells grown in 2D and 3D lrECM culture. “Common” refers to the gene name in use, “Genbank” column contains unique gene identifiers, “Normalized” values refer to the mean value of the ratio of 3D/2D across four microarray experiments in which Lowess normalization was performed, and t-test p-value is across four independent array experiments.
Supplemental Figure A. Phase contrast images of T4-2 cells on tissue culture plastic (2D) or within lrECM (3D). Scale bars, 50 μm.
Supplemental Figure B. Bar graph of mean projected cell area for T4-2 cells cultured on unpatterned or patterned substrata.
Supplemental Figure C. Table showing flow cytometric analysis of cell cycle distribution of S1 cells in 2D or 3D lrECM.


