Abstract
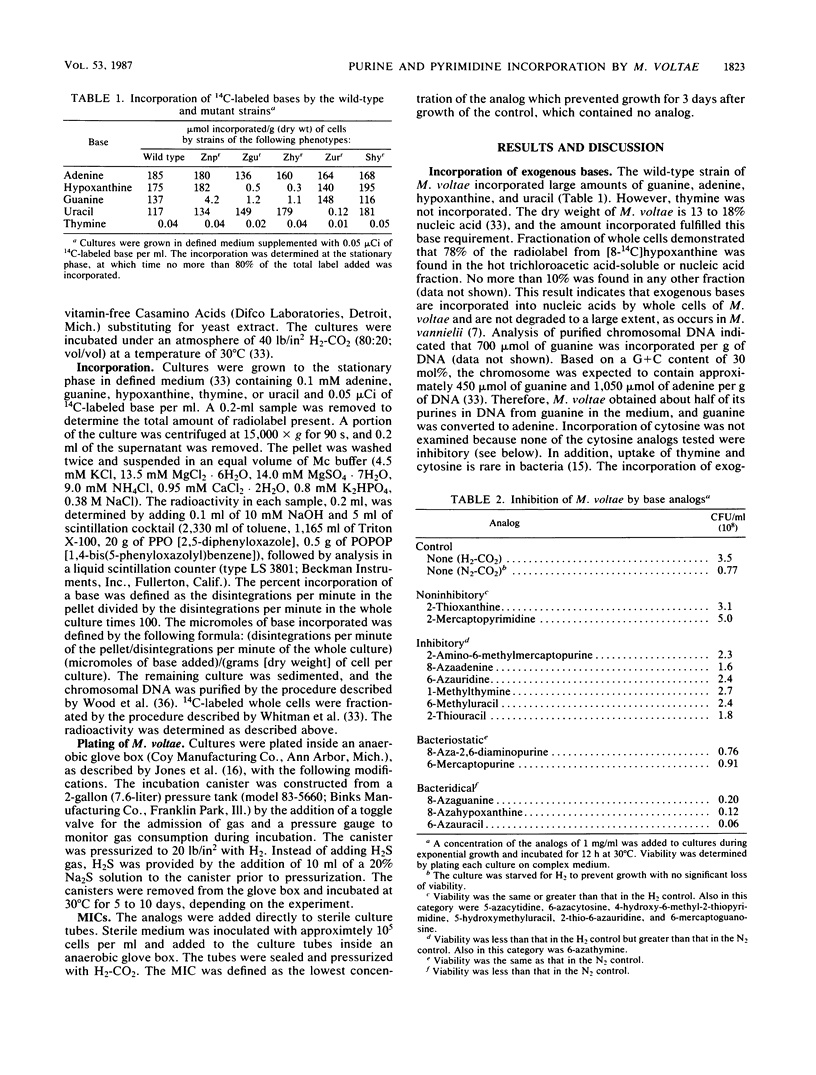
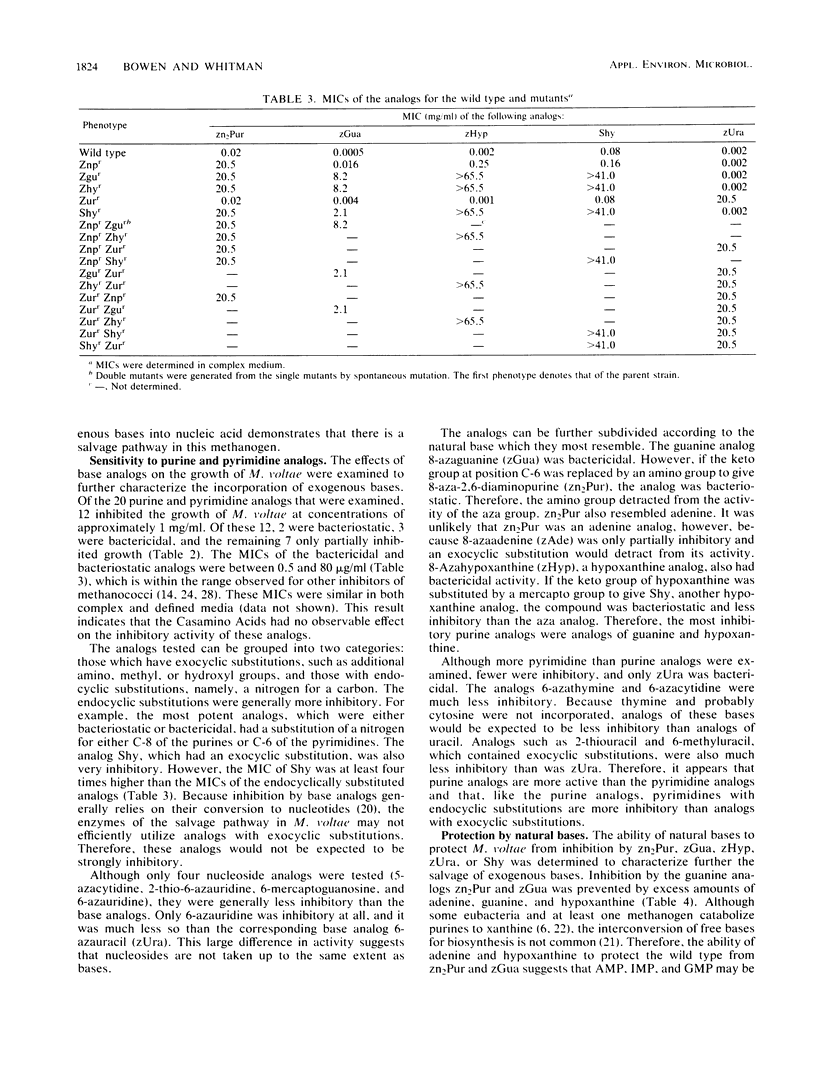
Methanococcus voltae incorporated exogenous adenine, guanine, hypoxanthine, and uracil, but not thymine. Growth of M. voltae was also sensitive to purine and pyrimidine analogs. Of the 20 analogs tested, 12 were inhibitory at 1 mg/ml. The most effective inhibitors were purine analogs with endocyclic substitutions. Nucleoside analogs and analogs with exocyclic substitutions or additions were less effective. Four purine analogs, 8-aza-2,6-diaminopurine, 8-azaguanine, 8-azahypoxanthine, and 6-mercaptopurine and one pyrimidine analog, 6-azauracil, were especially toxic. The MICs were 20, 0.5, 2.0, 80, and 10 μg/ml, respectively. Spontaneous resistance mutants were isolated for these five analogs. The MICs for these mutants were 20.5, 8.2, >65, >41, and 20.5 mg/ml, respectively. These concentrations far exceeded the solubilities of the analogs and represented an increase in resistance of at least three orders of magnitude. In addition to demonstrating cross resistance to several of the analogs, four of these mutants lost the ability to incorporate exogenous bases. These appeared to be mutations in the salvage pathways for purines and pyrimidines. In contrast, the mutant resistant to 6-mercaptopurine was not defective in purine uptake. Instead, it degraded 6-mercaptopurine. In the presence or absence of high concentrations of the analogs, the growth rates of the resistant mutants were no less than one-half of the growth rate of the wild type in the absence of the analog. The high level of resistance and rapid growth are very desirable properties for the application of the mutants in genetic experiments.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brown J. W., Reeve J. N. Polyadenylated, noncapped RNA from the archaebacterium Methanococcus vannielii. J Bacteriol. 1985 Jun;162(3):909–917. doi: 10.1128/jb.162.3.909-917.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coggin J. H., Loosemore M., Martin W. R. Metabolism of 6-Mercaptopurine by Resistant Escherichia coli Cells. J Bacteriol. 1966 Aug;92(2):446–454. doi: 10.1128/jb.92.2.446-454.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cue D., Beckler G. S., Reeve J. N., Konisky J. Structure and sequence divergence of two archaebacterial genes. Proc Natl Acad Sci U S A. 1985 Jun;82(12):4207–4211. doi: 10.1073/pnas.82.12.4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMoll E., Tsai L. Conversion of purines to xanthine by Methanococcus vannielii. Arch Biochem Biophys. 1986 Nov 1;250(2):440–445. doi: 10.1016/0003-9861(86)90747-2. [DOI] [PubMed] [Google Scholar]
- DeMoll E., Tsai L. Utilization of purines or pyrimidines as the sole nitrogen source by Methanococcus vannielii. J Bacteriol. 1986 Aug;167(2):681–684. doi: 10.1128/jb.167.2.681-684.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HANDSCHUMACHER R. E. Orotidylic acid decarboxylase: inhibition studies with azauridine 5'-phosphate. J Biol Chem. 1960 Oct;235:2917–2919. [PubMed] [Google Scholar]
- HANDSCHUMACHER R. E. Studies of bacterial resistance to 6-azauracil and its riboside. Biochim Biophys Acta. 1957 Feb;23(2):428–430. doi: 10.1016/0006-3002(57)90348-7. [DOI] [PubMed] [Google Scholar]
- Hamilton P. T., Reeve J. N. Sequence divergence of an archaebacterial gene cloned from a mesophilic and a thermophilic methanogen. J Mol Evol. 1985;22(4):351–360. doi: 10.1007/BF02115691. [DOI] [PubMed] [Google Scholar]
- Hamilton P. T., Reeve J. N. Structure of genes and an insertion element in the methane producing archaebacterium Methanobrevibacter smithii. Mol Gen Genet. 1985;200(1):47–59. doi: 10.1007/BF00383311. [DOI] [PubMed] [Google Scholar]
- Hochstadt J. The role of the membrane in the utilization of nucleic acid precursors. CRC Crit Rev Biochem. 1974 Mar;2(2):259–310. doi: 10.3109/10409237409105449. [DOI] [PubMed] [Google Scholar]
- Jones W. J., Whitman W. B., Fields R. D., Wolfe R. S. Growth and plating efficiency of methanococci on agar media. Appl Environ Microbiol. 1983 Jul;46(1):220–226. doi: 10.1128/aem.46.1.220-226.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KALLE G. P., GOTS J. S. Alterations in purine nucleotide pyrophosphorylases and resistance to purine analogues. Biochim Biophys Acta. 1961 Oct 14;53:166–173. doi: 10.1016/0006-3002(61)90803-4. [DOI] [PubMed] [Google Scholar]
- MOAT A. G., FRIEDMAN H. The biosynthesis and interconversion of purines and their derivatives. Bacteriol Rev. 1960 Sep;24(3):309–339. doi: 10.1128/br.24.3.309-339.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeve J. N., Trun N. J., Hamilton P. T. Beginning genetics with methanogens. Basic Life Sci. 1982;19:233–244. doi: 10.1007/978-1-4684-4142-0_19. [DOI] [PubMed] [Google Scholar]
- Santoro N., Konisky J. Characterization of bromoethanesulfonate-resistant mutants of Methanococcus voltae: evidence of a coenzyme M transport system. J Bacteriol. 1987 Feb;169(2):660–665. doi: 10.1128/jb.169.2.660-665.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuber A. P., Orr E. C., Recny M. A., Schendel P. F., May H. D., Schauer N. L., Ferry J. G. Cloning, expression, and nucleotide sequence of the formate dehydrogenase genes from Methanobacterium formicicum. J Biol Chem. 1986 Oct 5;261(28):12942–12947. [PubMed] [Google Scholar]
- Whitman W. B., Ankwanda E., Wolfe R. S. Nutrition and carbon metabolism of Methanococcus voltae. J Bacteriol. 1982 Mar;149(3):852–863. doi: 10.1128/jb.149.3.852-863.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Fox G. E. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci U S A. 1977 Nov;74(11):5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood A. G., Redborg A. H., Cue D. R., Whitman W. B., Konisky J. Complementation of argG and hisA mutations of Escherichia coli by DNA cloned from the archaebacterium Methanococcus voltae. J Bacteriol. 1983 Oct;156(1):19–29. doi: 10.1128/jb.156.1.19-29.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood A. G., Whitman W. B., Konisky J. A newly-isolated marine methanogen harbors a small cryptic plasmid. Arch Microbiol. 1985 Aug;142(3):259–261. doi: 10.1007/BF00693400. [DOI] [PubMed] [Google Scholar]


