Abstract
A major pathway of eukaryotic mRNA turnover begins with deadenylation, followed by decapping and 5′→3′ exonucleolytic degradation. A critical step in this pathway is decapping, which is carried out by an enzyme composed of Dcp1p and Dcp2p. The crystal structure of Dcp1p shows that it markedly resembles the EVH1 family of protein domains. Comparison of the proline-rich sequence (PRS)-binding sites in this family of proteins with Dcp1p indicates that it belongs to a novel class of EVH1 domains. Mapping of the sequence conservation on the molecular surface of Dcp1p reveals two prominent sites. One of these is required for the function of the Dcp1p-Dcp2p complex, and the other, corresponding to the PRS-binding site of EVH1 domains, is probably a binding site for decapping regulatory proteins. Moreover, a conserved hydrophobic patch is shown to be critical for decapping.
The process of mRNA turnover is important in numerous aspects of eukaryotic mRNA physiology. These roles include the control of gene expression, antiviral defenses1,2 and mRNA surveillance by recognizing and degrading the aberrant mRNAs3. Two major pathways of mRNA decay exist in eukaryotic cells4. In both yeast and mammals, mRNA degradation usually begins with the shortening of the poly(A) tail at the 3′ end of the mRNA. After deadenylation, the 5′ cap structure can be removed (decapping), thereby exposing the transcript to digestion by a 5′→3′ exonuclease Xrn1p5-8. Alternatively, after deadenylation in both yeast and mammals, mRNAs can be degraded in a 3′→5′ direction by the cytoplasmic exosome9-12. The resulting cap structure is hydrolyzed by the DcpS scavenger decapping enzyme, which is encoded by the DCS1 gene in yeast13.
Decapping is a key step in the 5′→3′ decay pathway because it induces degradation of the mRNA, and thus it is subject to numerous control inputs. Decapping is also critical in an aspect of mRNA surveillance in which aberrant mRNAs containing nonsense codons are decapped without a requirement for deadenylation and are degraded in a 5′→3′ direction14-16. Several observations suggest that Dcp1p and Dcp2p function together in vivo in a decapping holoenzyme with Dcp2p as the catalytic subunit. First, both Dcp1p and Dcp2p are required for decapping in vivo in yeast17,18. Second, yeast and human Dcp1 and Dcp2 proteins physically interact with each other in vivo and in vitro18-21. Evidence that Dcp2p is the catalytic subunit comes from the recent demonstration that recombinant yeast and human Dcp2 proteins have decapping activity20-23. Dcp2p contains a Nudix motif, which is found in a class of pyrophosphatases18,23-25, and mutations in the Dcp2p Nudix motif inactivate decapping activity in vivo and in vitro in both the yeast and mammalian enzymes18,20-23. Notably, recombinant human Dcp2 protein is a more robust enzyme than the yeast protein, which requires either manganese or high levels of magnesium to function21-23. Although recombinant Dcp1p has been reported to have decapping activity26, several groups have not detected such activity under a variety of conditions20-22. However, recombinant Dcp1p can clearly enhance the activity of yeast Dcp2p, allowing Dcp2p to decap efficiently under conditions in which it has little activity on its own21. The simplest interpretation of these observations is that Dcp2p is the catalytic subunit of the decapping complex, and Dcp1p primarily functions to enhance Dcp2p activity, although how Dcp1p does this is unknown. The Dcp1 protein is conserved in eukaryotes, and two human homologs, hDcp1a and hDcp1b, have been identified, with hDcp1a copurified with decapping activity20. Moreover, hDcp1a (also called SMIF) may have a role in SMAD-mediated TGFβ signaling because it interacts with Smad4 and partially translocates to the nucleus in response to transformation growth factor-β stimulation27.
To gain insight into the molecular mechanism of decapping carried out by Dcp1p and Dcp2p, we determined the crystal structure of yeast Dcp1p at a resolution of 2.3 Å. The overall fold of Dcp1p is markedly similar to that of EVH1/WH1 family of domains. Comparison of the PRS-binding sites in this family of proteins with Dcp1p indicates that Dcp1p belongs to a novel class of EVH1 domains. Mapping of the sequence conservation on the molecular surface reveals two prominent sites, one required for decapping activity and the other a possible regulatory site. Furthermore, we show that a conserved hydrophobic patch on the Dcp1p surface is critical for decapping.
RESULTS
Overall structure of Dcp1p
The crystal structure of Dcp1p was solved by single wavelength anomalous dispersion (SAD) using selenomethionine (SeMet)-substituted crystals. The final model contains two copies of the Dcp1p, designated as molecules A and B, respectively. Several regions of the polypeptide are not visible in the electron density map and are assumed to be disordered, namely residues 1-21, 74-134 and 229-231 for molecule A, and residues 1-16 and 81-134 for molecule B in the crystallographic asymmetric unit. The current refined model has an R-factor of 22.7% and a Rfree value of 26.8% at a resolution of 2.3 Å with very good stereochemistry (see Table 1 and Methods). m7GDP could not be located in the electron density map or in washed crystals, suggesting that m7GDP probably does not bind in the crystal lattice (data not shown).
Table 1.
X-ray data collection, phasing and refinement statistics
| Data collection | |
| Wavelength (Å) | 0.9798 Å |
| Resolution (Å) | 50-2.3 Å |
| Reflections | |
| Measured | 286,716 |
| Unique | 34,251 |
| Completeness (%) | 97.9 |
| I/σ (I)a | 8.3 (1.9) |
| Rmerge (%)a,b | 6.6 (37.1) |
| Phasing statistics | |
| Number of Se sites | 4 |
| Rcullisc | 0.70 |
| Figure of merit | |
| Before density modification | 0.29 |
| After density modification | 0.86 |
| Refinement | |
| Resolution range (Å) | 20.0-2.3 |
| Number of atoms | |
| Protein | 2,571 |
| Water | 156 |
| Rcryst (%)d | 22.7 |
| Rfree (%)e | 26.8 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.006 |
| Bond angles (°) | 1.3 |
Values in parentheses are for the highest-resolution shell.
, where Ij is the intensity of an individual reflection, and is the average intensity of that reflection.
.
, where Fo denotes the observed structure factor amplitude, and Fc the structure factor amplitude calculated from the model.
Rfree is as for Rcryst but calculated with 10% of randomly chosen reflections omitted from the refinement.
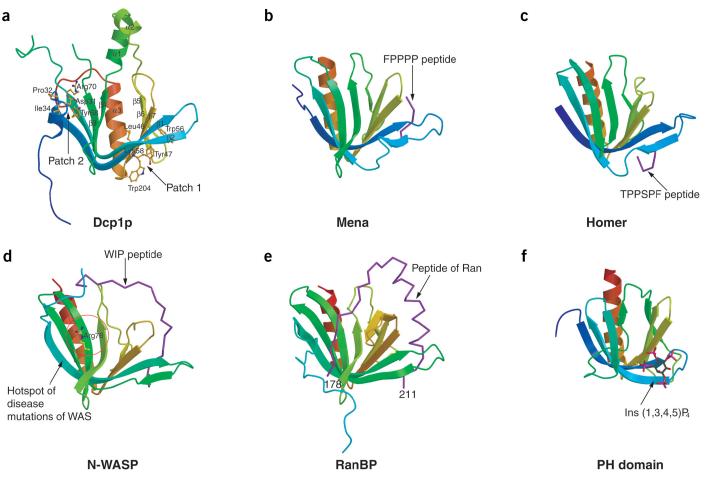
No substantial differences are observed between the structures of the two Dcp1p molecules in the asymmetric unit (r.m.s. deviation of 0.60 Å for all the equivalent Cα atoms). Because molecule B is more complete than molecule A, all subsequent analyses reported here refer to the coordinates of the molecule B. Dcp1p is composed of three α-helices and seven β-strands. A notable feature of the tertiary structure is the formation of a β-sandwich by two antiparallel β-sheets (Fig. 1a). A V-shaped concave groove is formed by the outside of β-strands 1, 2, 5, 6 and 7; another concave groove, which is opposite to the V-shaped groove, is formed by the N-terminal 310-helix and β-strands 1-4. A long helix (α3) is located on one side of the β-sandwich with two short helices (α1 and α2) running across the top of the β-sandwich.
Figure 1.
Crystal structure of Dcp1p in comparison with EVH1 domains from Mena, Homer, N-WASP and RanBP. (a) Ribbon diagram of Dcp1p with secondary structural elements labeled. Two clusters of conserved residues on opposite concave surfaces (patches 1 and 2) are shown as ball-and-stick models colored by atom type. (b) Structure of the Mena EVH1 domain in complex with an eight-residue peptide (purple) from ActA. (c) Structure of the Homer EVH1 domain in complex with a five-residue peptide (purple) from mGluR. (d) NMR structure of the N-WASP EVH1 domain in complex with WIP peptide (residues 461-485, purple). Residues 479-485 are not included in the drawing owing to a lack of resonance assignments. Arg76, the most frequently mutated residue in Wiskott-Aldrich syndrome (WAS) is shown in ball-and-stick form. (e) Structure of the RanBP EVH1 domain in complex with an extended >25-residue polypeptide from Ran (purple). (f) Structure of PH domain from DAPP1/PHISH in complex with Ins(1,3,4,5)P4. The bound inositol phosphate is shown in ball-and-stick form. Figures 1 and 5 were produced using MolScript48.
Dcp1p structurally resembles EVH1/WH1 domains
A search of the Protein Data Bank using the Dali server28 indicated that Dcp1p belongs to a structural family of EVH1/WH1 domains including Mena29 (PDB entry 1EVH; Z-score 12.9), and Ranbinding protein30 (RanBP; PDB entry 1RRP; Z-score 11.4). Dcp1p markedly resembles the structures of the EVH1/WH1 domains (Fig. 1). Superposition of the equivalent Cα atoms of Dcp1p and other EVH1 domains gives rise to r.m.s. deviation values of 1.70 Å, 1.6 Å, 1.7 Å and 1.8 Å for Mena, Homer31 (PDB entry 1DDV), N-WASP32 (PDB entry 1MKE) and RanBP, respectively. The core regions for these structures are very similar. The best matches are found in the central β-sandwich, with the largest deviations in the C-terminal α-helix (α3). Consistent with this analysis, human Dcp1a/SMIF, which shares ∼17% identity with yeast Dcp1p in the N terminus (Fig. 2), has been predicted to have an EVH1/WH1 domain at its N terminus33. EVH1/WH1 domains are distinct members of the PH domain superfamily, including PH, PTB and EVH1 domains29. Consistent with these observations, a Dali28 search revealed that Dcp1p is structurally homologous to PH and PTB domains as well, with Z-scores of 9.8 and 9.5, respectively. Superposition of Dcp1p and PH domain results in an r.m.s. deviation of 1.3 Å for 59 equivalent Cα atoms (Fig. 1f).
Figure 2.
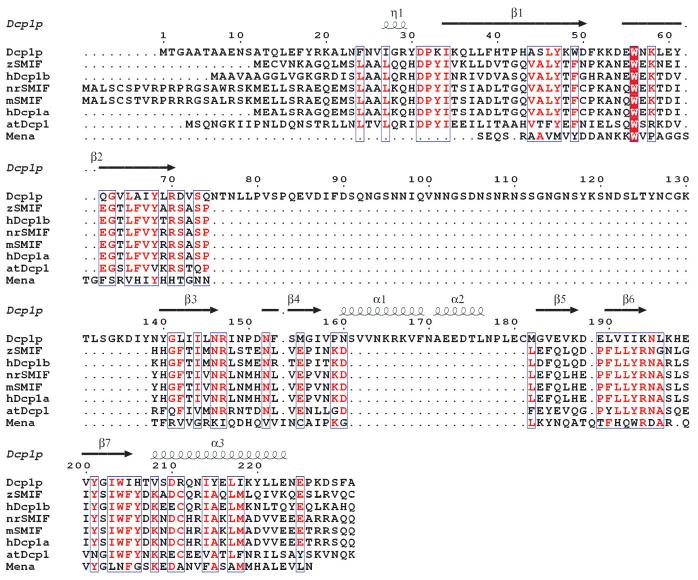
Structure-based sequence alignments of Dcp1 proteins with the Mena EVH1 domain. Sequences of Dcp1p from Saccharomyces cerevisiae and the N-terminal domains (residues 1-140) of hDcp1a and hDcp1b from Homo sapiens, nrSMIF from Rattus norvegicus, mSMIF from Mus musculus, zSMIF from Danio rerio and atDcp1 from Arabidopsis thaliana were aligned with ClustalW49. The alignment of the Mena EVH1 domain sequence is based on its structural similarity with Dcp1p. The secondary structural elements were assigned from the X-ray structure of Dcp1p. Invariant residues are red.
Despite the similarities, some structural differences exist between the Dcp1p structure and these related structures. The most notable differences are the two short helices, α1 and α2, which have no counterparts in the EVH1 domains of Mena, Homer, N-WASP and RanBP. Sequence comparison of Dcp1p with other eukaryotic Dcp1 proteins indicates that this region is a unique feature of yeast Dcp1p (Fig. 2). The N-terminal extended peptide of Dcp1p is reminiscent of the N-terminal extension of RanBP, but such an N-terminal extension is nonexistent in other EVH1 domains (Fig. 1). This N-terminal extension seems to be functionally important, because mutations in this region can affect decapping in vivo34. All known EVH1 domains consist of contiguous stretches of amino acids found at the N terminus35. In contrast, the EVH1 fold in yeast Dcp1p has an insert (residues 80-135) that is largely disordered in the structure. Because this insert is only present in the yeast Dcp1p (Fig. 2), it is not likely to be central to Dcp1p function and is probably involved in yeast-specific functions.
Two patches as putative ligand-binding sites
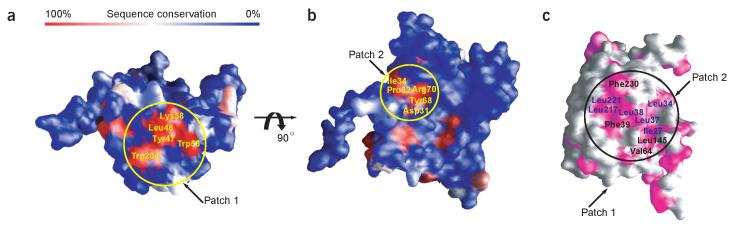
Previous analyses of Dcp1p have indicated that this protein interacts with the mRNA substrate and several yeast proteins involved in decapping including Dcp2p, Dhh1p, Edc1p, Edc2p and Edc3p17-19,36-38, although only in the case of Dcp2p is there clear evidence that the interaction is direct. To identify key domains on the surface of Dcp1p, we mapped the degree of amino acid conservation shared between Dcp1p and other eukaryotic Dcp1 sequences on the Dcp1p molecular surface. This analysis revealed two prominent regions of structural conservation, which we discuss below (Fig. 3a,b).
Figure 3.
Molecular surface views of Dcp1p. (a) Surface representation of Dcp1p showing the regions of high to low sequence conservation shared by the eukaryotic Dcp1 proteins, corresponding to a color ramp from red to blue, respectively. The conserved patch 1, corresponding to the PRS-binding site in other EVH1 domains, is shown. Invariant residues are labeled. (b) The conserved patch 2, which is critical for Dcp1p function. Invariant residues are labeled. The coloring scheme is as in a. The view is similar to that in Figure 1a. (c) Molecular surfaces of Dcp1p colored according to residue property, with hydrophobic residues magenta and other residues gray. The hydrophobic patch is marked by a black circle and residues forming the hydrophobic patch on Dcp1p surface are labeled, with conserved residues blue and variant residues black. Figures were produced using GRASP50.
One of the regions (designated as conserved patch 1) is situated in a concave V-shaped groove formed by β-strands 1, 2, 5, 6 and 7 (Figs. 1a and 3a). This region corresponds structurally to the PRS-binding sites on other EVH1 domains (Fig. 1a-e). This region of EVH1 domains forms a hydrophobic platform that interacts with proteins containing PRSs in actin cytoskeleton and postsynaptic signaling pathways33. Thus, the conserved patch 1 of Dcp1p may be a PRS-binding site. Consistent with this conclusion, the N-terminal domain of hDcp1a/SMIF has been found to interact specifically with the proline-rich region (residues 275-308) of SMAD4 so that hDcp1a/SMIF can function as a coactivator in TGFβ signaling27. Also consistent with a functional role for conserved patch 1, alanine-scanning mutagenesis has shown that three mutations in this patch, W56A, Y47A and W204A, cause a partial loss-of-function phenotype in vivo34.
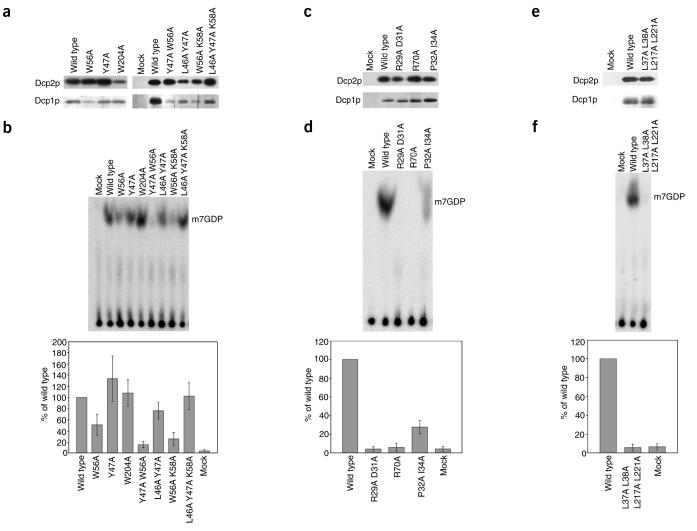
To probe the function of this region biochemically, we constructed a series of mutations in this region and assayed Dcp1p function using a system where Dcp1p is coexpressed in Escherichia coli with the N-terminal portion of Dcp2p (residues 1-300)21. This system allowed us to assess simultaneously whether Dcp2p copurified with the mutant Dcp1 proteins and whether the mutant Dcp1 proteins enhanced the activity of the copurified Dcp2p. The mutations we tested include single and multiple amino acid changes of the conserved residues in this region to alanine. None of these mutations substantially reduced the amount of Dcp2p that copurified with Dcp1p relative to wild-type Dcp1p (Fig. 4a). The finding that Dcp2p copurified with the mutants indicates that patch 1 is not absolutely required for Dcp1p-Dcp2p binding.
Figure 4.
Biochemical analysis of mutations in conserved surface patches of Dcp1p. (a) Analysis of Patch 1 mutants. Wild-type and Patch 1 mutant alleles of Flag-tagged Dcp1p were coexpressed with His-tagged N-terminal 30-kDa fragment of Dcp2p and purified by Flag affinity chromatography. Patch 1 mutant alleles analyzed were W56A, Y47A, W204, Y47A W56A, L46A Y47A, W56A K58A, and L46A Y47A K58A. Top, western blot analysis of Dcp2p that copurified with Dcp1p using anti-His monoclonal antibody. Bottom, silver stain analysis of Dcp1p. Mock represents a sample purified under the same conditions from E. coli containing the empty expression vector. (b) Analysis of decapping activity of Dcp2p copurified with wild type and Patch 1 mutants of Dcp1p. Migration of decapping product, m7GDP, is indicated on right. Bottom, percentage of decapping activity of each mutant relative to decapping activity of wild-type Dcp1p control. The values plotted are the mean and the standard deviation of at least four independent experiments. (c) Analysis of Patch 2 mutants. Wild-type and Patch 2 mutant alleles of Flag-tagged Dcp1p were analyzed as described in a. Patch 2 mutant alleles analyzed were R29A D31A, R70A, and P32A 134A. (d) Analysis of decapping activity of Dcp2p copurified with wild-type and Patch 2 mutants of Dcp1p as described in b. The values plotted are the mean and the standard deviation of at least six independent experiments. (e) Analysis of hydrophobic patch mutant. Wild type and the hydrophobic patch mutant allele of Flag-tagged Dcp1p were analyzed as described in a. The hydrophobic patch mutant allele analyzed was L37A L38A L217A L221A. (f) Analysis of decapping activity of Dcp2p copurified with wild type and the hydrophobic patch mutant of Dcp1p as described in b. The values plotted are the mean and the standard deviation of at least two independent experiments.
To determine whether these mutations caused a biochemical defect in the Dcp1p-Dcp2p complex, we assayed the decapping activity of Dcp2p copurified with the mutant proteins in vitro in the absence of manganese, a condition under which yeast Dcp2p is dependent on Dcp1p for measurable activity21 (C.J.D. and R.P., unpublished data). In general, most of the mutations in patch 1, including a triple mutant (L46A Y47A K58A), did not substantially affect the decapping activity of the Dcp1p-Dcp2p complex (Fig. 4b). Changing Trp56 to alanine, alone or in combination with the Y47A or K58A mutations, did however reduce decapping activity (Fig. 4b). The reduction in decapping in these mutants is most likely due to the W56A mutation, because Y47A or K58A mutation alone or in combination with mutations other than W56A did not reduce decapping. The effect of the W56A mutation on decapping suggests that the conserved patch 1 affects some intrinsic function of Dcp1p. Alternatively, because Trp56 is absolutely conserved in all EVH1 folds (Fig. 2) and mutation of the corresponding residue (Trp23) in the Mena EVH1 protein alters its structure39, Trp56 may have some structural role in Dcp1p. Given that most of the mutations in patch 1 did not affect decapping, one possible function of patch 1 is to enhance some biochemical properties of the enzyme not assessed in our assays. Alternatively, the lack of a strong biochemical phenotype for such marked changes in this conserved surface of the protein suggests that this surface may have a role in contacting regulatory proteins not present in our purified system.
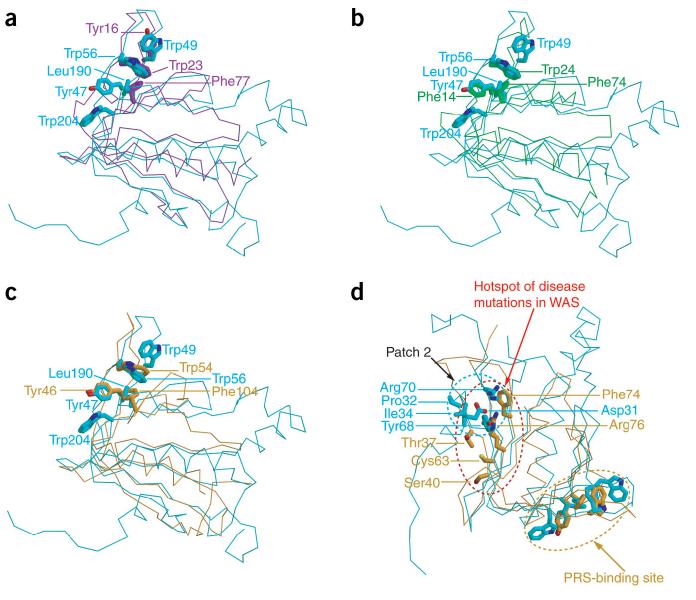
Conserved patch 2 is located on a concave surface directly opposite the putative PRS-binding site formed by the N-terminal 310-helix and β-strands 1-4 (Figs. 1a and 3b). Structurally, this region is analogous to the hotspot of disease mutations found in the N-WASP EVH1 domain32 and to the region where residues 178-181 of Ran interact with the RanBP EVH1 domain30 (Fig. 1d,e). In Dcp1p, a highly conserved cluster of two charged residues, Arg70 and Asp31, and three hydrophobic residues, Tyr68, Pro32 and Ile34, is located in this region (Figs. 1a and 3b). In support of this region being functionally important for Dcp1p, mutation of Arg70 or Asp31 to alanine inhibits decapping in vivo and leads to an inhibition of the decapping activity purified from yeast cells34.
To study the function of patch 2 in more detail, we constructed a series of mutations in this region and purified them from E. coli coexpressing Dcp2p21. We tested two alleles with mutations in the charged residues in this region, R29A D31A double mutant and R70A, as well as an allele in which two of the conserved uncharged residues in this region were mutated (P32A I34A). All the mutant Dcp1 proteins copurified with approximately equal amounts of Dcp2p relative to wild-type Dcp1p (Fig. 4c). This suggests that this region is not required for Dcp1p and Dcp2p to interact, although it could affect the strength of the Dcp1p-Dcp2p interaction.
The mutations of the charged residues in this region caused strong defects in the decapping activity of purified Dcp1p-Dcp2p (Fig. 4d). Similarly, changing the two hydrophobic residues in this patch led to a partial defect in decapping activity. These results indicate that patch 2 is required for the function of the decapping enzyme Dcp1p-Dcp2p. In addition, because Dcp2p still copurified with these point mutants, these results indicate that at least one function of this region is independent of its role as a binding site for Dcp2p. In this regard, possible functions for patch 2 would include directly binding substrate, assisting in Mg2+ coordination, or promoting an activating conformational change in Dcp2p.
The defects in decapping caused by mutations in patches 1 and 2 observed in the coexpression system were examined by reconstituting the decapping enzyme with Dcp1p (either wild-type or mutant proteins) and Dcp2p that had been expressed and purified separately. The results are consistent with the coexpression data (see Supplementary Fig. 1 online).
A hydrophobic patch is critical for decapping
Mapping of the side chains of hydrophobic residues on the molecular surface of Dcp1p reveals that a large hydrophobic patch is located close to patches 1 and 2 (Fig. 3c). This hydrophobic patch is predicted to be conserved between yeast and human Dcp1 proteins, as the structure models of human Dcp1a and Dcp1b have similar surface properties (data not shown). The distance from the hydrophobic patch to patch 1 is about three to four residues, consistent with the finding that the two aromatic residues (Tyr301 and Trp302) from the PRS-containing peptide (residues 275-308) of SMAD4 that are essential for hDcp1a binding are four residues away from the PRS27. Notably, in the Dcp1p crystal lattice, Val168 and Phe169 from symmetry-related molecules bind to this region through hydrophobic interactions, suggesting that this region indeed is a hydrophobic groove and is possibly involved in binding Dcp2p or other factors.
To examine the role of the hydrophobic patch in Dcp2p binding and decapping, we constructed a quadruple mutant in which Leu37, Leu38, Leu217 and Leu221 were all mutated to alanine. The quadruple mutant was coexpressed with Dcp2p in E. coli and purified. Approximately equal amounts of Dcp2p copurified with the mutant protein as with the wild-type Dcp1 protein (Fig. 4e). This result suggests that this hydrophobic surface is not absolutely required for Dcp1p to interact with Dcp2p. However, it is still possible that the hydrophobic surface affects the strength of the interaction. Although the L37A L38A L217A L221A mutation did not substantially affect Dcp2p binding, it did cause a strong defect in decapping (Fig. 4f) indicating that this surface is required for Dcp1p to enhance decapping. Given the closeness of the hydrophobic patch to patch 2 and the fact that both patches are required for Dcp1p function, these two features of the protein may function together to promote decapping by Dcp2p.
DISCUSSION
The crystal structure of yeast Dcp1p presented here resembles the structures of EVH1 domains. EVH1 domains can be divided into three general classes based on their target PRS ligands35. All previously characterized EVH1 domains use a highly conserved cluster of three surface-exposed aromatic residues (Tyr16, Trp23 and Phe77; Mena numbering) to recognize their target PRS ligands with remarkably low affinity but high specificity40.
Sequence comparison of all the Dcp1 proteins revealed that four out of five conserved aromatic residues, namely Tyr47, Trp49, Trp56 and Trp204, are clustered in the putative PRS-binding site, with the fifth, Tyr68, located in the conserved patch 2 (Figs. 2 and 3a,b). Comparison of the putative PRS-binding site of Dcp1p with those of other EVH1 domains revealed some similarities and marked differences as well (Fig. 5a-c). In Dcp1p, the potential PRS-binding site is lined with conserved aromatic residues including Trp49 (which is a phenylalanine in all other eukaryotic Dcp1 proteins), Trp56, Leu190 (which is a phenylalanine in all other eukaryotic Dcp1 proteins), Tyr47 and Trp204. Specifically, beginning at the upper right of the pocket, Trp49 in Dcp1p is analogous to Tyr16 in Mena, but Wasp and Homer lack an aromatic residue at this position. The next aromatic residue, Trp56, is structurally invariant in all EVH1 domains, corresponding to Trp23 in Mena, Trp24 in Homer and Trp54 in Wasp. Next comes a pair of hydrophobic residues in Dcp1p, Leu190 and Tyr47. Tyr47 matches well with Tyr46 of the EVH1 domain of N-WASP and Phe14 of the Homer EVH1 domain, but this aromatic residue is not present in Mena. Leu190 in Dcp1p corresponds to Phe77 in Mena, Phe104 in N-WASP, and Phe74 in Homer. Notably, this position does show additional variability, as it corresponds to Ser109 in RanBP, although the ligand of this site is not proline-rich. A unique feature of the Dcp1p pocket is Trp204, an invariant residue in all the Dcp1 proteins, that extends the hydrophobic surface, but has no aromatic counterpart in other classes of EVH1 domains. The unique structural arrangement of these key aromatic residues (Tyr47, Trp49, Trp56 and Trp204) in Dcp1p suggests that Dcp1p may have a specificity distinct from that of other EVH1 domains toward the PRS-containing ligands. Based on these comparisons, we suggest that the binding pocket for PRS ligands of EVH1 domains consists of the surface of hydrophobic and aromatic residues and can be substantially extended, as in Dcp1p (five conserved positions), or quite short, as in Mena or Homer, which both have a different set of three of these conserved positions.
Figure 5.
Comparison of the potential ligand-binding sites of Dcp1p with those of EVH1 domains from Mena, Homer and N-WASP. (a) Comparison of the PRS-binding site in Dcp1p and Mena EVH1 domain. The backbone of Cα traces are cyan and purple for Dcp1p and Mena, respectively. Trp23, Tyr16 and Phe77 from Mena and Tyr47, Trp56, Trp204, Trp49 and Leu190 from Dcp1p are shown as sticks. (b) Comparison of the PRS-binding site in Dcp1p and Homer EVH1 domain. The backbone of Cα traces are cyan and green for Dcp1p and Homer, respectively. Trp24, Phe14 and Phe74 from Homer and Tyr47, Trp56, Trp204, Trp49 and Leu190 from Dcp1p are shown as sticks. (c) Comparison of the PRS-binding site in Dcp1p and N-WASP EVH1 domain. The backbone of Cα traces are cyan and orange for Dcp1p and N-WASP, respectively. Trp54, Tyr46 and Phe104 from N-WASP and Tyr47, Trp56, Trp204, Trp49 and Leu190 from Dcp1p are shown as sticks. (d) Comparison of conserved patch 2 of Dcp1p with the hotspot of disease mutations in N-WASP, revealing the overlap of these two regions. The backbone of Cα traces are cyan and orange for Dcp1p and N-WASP, respectively. The most frequently mutated residues, Arg76, Thr37, Cys63, Ser40 and Phe74 from N-WASP, and the conserved residues Pro32, IIe34, Asp31, Arg70 and Tyr68 of Dcp1p are shown as sticks. The view is as in Figure 1a.
Notably, the conserved patch 2 on Dcp1p corresponds to the hotspot of disease mutations in the EVH1 domain of N-WASP (Fig. 5d). The hotspot of disease mutations in N-WASP includes the most frequently mutated residue Arg76 and other surface residues that flank Arg76, including Leu31, Thr37, Ser40, Cys63 and Phe74 (ref. 32). This region has been speculated to be involved in WIP peptide binding to N-WASP based on the NMR structure, but exactly how WIP peptide interacts with the mutational hotspot centered around Arg76 is unknown32. Thus, the functional role of residues located at the hotspot of disease mutations of N-WASP remains elusive. The observation that the corresponding region of Dcp1p is critical for function in vivo and in vitro indicates that this surface is functionally important in multiple EVH1 domains34 (see above). Given the functional importance of this region in two members of the class III family of EVH1 domains, we suggest that the surface corresponding to patch 2 in Dcp1p is probably a critical region for the function of class III EVH1 domains. This region either could function to enhance the binding of ligands in the PRS site, as in WASP and RanBP1, or may interact with ligands distinct from those in the PRS-binding pocket and thereby allow EVH1 domains to link proteins together.
In conclusion, Dcp1p has two structurally conserved sites. One of these sites corresponds to the PRS-binding sites of other EVH1 proteins and may belong to a novel class of EVH1 domains. The second conserved site is required for decapping by the Dcp1p-Dcp2p complex. This site is structurally analogous to the hotspot of disease mutations in the EVH1 domain of N-WASP. This similarity suggests that certain classes of EVH1 domains may bear a second functionally key region in addition to their PRS-binding sites. Our work provides a starting point for further crystallographic, biochemical and genetic studies of Dcp1 proteins to elucidate their role in mRNA decapping as well as understanding the role of this second functional region of EVH1 domains.
METHODS
Cloning, expression and purification
The full-length Dcp1 gene was cloned from S. cerevisiae genomic DNA by PCR amplification using oligonucleotides Dcp1-F (GCGCGGATCCATGACCGGAGCAGCAACTGC) and Dcp1-R (GCGCCTCGAGTTAAGCAAAAGAATCTTTTGGCTC). The DNA fragment was digested with restriction enzymes BamHI and XhoI and ligated into the same site of pGEX-6p-1 (Pharmacia Biotech) to generate pGEX-dcp1. The construct was verified by DNA sequencing. E. coli strain BL21(DE3) star (Novagen) harboring the pGEX-dcp1 was grown in 4 L of LB medium at 37 °C and induced with 0.1 mM IPTG at 30 °C for 3 h upon reaching log phase. The cells were harvested by centrifugation, resuspended in lysis buffer (20 mM Tris-HCl, pH 7.6, 500 mM NaCl, 0.1 mM PMSF, 2 mM benzamidine, 2 mM DTT), and disrupted by sonication. Clarified cell lysate was applied to a glutathione Sepharose 4B column (Amersham). The eluted fractions were subjected to PreScission protease (Amersham) digestion at 4 °C overnight to cleave the GST tag. The desalted fractions were further purified by a second glutathione Sepharose 4B column and gel filtration (HiLoad 20/60 Superdex75, Amersham) in the ÄKTA FPLC system (Amersham). Monomeric Dcp1 protein was purified to homogeneity and concentrated to 12 mg ml−1 in storage buffer (20 mM Tris-HCl, pH 7.6, 100 mM NaCl, 2 mM MgCl2 and 2 mM DTT). Protein identity was confirmed by Edman degradation N-terminal sequencing and mass spectrometry molecular mass determination. SeMet-substituted Dcp1p was purified in the same way except the DTT concentration was 10 mM.
Crystallization and data collection
Dcp1 crystals were obtained by hanging-drop vapor diffusion by mixing 5 μl protein solution with 5 μl crystallization buffer (0.76 M (NH4)2SO4, 1.50 M NaCl, 5 mM DTT, 0.1 M Tris-HCl, pH 8.5, 6 mM EDTA, 2% (v/v) isopropanol). For cocrystallization with ligands, 0.5 mM m7GDP (Sigma) or m7GpppG (EPICENTER) was added to protein solution. Crystals appeared after 3 d at 20 °C. For data collection at 100 K, crystals were transferred to cryobuffer (25% (v/v) glycerol, 0.80 M (NH4)2SO4, 1.50 M NaCl, 5 mM DTT, 0.1 M Tris-HCl pH 8.5, 6 mM EDTA, 2% (v/v) isopropanol), and flash-frozen in liquid nitrogen. Data were collected on BL41XU at Spring-8 (Hyogo, Japan) using Mar CCD detector and processed with the HKL package41. A complete SAD dataset for Se-Dcp1p was collected on Se peak wavelength (λ = 0.9798 Å). Crystals of Dcp1p belong to space group P321 with cell parameters a = b = 123.56 Å, c = 77.90 Å, α = β = 90°, γ = 120°, with two molecules per asymmetric unit.
Structure determination and refinement
Four Se sites were found using SHELXD42. Refinement of the heavy-atom sites and phase estimation were done using SHARP43. The resulting phases were further improved by solvent flattening using SOLOMON44. Model building was done using O45. Crystallographic refinement was carried out with CNS46. σA-weighted 2Fo - Fc and Fo - Fc maps were used to guide rebuilding of the model using O through-out the refinement. Water molecules were automatically included with CNS and manually edited with electron densities. The final refinement statistics are summarized in Table 1.
Modeling of hDcp1a and hDcp1b
Dcp1p structure was used as a template to model the N-terminal domains of hDcp1a and hDcp1b with O45. The peptide gaps resulting from the two insertions in Dcp1p sequences were fixed by comparing the models with the Mena EVH1 structure. The resulting models were subjected to energy minimization using CNS46.
Biochemical analysis of mutant Dcp1 proteins
The mutations in Dcp1p were introduced into pRP1071, a plasmid designed to express both Flag-tagged full-length Dcp1p and a His6-tagged N-terminal 30-kDa fragment of Dcp2p in E. coli21. The R29A D31A, W56A Y47A W204, and R70A alleles were introduced by PCR-amplifying the mutant allele from the corresponding yeast expression plasmid containing the mutant DCP1 gene34 using oligonucleotides oRP1162 (GGAGATATACATATGGACTACAAGGACGACGATGACAAGATGACCGGAGCAGCAACTG) and oRP1163 (CGCGGATCCTCAAGCAAAAGAATCTTTTGGCT), which introduced the Flag-tag at the N terminus of Dcp1p and a BamHI site just 3′ of the stop codon. The resulting PCR products were amplified with oRP1161 (CGGTTTCCCTCTAGAAATAATTTTGTTTAACTTTAAGAAGGAGATATACATATGGACTACAAGG) and oRP1163 to introduce a ribosome-binding site and XbaI site 5′ of the Flag-tag. The resulting PCR products were cut with XbaI and BamHI and used to replace the wild-type Dcp1 gene in pRP1071. The additional mutant alleles were created using the QuikChange site-directed mutagenesis kit (Stratagene). Y47A W56A was generated using the W56A plasmid, oRP1164 (CACACACCACATGCGTCACTGGCCAAATGGGACTTCAAGAAGGATGAAGC) and oRP1165 (GCTTCATCCTTCTTGAAGTCCCATTTGGCCAGTGACGCATGTGGTGTGTG). L46AY47A was generated using the Y47A plasmid, oRP1166 (CACACACCACATGCGTCAGCGGCCAAATGGGACTTCAAG) and oRP1167 (CTTGAAGTCCCATTTGGCCGCTGACGCATGTGGTGTGTG). W56A K58A was generated using the W56A plasmid, oRP1168 (GGACTTCAAGAAGGATGAAGCTAATGCACTAGAATATCAAGGTGTTTTGG) and oRP1169 (CCAAAACACCTTGATATTCTAGTGCATTAGCTTCATCCTTCTTGAAGTCC). L46A Y47A K58A was generated using the L46A Y47A plasmid, oRP1174 (CACACACCAATGCGTCAGCGGCCAAATGGGACTTCAAGAAGGACGAATGGAATGC CCTAGAATATCAAGGTGTTTTGG) and oRP1175 (CCAAAACACCTTGATATTCTAGGGCATTCCATTCGTCCTTCTTGAAGTCCCATTTGGCCGCT GACGCATGTGGTGTGTG). P32A I34A was generated using pRP1071, oRP1178 (GAATTTCAACGTTATTGGGAGATACGATGCAAAAGCAAAGCAACTACTTTTTCACACACC) and oRP1179 (CGTGTGTGAAAAAGTAGTTGCTTTGCTTTTGCATCGTATCTCCCAATAACGTTGAAATTC). L37A L38A L217A L221A was generated in two steps. First, pRP1071 was used as the template with oRP1204 (CGTTATTGGGAGATACGATCCAAAAATAAAGCAAGCAGCTTTTCACACACCACATGCGTCACTG) and oRP1205 (CAGTGACGCATGTGGTGTGTGAAAAGCTGCTTGCTTTATTTTTGGATCGTATCTCCCAATAACG) to introduce L37A L38A. Then, the resulting mutant plasmid was used as the template with oRP1206 (CAGTCAGTGATAGACAAAATATCTACGAAGCAATAAAATATGCTCTGGAAAATGAGCCAAAAGATTC) and oRP1207 (GAATCTTTTGGCTCATTTTCCAGAGCATATTTTATTGCTTCGTAGATATTTTGTCTATCACTGACTG) to introduce L217A L221A. All mutant alleles were verified by sequencing.
The wild-type and mutant Flag-tagged Dcp1 and His6-tagged Dcp2 proteins were coexpressed in DH5α and purified as described21. Western blot analysis against Dcp2p was carried out using standard protocols with monoclonal anti-His (Novagen).
Cap-labeled 99 nucleotide MFA2 mRNA substrate was prepared as described47. Decapping reactions were done in 20-μl volumes containing 50 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 50 mM NH4Cl, 1 mM DTT, 10 units RNasin (Promega), 10 fmol substrate RNA and 0.12-1.2 μg total protein. After incubation at 30 °C for 15 min, a 2-μl aliquot was removed and reaction was stopped in this aliquot using 1 μl of 0.5 M EDTA on ice. Products were separated on a PEI cellulose thin layer chromatography plate (Selecto) in 0.75 M LiCl2 and detected with a phosphorimager (Molecular Dynamics).
Coordinates
The coordinates and structure factors for Dcp1p have been deposited in the Protein Data Bank (accession code 1Q67).
ACKNOWLEDGMENTS
We are grateful to the staffs at beamline BL41XU at SPRING-8, Japan for their support during data collection. This work is financially supported by the Agency for Science, Technology and Research (A*STAR) in Singapore (H.S.) and by the Howard Hughes Medical Institute (R.P.).
Footnotes
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
References
- 1.van Hoof A, Parker R. The exosome: a proteasome for RNA? Cell. 1999;99:347–350. doi: 10.1016/s0092-8674(00)81520-2. [DOI] [PubMed] [Google Scholar]
- 2.Waterhouse PM, Wang MB, Lough T. Gene silencing as an adaptive defence against viruses. Nature. 2001;411:834–842. doi: 10.1038/35081168. [DOI] [PubMed] [Google Scholar]
- 3.Frischmeyer PA, Dietz HC. Nonsense-mediated mRNA decay in health and disease. Hum. Mol. Genet. 1999;8:1893–1900. doi: 10.1093/hmg/8.10.1893. [DOI] [PubMed] [Google Scholar]
- 4.Tucker M, Parker R. Mechanisms and control of mRNA decapping in Saccharomyces cerevisiae. Annu. Rev. Biochem. 2000;69:571–595. doi: 10.1146/annurev.biochem.69.1.571. [DOI] [PubMed] [Google Scholar]
- 5.Muhlrad D, Parker R. Mutations affecting stability and deadenylation of the yeast MFA2 transcript. Genes Dev. 1992;6:2100–2111. doi: 10.1101/gad.6.11.2100. [DOI] [PubMed] [Google Scholar]
- 6.Decker CJ, Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- 7.Hsu CL, Stevens A. Yeast cells lacking 5′→3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol. Cell. Biol. 1993;13:4826–4835. doi: 10.1128/mcb.13.8.4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muhlrad D, Decker CJ, Parker R. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′→3′ digestion of the transcript. Genes Dev. 1994;8:855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- 9.Jacobs-Anderson JS, Parker R. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen CY, et al. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107:451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 11.Wang Z, Kiledjian M. Functional link between the mammalian exosome and mRNA decapping. Cell. 2001;107:751–762. doi: 10.1016/s0092-8674(01)00592-x. [DOI] [PubMed] [Google Scholar]
- 12.Mukherjee D, et al. The mammalian exosome mediates the efficient degradation of mRNAs that contain AU-rich elements. EMBO J. 2002;21:165–174. doi: 10.1093/emboj/21.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu H, Rodgers ND, Jiao X, Kiledjian M. The scavenger mRNA decapping enzyme DcpS is a member of the HIT family of pyrophosphatases. EMBO J. 2002;21:4699–4708. doi: 10.1093/emboj/cdf448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muhlrad D, Parker R. Premature translation termination triggers mRNA decapping. Nature. 1994;370:578–581. doi: 10.1038/370578a0. [DOI] [PubMed] [Google Scholar]
- 15.Hagan KW, Ruiz-Echevarria MJ, Quan Y, Peltz SW. Characterization of cis-acting sequences and decay intermediates involved in nonsense-mediated mRNA turnover. Mol. Cell. Biol. 1995;15:809–823. doi: 10.1128/mcb.15.2.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cao D, Parker R. Computational modeling and experimental analysis of nonsense-mediated decay in yeast. Cell. 2003;113:533–545. doi: 10.1016/s0092-8674(03)00353-2. [DOI] [PubMed] [Google Scholar]
- 17.Beelman CA, et al. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature. 1996;38:642–646. doi: 10.1038/382642a0. [DOI] [PubMed] [Google Scholar]
- 18.Dunckley T, Parker R. The Dcp2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J. 1999;18:5411–5422. doi: 10.1093/emboj/18.19.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dunckley T, Tucker M, Parker R. Two related proteins, Edc1p and Edc2p, stimulate mRNA decapping in Saccharomyces cerevisiae. Genetics. 2001;157:27–37. doi: 10.1093/genetics/157.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lykke-Andersen J. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol. Cell. Biol. 2002;22:8114–8121. doi: 10.1128/MCB.22.23.8114-8121.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steiger M, Carr-Schmid A, Schwartz DC, Kiledjian M, Parker R. Analysis of recombinant yeast decapping enzyme. RNA. 2003;9:231–238. doi: 10.1261/rna.2151403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Dijk E, et al. Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 2002;21:6915–6924. doi: 10.1093/emboj/cdf678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Z, Jiao X, Carr-Schmid A, Kiledjian M. The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc. Natl. Acad. Sci. USA. 2002;99:12663–12668. doi: 10.1073/pnas.192445599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koonin EV. A highly conserved sequence motif defining the family of MutT-related proteins from eubacteria, eukaryotes and viruses. Nucleic Acids Res. 1993;21:4847. doi: 10.1093/nar/21.20.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bessman MJ, Frick DN, O’Handley SF. The MutT proteins or ‘Nudix’ hydrolases, a family of versatile, widely distributed, ‘house cleaning’ enzyme. J. Biol. Chem. 1996;271:25059–25062. doi: 10.1074/jbc.271.41.25059. [DOI] [PubMed] [Google Scholar]
- 26.Vilela C, Velasco C, Ptushkina M, McCarthy JE. The eukaryotic mRNA decapping protein Dcp1 interacts physically and functionally with the elF4F translation initiation complex. EMBO J. 2000;19:4372–4382. doi: 10.1093/emboj/19.16.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bai RY, et al. SMIF, a Smad4-interacting protein that functions as a co-activator in TGFβ signalling. Nat. Cell Biol. 2002;4:181–190. doi: 10.1038/ncb753. [DOI] [PubMed] [Google Scholar]
- 28.Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 29.Prehoda KE, Lee DJ, Lim WA. Structure of the enabled/VASP homology 1 domain-peptide complex: a key component in the spatial control of actin assembly. Cell. 1999;97:471–480. doi: 10.1016/s0092-8674(00)80757-6. [DOI] [PubMed] [Google Scholar]
- 30.Vetter IR, Nowak C, Nishimoto T, Kuhlmann J, Wittinghofer A. Structure of a Ran-binding domain complexed with Ran bound to a GTP analogue: implications for nuclear transport. Nature. 1999;398:39–46. doi: 10.1038/17969. [DOI] [PubMed] [Google Scholar]
- 31.Beneken J, et al. Structure of the Homer EVH1 domain-peptide complex reveals a new twist in polyproline recognition. Neuron. 2000;26:143–154. doi: 10.1016/s0896-6273(00)81145-9. [DOI] [PubMed] [Google Scholar]
- 32.Volkman BF, Prehoda KE, Scott JA, Peterson FC, Lim WA. Structure of the N-WASP EVH1 domain-WIP complex: insight into the molecular basis of Wiskott-Aldrich syndrome. Cell. 2002;111:565–576. doi: 10.1016/s0092-8674(02)01076-0. [DOI] [PubMed] [Google Scholar]
- 33.Callebaut I. An EVH1/WH1 domain as a key actor in TGFβ signalling. FEBS Lett. 2002;519:178–180. doi: 10.1016/s0014-5793(02)02751-5. [DOI] [PubMed] [Google Scholar]
- 34.Tharun S, Parker R. Analysis of mutations in the yeast mRNA decapping enzyme. Genetics. 1999;151:1273–1285. doi: 10.1093/genetics/151.4.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Renfranz PJ, Beckerle MC. Doing (F/L) PPs: EVH1 domains and their proline-rich partners in cell polarity and migration. Curr. Opin. Cell Biol. 2002;14:88–103. doi: 10.1016/s0955-0674(01)00299-x. [DOI] [PubMed] [Google Scholar]
- 36.Kshirsagar M, Parker R. Identification of Edc3p as an enhancer of mRNA decapping in Saccharomyces cerevisiae. Genetics. doi: 10.1534/genetics.166.2.729. (in the press).
- 37.Coller JM, Tucker M, Sheth U, Valencia-Sanchez MA, Parker R. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA. 2001;7:1717–1727. doi: 10.1017/s135583820101994x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fischer N, Weis K. The DEAD box protein Dhh1 stimulates the decapping enzyme Dcp1. EMBO J. 2002;21:2788–2797. doi: 10.1093/emboj/21.11.2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ball LJ, et al. Dual epitope recognition by the VASP EVH1 domain modulates polyproline ligand specificity and binding affinity. EMBO J. 2000;19:4903–4914. doi: 10.1093/emboj/19.18.4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ball LJ, Jarchau T, Oschkinat H, Walter U. EVH1 domains: structure, function and interactions. FEBS letter. 2002;513:45–52. doi: 10.1016/s0014-5793(01)03291-4. [DOI] [PubMed] [Google Scholar]
- 41.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 42.Sheldrick GM. In: Direct Methods for Solving Macromolecular Structures. Fortier S, editor. Kluwer Academic; Dordrecht, The Netherlands: 1998. pp. 401–411. [Google Scholar]
- 43.de la Fortelle E, Bricogne G. Maximum-likelihood heavy-atom parameters refinement in the MIR and MAD methods. Methods Enzymol. 1997;276:472–494. doi: 10.1016/S0076-6879(97)76073-7. [DOI] [PubMed] [Google Scholar]
- 44.Abrahams JP. Bias reduction in phase refinement by modified interference functions: introducing the γ correction. Acta Crystallogr. D. 1997;53:43–48. doi: 10.1107/S0907444996015272. [DOI] [PubMed] [Google Scholar]
- 45.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 46.Brünger AT, et al. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 47.LaGrandeur TE, Parker R. Isolation and characterization of Dcp1p, the yeast mRNA decapping enzyme. EMBO J. 1998;17:1487–1496. doi: 10.1093/emboj/17.5.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kraulis PJ. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. App. Crystallogr. 1991;24:946–950. [Google Scholar]
- 49.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nicholls A, Sharp KA, Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]