Abstract
Mycoplasma genitalium, a small bacterium having minimal genome size, has only one identified exoribonuclease, RNase R (MgR). We have purified MgR to homogeneity, and compared its RNA degradative properties to those of its Escherichia coli homologs RNase R (EcR) and RNase II (EcII). MgR is active on a number of substrates including oligoribonucleotides, poly(A), rRNA, and precursors to tRNA. Unlike EcR, which degrades rRNA and pre-tRNA without formation of intermediate products, MgR appears sensitive to certain RNA structural features and forms specific products from these stable RNA substrates. The 3′-ends of two MgR degradation products of 23S rRNA were mapped by RT-PCR to positions 2499 and 2553, each being 1 nucleotide downstream of a 2′-O-methylation site. The sensitivity of MgR to ribose methylation is further demonstrated by the degradation patterns of 16S rRNA and a synthetic methylated oligoribonucleotide. Remarkably, MgR removes the 3′-trailer sequence from a pre-tRNA, generating product with the mature 3′-end more efficiently than EcII does. In contrast, EcR degrades this pre-tRNA without the formation of specific products. Our results suggest that MgR shares some properties of both EcR and EcII and can carry out a broad range of RNA processing and degradative functions.
Keywords: RNase R, Mycoplasma, RNA degradation, ribose methylation, tRNA processing
INTRODUCTION
Exoribonucleases play important roles in RNA metabolism, including RNA degradation, maturation, and end-turnover (Mitchell and Tollervey 2000; Kushner 2004; Li and Deutscher 2004; Deutscher 2006). In Escherichia coli, seven distinct exoribonucleases have been identified (Li and Deutscher 2004; Ezraty et al. 2005). Among them, RNase II and polynucleotide phosphorylase (PNPase) are most efficient in degrading nonstructured RNA and are thought to be the major activities for degrading mRNA decay intermediates (Regnier and Arraiano 2000; Kushner 2002). RNase T, PH, and D are responsible for the maturation of stable RNA species (Li and Deutscher 2004). Oligoribonuclease, the enzyme most active on short oligonucleotides, acts as a scavenger to remove the end products generated by other exonucleases (Ghosh and Deutscher 1999). RNase R was originally discovered as a residual hydrolytic exoribonuclease activity in strains lacking RNase II (Gupta et al. 1977; Kasai et al. 1977). The rnr gene encoding RNase R was subsequently identified based on its homology with RNase II (Cheng et al. 1998). Recent studies have revealed important roles for RNase R in many aspects of RNA metabolism and in protection of cells against stress conditions (Cairrao et al. 2003; Cheng and Deutscher 2003, 2005; Z. Li, X. Gong, S. Wu, and R. Tao, unpubl.).
E. coli RNase R (EcR) is structurally and catalytically similar to RNase II (EcII), although the two enzymes differ in their substrate specificity (Cheng and Deutscher 2002). Both enzymes are nonspecific processive exoribonucleases that degrade RNA from the 3′-end. Purified RNase R releases 5′-nucleoside monophosphates from RNA, leaving behind an undigested core 2–3 nucleotides (nt) in length. In contrast, RNase II becomes more distributive as substrate length becomes ∼10 nt or shorter and leaves an undigested core 3–5 nt in length. Among the substrates tested in vitro, poly(A) is most efficiently degraded by both RNase R and II. Interestingly, RNase R is much more active than RNase II on structured RNAs, such as rRNA and tRNA. While both enzymes are active on synthetic homopolymers, neither can degrade a fully double-stranded RNA–RNA duplex or RNA–DNA hybrid (Cheng and Deutscher 2002).
Disruption of the rnr gene in E. coli causes little defect in growth under normal laboratory conditions. However, a double mutant lacking both RNase R and PNPase activity is inviable (Cheng et al. 1998). Fragments of rRNA accumulate to high levels in temperature-sensitive double-mutant cells (Cheng and Deutscher 2003). RNase R also is responsible for the degradation of structured regions of mRNA, such as REP sequences (Cheng and Deutscher 2005). Therefore, RNase R is an important enzyme for degrading structured RNA in E. coli, a function that is important for removing RNA fragments generated from mRNA decay or degradation of stable RNAs. Such activities may be important for cell survival under stress conditions such as cold shock, starvation, or oxidative stress (Cairrao et al. 2003; Chen and Deutscher 2005; Z. Li, X. Gong, S. Wu, and R. Tao, unpubl.). Consistent with this notion, it has recently been discovered that RNase R is highly expressed under stress conditions in E. coli (Chen and Deutscher 2005).
Similar to what was found for RNase R, inactivation of the rnb gene encoding RNase II alone does not affect growth of E. coli cells. However, a deficiency in both RNase II and PNPase renders cells inviable and leads to accumulation of mRNA fragments at high levels (Donovan and Kushner 1986). RNase II generates mature 3′-ends of tRNA, albeit poorly compared to the activities of other tRNA 3′-maturation exoribonucleases (Li and Deutscher 1996).
RNase R is more widespread in eubacteria than is RNase II (Zuo and Deutscher 2001; Li et al. 2005). Interestingly, only one putative exoribonuclease has been identified in Mycoplasma genitalium based on extensive sequence analysis. This protein belongs to the RNase R/II family and was previously assigned as RNase R (Zuo and Deutscher 2002). The Mycoplasma RNase R (MgR) shows 27% identity and 48% similarity to EcR, and 27% identity and 43% similarity to EcII. All conserved regions of EcR are present in MgR. Based on exhaustive data mining, including PHI-BLAST, multiple sequence alignment and motif identification, no other exoribonuclease is identifiable in Mycoplasma (Zuo and Deutscher 2001; Li et al. 2005). Although it is possible that other exoribonuclease(s) may exist without significant sequence similarity to previously characterized nucleases, it is clear that homologs of most exoribonucleases are not encoded in the small genomes of Mycoplasma, and that RNase R likely functions in multiple aspects of RNA metabolism in this organism. In a genome-wide mutagenesis study, the RNase R gene in M. genitalium could not be interrupted, indicating that the enzyme is essential in this organism (Hutchison et al. 1999).
To understand how a single exoribonuclease can carry out RNA degradation as well as RNA processing functions, we characterized the catalytic properties and in vitro substrate specificity of RNase R from M. genitalium, and compared its properties to that of E. coli RNase R and II. Our results suggest that this single exoribonuclease can act on multiple RNA substrates in Mycoplasma and may play a role in a broader range of RNA metabolism as compared to its E. coli counterparts.
RESULTS
Cloning, overexpression, and purification of M. genitalium RNase R
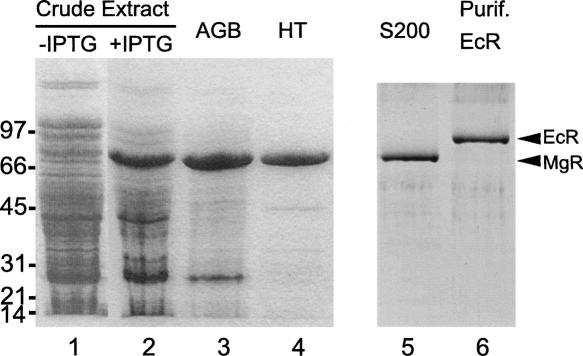
The PCR product from the predicted coding sequence of the rnr gene of M. genitalium G37 (MG104) was cloned into an expression vector pET15b as described in Materials and Methods. Upon IPTG induction of strain Rosetta-gami(DE3)/pLysS harboring plasmid pETmgR, MgR was expressed to the extent that it was the most abundant protein present in the cell extract (Fig. 1, lane 2). Starting from the induced cell extract, MgR was purified to near homogeneity by passage through Affi-gel Blue (AGB) and Hydroxyapatite (HT) columns (Fig. 1, lanes 3,4). The enzyme was further purified by MonoQ anion exchange and Superdex S200 gel filtration to apparent homogeneity (Fig. 1, lane 5). The purified protein has an apparent molecular mass of ∼80 kDa by SDS-PAGE, in close agreement with the predicted size of 82.9 kDa. Gel filtration of the purified protein shows a single peak of ∼80 kDa, suggesting a globular and monomeric architecture. Considering that this protein can only be purified from the pETmgR harboring Rosetta-gami(DE3)/pLysS strain after IPTG induction (Fig. 1, lanes 1,2) and that it displays molecular weight and chromatographic behavior distinct from any known E. coli nuclease, the purified protein and its attendant nuclease activity (shown below) are, therefore, not a manifestation of an endogenous E. coli nuclease. The identity of MgR was further verified by electrospray ionization-based mass spectroscopy analysis using the QSTAR LC/MS facility at Florida Atlantic University (data not shown).
FIGURE 1.
Purification of recombinant M. genitalium RNase R (MgR) from E. coli. About 5 μg of protein from each fraction were denatured, separated on a 4%–15% SDS-polyacrylamide gel, and stained with Coomassie blue. (lane 1) Crude cell extract from culture of Rosetta-gami (DE3)/pLysS harboring pETmgR before IPTG induction; (lane 2) crude extract from culture after IPTG induction; (lane 3) pooled Affi-gel Blue (AGB) peak fractions; (lane 4) flowthrough of hydroxyapatite (HT) column; (lane 5) peak fractions of Sephadex S200 column. In addition, E. coli RNase R (EcR) was purified as described previously (Cheng and Deutscher 2002; Zuo et al. 2006) and is shown in lane 6. The migration positions of molecular mass standards are shown on the left (in kilodaltons). The positions of MgR and EcR are indicated by arrows on the right.
E. coli RNase R (EcR) and RNase II (EcII) were purified to apparent homogeneity as described previously (Cheng and Deutscher 2002; Zuo et al. 2006). The purity of EcR preparation was confirmed by SDS-PAGE (Fig. 1, lane 6).
Degradation of polyadenylates
EcR and EcII degrade poly(A) over a broad pH range and require divalent cations for nuclease activity (Cheng and Deutscher 2002; Cheng 2003). Purified MgR was assayed on poly(A) under various conditions to compare the assay requirements of these enzymes. Poly(A) of the same average length (∼150 nt) and concentration as the assays for EcR and EcII activities (Cheng and Deutscher 2002; Cheng 2003) was used. Our results indicate that MgR also degrades poly(A). The specific activity of MgR on poly(A) is ∼4000 nmol/min/mg protein under optimal conditions. Higher activities of EcR (26,000 nmol/min per mg) and EcII (110,000 nmol/min/mg) on poly(A) were observed in our experiments using the same batch of poly(A) substrate, which agree well with its activity reported previously (Cheng and Deutscher 2002). Thus, the poly(A) degradation-specific activity of MgR is only ∼15% of the activity seen in EcR or ∼4% of that of EcII under similar conditions. MgR is active under conditions similar to those observed for EcR; however, MgR does display some differences. Differences in enzymatic parameters of MgR and EcR that may account for the observed differences in activity on poly(A) remain to be elucidated.
MgR activity peaks at pH 8.5. At pH 7.5 and 9.0, the activity drops by ∼20% (Fig. 2A). The enzyme is essentially inactive below pH 6.8. In contrast, EcR is active over a much broader pH range, displaying unchanged activity at pH values between 7.5 and 9.5, and ∼50% activity at pH 6.5 (Cheng 2003). EcII is most active between pH 8 and pH 9 (Cheng 2003).
FIGURE 2.
Degradation of polyadenylates by M. genitalium RNase R. RNase R activity on [H3]-poly(A) substrate was determined by acid soluble assay as described in Materials and Methods. Specific activities [in nanomoles of poly(A) per minute per milligram of enzyme] under various conditions are plotted as mean±standard error from three independent experiments. (A) pH range with 10 μM ZnCl2 and 100 mM KCl; (B) ZnCl2 or MgCl2, with 100 mM KCl at pH 8.5; (C) KCl, with 10 μM ZnCl2 at pH 8.5.
MgR requires a divalent cation at low concentrations, with Zn2+ being better than Mg2+ (Fig. 2B). When no divalent cation was added to the reaction, MgR displayed activity varying from 30% to 60% of the optimum, presumably due to contamination by trace levels of divalent metal ions. Consistent with this notion, 1 mM EDTA inhibited MgR activity completely, as previously reported for EcR and EcII (Cheng and Deutscher 2002). The highest activity of MgR is achieved using 5–10 μM Zn2+, nearly 50% higher than using Mg2+ at its optimal concentration of 0.05–0.25 mM (Fig. 2B). Higher concentrations of Zn2+ or Mg2+ inhibit MgR activity. Similarly, EcR was found to be most active with Mg2+ between 0.1 and 0.5 mM (Cheng and Deutscher 2002). In our experiments, EcR works equally well with 10 μM Zn2+ or 0.25 mM Mg2+. Very low MgR activity, if any, was observed when Mn2+ or Ca2+ was used at various concentrations (data not shown). In contrast to MgR and EcR, EcII is most active at 10 mM MgCl2 (Cheng 2003). Compared to EcII and other well-studied exoribonucleases in E. coli (Li and Deutscher 2004), MgR and EcR seem to require much lower concentrations of divalent metal ions.
Little MgR activity was observed when monovalent cations were absent. Addition of KCl increases activity with an optimal concentration of 100 mM (Fig. 2C). To our surprise, MgR exhibited no poly(A) degradation activity in the presence of the monovalent cations, NH4Cl or NaCl, at concentrations 50–300 mM (data not shown). It should be noted that both EcR and EcII are active in the absence of monovalent cations. In the presence of 50–500 mM KCl, EcR's activity is stimulated approximately twofold, whereas the activity of EcII is increased five- to sixfold (Cheng 2003).
Dithiothreitol (DTT) does not increase the activity of MgR or EcR (data not shown), indicating that these two enzymes are not sensitive to oxidation of sulfhydryl groups.
Degradation of RNA oligoribonucleotides
Considering MgR is the only identifiable exoribonuclease in the M. genitalium genome, we examined MgR activity on oligonucleotides to see whether it is capable of degrading short RNA fragments. As shown in Figure 3, MgR can efficiently degrade 14-mer (CA14) and 17-mer (C17) oligoribonucleotides from 3′ to 5′ in a manner similar to EcR. As revealed by reactions with different amounts of RNase R, both enzymes act processively without accumulation of intermediates. MgR action on longer substrates appears to be at least as efficient as EcR and accumulates mostly trinucleotides, with smaller amounts of di- and tetranucleotides. In contrast, EcR generates dinucleotides as the major product with lower amounts of trinucleotides. Neither enzyme displays nucleotide specificity since no difference is observed between substrates containing only C (C17) or containing both A and C (CA14). However, the activity of MgR is weaker than that of EcR on an RNA tetranucleotide (C4), compared to their activities on C17 and CA14 (Fig. 3, left panel). Our data on EcR degradation of oligoribonucleotides agree very well with those described previously (Cheng and Deutscher 2002). A previous study showed that EcII is also active on oligo RNA substrates of various lengths, producing short oligo RNAs that are predominantly tetranucleotides (Cheng and Deutscher 2002).
FIGURE 3.
Degradation of oligoribonucleotides. Oligonucleotides were labeled at their 5′-ends with 32P. Assays were carried out at 37°C in 40 μL reaction mixtures containing 25 μM of the indicated substrate; 1.0 μg of enzyme (EcR or MgR) was used in these analyses. Lanes labeled as “Buffer” indicate control reactions with no enzymes added. Five-microliter aliquots were taken at the times indicated, and the reactions were stopped with 2 volumes of RNA loading buffer. Products were resolved on 22.5% denaturing polyacrylamide gels. The sequences of the oligonucleotides used in this experiment are as follows: C4, CA14 (5′-CCCCACCACCAACA-3′), and C17.
Degradation of rRNA
The fact that E. coli RNase R is able to digest both 16S and 23S rRNA molecules demonstrates that the enzyme is capable of working through the extensive secondary structures present in rRNA (Cheng and Deutscher 2002, 2003). In contrast, RNase II has very limited activity on these substrates (Cheng and Deutscher 2002) and is unable to digest through structured RNA (Vincent and Deutscher 2006; Zuo et al. 2006). Here, we compared the degradation of E. coli rRNA by MgR and EcR. Ribosomal RNAs were isolated from E. coli as described in Materials and Methods.
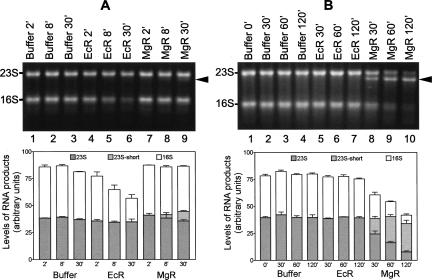
After treatment with EcR or MgR, the RNAs were separated on agarose gels and visualized by staining with SYBR Gold. The extent of degradation was measured by quantifying the reduction in the amount of, or shortening of, full-length rRNA. When equal amounts of protein (150 ng) were used, EcR demonstrated a higher activity than MgR for degrading 23S and 16S RNA (Fig. 4A), consistent with their respective poly(A) degrading activities. EcR digestion results in the reduction of full-length 16S rRNA to ∼50% in 30 min (Fig. 4A, lanes 4–6, and lower panel). Little 23S rRNA was degraded by EcR under this condition. No specific degradation product accumulates to a detectable level, suggesting that degradation of rRNAs by EcR does not stop at specific positions. Compared to EcR, the same amount of MgR showed very limited activity on 23S and 16S rRNA. However, a hint of a minor degradation intermediate of 23S rRNA was seen (Fig. 4A, lane 9).
FIGURE 4.
Degradation of E. coli rRNA by MgR and EcR. Isolated rRNA was treated with RNases in Zn2+-containing buffer for the length of time indicated on the top of each lane. Agarose gels (1.2%) were used to separate the RNA. A photograph was taken under UV lights after staining with SYBR Gold. Intermediate 23S RNA degradation products are labeled on the right side by arrows. The RNA products were quantified using the integrated optical density of the bands detected by the Epi Chemi II Darkroom (UVP Laboratory Products). Gels from three independent runs were quantified, and average values with standard errors are shown in the bottom panels. (A) Reactions with equal amounts of MgR and EcR (0.15 μg each). (B) Reactions with MgR and EcR of similar poly(A) degradation activity (0.5 μg of MgR or 0.038 μg of EcR).
When the amount of EcR and MgR of equal poly(A) degradation activity (500 ng of MgR and 37.5 ng of EcR) was used, MgR exhibits greater activity than EcR on rRNA substrates (Fig. 4B). Remarkably, degradation of 23S rRNA by MgR, but not by EcR, forms an intermediate product several hundred nucleotides shorter than 23S RNA (Fig. 4B, lanes 8–10). Little 23S RNA, if any, is degraded by MgR beyond this position. The majority of 23S RNA was converted to the shorter product after 120-min incubation with MgR (Fig. 4B, lane 10, and lower panel). In contrast, 16S RNA is almost completely degraded by MgR in 120 min without apparent accumulation of a specific intermediate. EcR showed little activity under this condition. The results indicate that MgR is more active on rRNA than on poly(A) when compared to EcR.
The reactions shown in Figure 4 were carried out in the presence of 10 μM ZnCl2, which supports optimum poly(A) degradation activity by both MgR and EcR. When 0.25 mM MgCl2 was used instead, EcR displays the same degradation activity on rRNAs, whereas MgR degrades rRNA at a slower rate (data not shown). This behavior is consistent with their respective poly(A) degradation activities. In conclusion, MgR is very active on structured RNA, but its activity is sensitive to certain specific features in the RNA (see below).
MgR is sensitive to 2′-O-ribose methylations in rRNA substrates
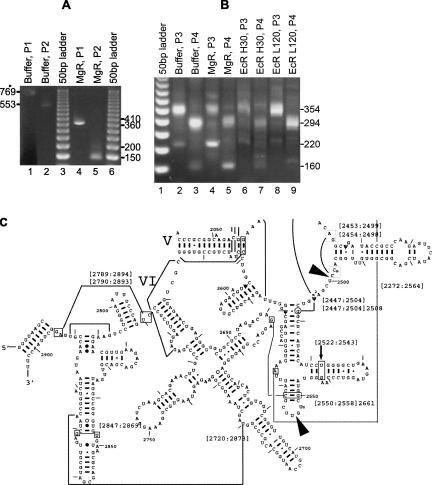
To look for sequence or structural features in 23S RNA that may cause degradation to stop, we determined the 3′-ends of the 23S RNA degradation intermediates formed by MgR treatment. Mapping of the 3′-end was carried out by ligation of a DNA linker to the 3′-end of the RNA. The RNA was then reverse-transcribed using a primer complementary to the linker. PCR was next carried out using the same forward primer complementary to the linker, and reverse primers annealing to two downstream locations in the cDNA. The 3′-ends of degradation intermediates of 23S RNA can be estimated by the sizes of the PCR products. The exact locations of RNA 3′-ends were determined by sequencing of the PCR products. Using this procedure, MgR was shown to stall at two close positions (Fig. 5A, lanes 4,5). The major PCR product from MgR-treated RNA is ∼400 nt shorter than that from the full-length 23S RNA. Another minor PCR product is ∼50 base pairs (bp) longer than the major one. Products independently formed using two pairs of primers from full-length and truncated 23S RNA agree with each other based on their size differences. Products corresponding to the full-length 23S RNA were not detectable with MgR-treated RNA, although they would have also been produced. Sequencing of the PCR products located the 3′-ends of the degradation products at positions 2499 and 2553 of 23S rRNA, respectively. After these positions, each sequence is followed by a clean linker sequence (data not shown), indicating that a single stop at each position was produced by MgR. These positions in the E. coli 23S RNA and nearby secondary structures are shown in Figure 5C.
FIGURE 5.
Determination of the 3′-end of the 23S and 16S rRNA degradation products by MgR. RNA was purified by phenol extraction from reactions with buffer, MgR (RNA from reactions in Fig. 4B, lanes 3,9), or EcR (reactions in Fig. 4A, lane 6, and Fig. 4B, lane 7). Linker was ligated to RNA of both samples. RT-PCR was carried out as described in Materials and Methods. PCR products were separated on agarose gel. (A) RT-PCR products from 23S rRNA. (Lane 1) RT-PCR from RNA treated with buffer using primer pair P1 (RP + 23S-LP2). A product with an expected size of 769 bp from full-length 23S RNA was formed. (Lane 2) RT-PCR using the same RNA and primer pair P2 (RP + 23S-LP1). An expected product of 553 bp from full-length 23S RNA is shown. (Lanes 4,5) RT-PCR products from RNA treated with MgR using P1 and P2, respectively. Estimated sizes of the products are indicated on the right side of the gel. (Lanes 3,6) Fifty-base pair DNA Step Ladder (Promega). (B) RT-PCR products from 16S rRNA. cDNA samples were prepared using RNA from reactions indicated on the top of each lane. RT-PCR was carried out using either primer pair P3 (RP + 16S-LP1, producing 354-bp DNA from full-length 16S rRNA) or primer pair P4 (RP + 16S-LP2, producing 294-bp DNA from full-length 16S rRNA). (Lane 1) Fifty-base pair DNA Step Ladder (Promega). (Lanes 2,3) RT-PCR from RNA treated with buffer (Fig. 4B, lane 3), showing the expected products of (lane 2) 354 bp and (lane 3) 294 bp from full-length 16S rRNA. In addition, less abundant products of (lane 2) ∼220 bp and (lane 3) ∼160 bp were also detected. (Lanes 4,5) RT-PCR from RNA treated with 500 ng of MgR for 60 min (Fig. 4B, lane 9). (Lanes 6,7) RT-PCR from RNA treated with 150 ng of EcR for 30 min (Fig. 4A, lane 6). (Lanes 8,9) RT-PCR from RNA treated with 37.5 ng of EcR for 120 min (Fig. 4B, lane 7). (C) RNA secondary structure representation of the 3′-region of E. coli 23S rRNA (adapted from Cannone et al. 2002a, with permission from BioMed Central, ©2000; kindly made available by Dr. Robin Gutell's group at http://www.rna.icmb.utexas.edu/). (Arrows) MgR stop sites; (numbers, boxes, and lines) various tertiary interactions between the numbered nucleotides.
Interestingly, the 3′-ends for both these degradation intermediates are located 1 nt downstream from a ribose-methylated residue, 2′-O-methyluridine (Um) at 2552 and 2′-O-methylcytidine (Cm) at 2498, respectively (Limbach et al. 1994; Cannone et al. 2002a,b). Apart from these two modifications, there is also a base modification (m2A at 2503) between these sites and the 3′-end of mature 23S RNA. This base methylation does not appear to inhibit degradation by MgR. Note that a stable stem–loop is present 8 nt upstream of 2552 and 3 nt upstream of 2499. Since the abundance of each PCR product is affected by ligation efficiency of the linker to different RNA 3′-ends, the relative abundance of the two RNA products can be somewhat different, although optimal RNA ligation conditions and an excessive amount of linkers were used.
A plausible explanation for the formation of the intermediates is that MgR stalls at ribose methylations since these modifications are very close to the site of hydrolytic attack. These findings prompted further investigation to determine if other ribose methylations in rRNA also inhibit MgR digestion. There is also a ribose methylation upstream at position 2251 (2′-O-methylguanosine, or Gm). MgR does not appear to proceed beyond position 2499, since no intermediate corresponding to the ribose methylation at position 2251 was observed. All other methylations on the E. coli 23S rRNAs are located on the base (Limbach et al. 1994; McCloskey and Crain 1998).
We also examined if the sole base and ribose methylation in 16S rRNA (m4Cm 1402) (Limbach et al. 1994) would stop MgR digestion, although no apparent intermediate was originally observed in the 16S degradation experiments (Fig. 4B). As anticipated, the PCR products expected for a stop at this location were produced (Fig. 5B, lanes 4,5, DNA bands of ∼220 bp from an upstream forward primer and ∼160 bp from a downstream forward primer, respectively), indicating that MgR treatment, indeed, creates a 16S RNA fragment of this size. DNA sequencing of the products confirmed that the 3′-end is 1 nt downstream from m4Cm 1402. However, PCR products with the same 3′-end (verified by DNA sequencing) were also observed at lower levels in the untreated rRNA samples (Fig. 5B, lanes 2,3) and in an independent RNA preparation (data not shown). This suggests the existence in total RNA of a small amount of 16S RNA truncated at this position for reasons yet unknown. Such products were either present at low levels or undetectable in EcR treatment reactions (Fig. 5B, lanes 6–9). Overall, the results demonstrated sensitivity of MgR, but not EcR, to 2′-O-methylations. The efficiency and exact location of the MgR stop may also be affected by local RNA structure, as suggested by studies of EcR (Cheng and Deutscher 2005; Vincent and Deutscher 2006).
Degradation of RNA oligonucleotide containing a 2′-O-ribose methylation
To further confirm the sensitivity of MgR to ribose methylation in RNA, a synthetic 17-mer RNA oligonucleotide containing a 2′-O-methyluridine at the seventh position was treated with MgR, EcR, and EcII as described in Materials and Methods (Fig. 6). MgR digests this oligonucleotide substrate and stops at position 9, which is 2 nt downstream from the methyluridine (Fig. 6, lanes 10–13). After prolonged incubation, MgR converted almost all the substrate to this 9-nt product, with a just a few faint bands indicating negligible shorter products. In contrast, EcR and EcII degrade this substrate more efficiently and digest through the position of 2′-O-methylation. At shorter time points, the 9-nt product was produced at low levels by EcR and was degraded completely upon longer incubation (Fig. 6, lanes 5–9). EcII also can digest through this methyluridine but is slightly more sensitive than EcR to this modification (Fig. 6, lanes 14–17). When an RNA substrate of the same sequence but having no methylation was treated by these enzymes, the 9-nt product was not formed (data not shown). These data clearly demonstrated that MgR is very sensitive to 2′-O-ribose methylation, whereas the activity of EcR and EcII is only slightly affected by this modification. Further studies will be required to understand the structural differences between these enzymes, which may account for the different activity on ribose-methylated RNA.
FIGURE 6.
Degradation of a 2′-O-methylated oligoribonucleotide by MgR, EcR, and EcII. Reactions were carried out and products were separated using the same condition as in Figure 3. The sequence of this 17-mer oligoribonucleotide is 5′-CAGUUG(Um)GAUCGAUCCC-3′. The labeled 17-mer was treated with buffer or RNase in a time course indicated on the top of the gel. A [32P]-labeled RNA Decade Markers (Ambion) was included in the gel (not shown), and the sizes are shown on the left. The sizes of RNA products are shown on the right.
Processing/degradation of tRNA precursor
All tRNAs are transcribed as precursor molecules that require processing at both 3′- and 5′-ends before they become functional (Deutscher 1990; Hopper and Phizicky 2003). In E. coli, a major pathway for tRNA 3′-maturation involves shortening of the 3′-trailer sequences by endonucleolytic cleavages by RNase E, followed by exonucleolytic trimming reactions (mainly by RNase T, PH, or D, also by RNase II and BN at lower efficiency) to generate mature 3′-ends (Li and Deutscher 1996, 2002; Li et al. 2005). In some other bacteria, RNase Z cleavages remove the entire 3′-trailer from the 3′-end of tRNA or CCA-less tRNA (Pellegrini et al. 2003; Minagawa et al. 2004; Li et al. 2005). The CCA-containing tRNA precursors in Bacillus subtilis are matured exonucleolytically by RNase PH, assisted by several other exoribonucleases including RNase R (Wen et al. 2005). It is surprising that none of the major tRNA 3′ processing activities has been identified in Mycoplasma (Zuo and Deutscher 2001; Li et al. 2005). The degradation pattern of 23S rRNA by MgR suggests that this enzyme works differently from EcR by stopping at some specific locations on RNA substrates (see above), making MgR a possible candidate for tRNA 3′-maturation. Similar to EcII, MgR may be able to stop at the mature 3′-end of tRNA instead of fully degrading it. The CCA sequence is encoded in all tRNAs in M. genitalium, and any tRNA maturation by MgR will require the trimming reaction to stop precisely after the CCA sequence.
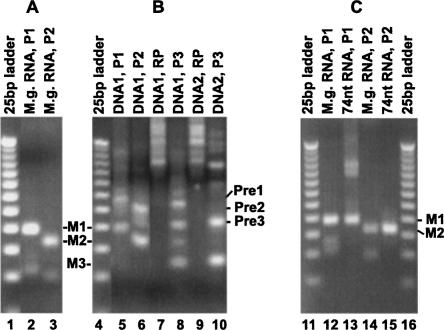
To examine if tRNAs in M. genitalium undergo 3′ processing, we carried out 3′-RACE to detect any tRNA precursor containing 3′-trailer sequence in vivo. For these tests, we chose to focus on one tRNA, tRNA1 Gly. Total RNA isolated from cultured M. genitalium was ligated to a linker, and PCR products were made from a primer complementary to the linker and other primers internal to tRNA1 Gly. As shown in Figure 7A, the major PCR products using two different pairs of primers (Fig. 7A, lanes 2,3, marked as M1 and M2) are about the sizes expected from the mature tRNA1 Gly. These PCR products were derived from tRNA1 Gly containing a mature 3′-end as confirmed by DNA sequencing. Interestingly, products a few tens to 100 bp longer were also produced in trace amounts and can be detected when larger amounts of PCR products were loaded on gel (data not shown). These longer products were isolated from the gel and were amplified by nested PCR (Fig. 7B). A major product nearly 40 bp longer than the one from mature tRNA was generated from each of the PCR reactions (Fig. 7B, lanes 5,6,8,10). Importantly, DNA sequencing showed that this product was produced from an RNA containing the encoded 43 extra nucleotides downstream from the 3′-end of tRNA1 Gly. In addition, some other minor products longer than the ones from mature tRNA were also observed as a smear, consistent with the notion of exonucleolytic processing of the 3′-end. These products were specific to tRNA sequence since they are not produced in reactions without the tRNA-specific primers (Fig. 7B, lanes 7,9). The existence of a small amount of precursor to tRNA1 Gly in total RNA suggests that tRNAs of M. genitalium undergo processing at the 3′-end in vivo.
FIGURE 7.
3′-RACE of M. genitalium tRNA1 Gly products. RNA was ligated to the DNA linker as described in Materials and Methods and in the legend of Figure 5. RT-PCR was carried out using primer pairs P1 (RP + tRNA-LP1), P2 (RP + tRNA-LP2), P3 (RP + tRNA-LP3), or a single primer RP. The expected PCR products from the mature 3′-end of tRNA1 Gly using P1, P2, or P3 are labeled as M1, M2, or M3, respectively. A 25-bp DNA Step Ladder (25bp ladder; Promega) was used as the size marker in each gel. (A) RT-PCR products from total RNA isolated from M. genitalium culture (M.g. RNA). (B) PCR products using DNA templates gel-isolated from A: (Lane 2) DNA1 products above M1, up to ∼100 bp longer than M1; (lane 3) DNA2 products above M2, up to ∼100 bp longer than M2. Major PCR products longer than the products from mature tRNA are labeled Pre1, Pre2, and Pre3 on the right. (C) RT-PCR products from the 74-nt RNA species generated from treatment of pre-tRNA1 Gly by MgR. Total RNA from M. genitalium (M.g. RNA) was used to show the sizes of PCR products from mature tRNA1 Gly.
To test if MgR is able to degrade 3′-trailer sequence of tRNA precursor and generate mature 3′-ends, a precursor to M. genitalium tRNA1 Gly was constructed and treated with purified RNase R or II enzymes in vitro. [32P]-labeled pre-tRNA1 Gly was made by in vitro transcription using a DNA template as described in Materials and Methods. The precursor RNA transcript starts from the 5′-end of the tRNA, followed by a 3′-trailer of 21 nt. The precursor was incubated with EcR, MgR, and EcII, at various Mg2 + concentrations. The products were analyzed on denaturing polyacrylamide gels. Upon EcR treatment, the majority of the substrate was degraded in the presence of Mg2+ (Fig. 8, lanes 9–13). In contrast, MgR treatment resulted in the formation of a major product 74 nt in length, which is the size of mature tRNA1 Gly. Several minor products ranging from 6 nt shorter to 5 nt longer than the mature tRNA1 Gly were also observed (Fig. 8, lanes 14–18). Interestingly, the 74-nt product is more abundant at higher concentrations of Mg2+. In addition, some precursor molecules must have been completely degraded by MgR, especially at lower concentrations of Mg2+, since the levels of the intermediate products are relatively low (Fig. 8, lanes 14,15) compared to the amount of the substrate used in the reactions. This result demonstrates that MgR can shorten the 3′-trailer of a tRNA precursor to generate the mature 3′-end. As expected, EcII converts the pre-tRNA to shorter products, the majority being 3 and 4 nt longer than 74 nt when Mg2+ is present (Fig. 8, lanes 20–23). In the absence of Mg2+, the major products are in the sizes of mature tRNA and 1 nt shorter (Fig. 8, lane 19). The amount of these products decreases as Mg2+ concentration increases.
FIGURE 8.
Processing and degradation of M. genitalium tRNA1 Gly precursor. Pre-tRNA1 Gly with a mature 5′-end and a 21-nt 3′-trailer was uniformly labeled with 32P and was treated with RNases as described in Materials and Methods. RNA products were detected by autoradiography after separation on an 8% denaturing polyacrylamide gel. (Lane 1) A [32P]-labeled RNA ladder (RNA Decade Markers; Ambion) was used as the size marker. (Lanes 2,3) In addition, 78-nt and 74-nt RNA transcripts containing the same sequence as tRNA1 Gly were used as size markers. These markers were generated using PCR templates from the tRNA gene, starting from the 5′-end of the tRNA and ending at the 3′-end (for 74 nt) or 4 nt downstream (for 78 nt). Pre-tRNA1 Gly was treated by buffer (containing 10 μM ZnCl2), EcR, MgR, or EcII for 30 min at 37°C in the presence of various concentrations of MgCl2 as indicated on the top of each lane. (P) The size of the precursor is 95 nt. (M) The size of mature tRNA1 Gly is 74 nt.
To ascertain that the 74-nt product generated by MgR contains a mature 3′-end, we have performed 3′-RACE to determine the 3′-end sequence of this tRNA product. The 74-nt RNA was purified from polyacrylamide gel and was ligated to the linker. PCR reactions using a primer complementary to the linker and two other primers internal to the tRNA revealed products expected from mature tRNA1 Gly (Fig. 7C, lanes 13,15), the same as the RT-PCR products obtained when total RNA from M. genitalium was used (Fig. 7C, lanes 12,14). Sequencing of the RT-PCR products from the 74-nt RNA demonstrated that the tRNA product contains a mature 3′-end.
DISCUSSION
RNase R is the only exoribonuclease identified in Mycoplasma. Although this enzyme from M. genitalium is named RNase R, its sequence is equally similar to those of RNase R and II from E. coli. One important distinguishing feature of RNase R is its ability to degrade structured RNA (Vincent and Deutscher 2006), unlike RNase II, which can only degrade single-strand RNA (Zuo et al. 2006). We show here that MgR, like EcR, can efficiently degrade structured RNA such as ribosomal RNA. However, MgR is sensitive to ribose modifications and stops at specific locations when digesting E. coli ribosomal RNA. MgR also possesses the ability, similar to EcII, to process tRNA and is a likely candidate for the maturation of tRNA 3′-ends. These features of MgR are distinct from those of EcR in terms of their biochemical and biological functions. These observations suggest that in addition to RNA degradation, RNase R in Mycoplasma may be responsible for several essential functions known to be carried out by other exoribonucleases in E. coli. The importance of MgR in Mycoplasma has been demonstrated by the finding that disruption of the gene encoding RNase R makes M. genitalium inviable (Hutchison et al. 1999). Consistent with the notion that RNase R may function in both RNA processing and degradation, it was recently reported that an enzyme belonging to the RNase R family is responsible for rRNA 3′-maturation in chloroplasts of Arabidopsis (Kishine et al. 2004; Bollenbach et al. 2005) and in Pseudomonas syringae (Purusharth et al. 2007). RNase R has been known to participate in mRNA turnover in E. coli (Cheng and Deutscher 2005; Andrade et al. 2006) and B. subtilis (Oussenko et al. 2005). Much more remains to be learned about how the activities of Mycoplasma RNase R on various substrates are determined.
Little information is available on the processing of stable RNAs in Mycoplasma. In this study, we have detected precursors to tRNA1 Gly in vivo that contain extra 3′-trailer sequence, suggesting that tRNA in M. genitalium may undergo 3′-maturation. We have shown that MgR shortens a tRNA precursor of M. genitalium to the mature 3′-ends, suggesting a possible role of MgR in tRNA 3′-maturation. In vitro, MgR generates the mature 3′-end of tRNA as the major product, accompanied by other minor products and complete degradation of some precursor molecules. More efficient processing of the tRNA 3′-end may be achieved by MgR under in vivo conditions with factors that are missing in vitro. For instance, tRNA precursors may be modified in cells, and 5′-processing may affect 3′-maturation. It is known that the E. coli RNase R is not involved in 3′-maturation of tRNA, and RNase II does it poorly (Li and Deutscher 1994, 1996; Deutscher 2006). Since no known tRNA 3′-maturation activities have been identified in Mycoplasma, it is likely that MgR is adapted to carry out this function by a mechanism that remains to be fully elucidated (Li et al. 2005). It is interesting to note that all 36 tRNAs in M. genitalium contain encoded CCA sequences at their 3′-ends. tRNA nucleotidyl transferase normally adds CCA to tRNAs lacking this sequence. This enzyme was not found in Mycoplasma by BLAST search (data not shown). Therefore, if MgR is responsible for generation of the mature 3′-ends of tRNAs, it would have to stop precisely after CCA. In our in vitro treatment, more tRNA of mature size and sequence was generated by MgR at higher concentrations of Mg2+ (Fig. 8). Since Mg2+ is not required for MgR activity on poly(A) and rRNA, its role is probably to keep pre-tRNA in a conformation for MgR to stop at the mature 3′-end. Similarly, Mg2+ seems to cause EcII to stop, but at positions 3–4 nt downstream from the mature tRNA.
Our results suggest several specific points that deserve further study. First, MgR degrades poly(A) and rRNA more efficiently in the presence of Zn2+ than Mg2+, whereas EcR works equally well with both divalent cations. It is known that the base form of histidine residues is responsible for Zn2+ binding. The pK a of histidine residues in proteins is 6.5, but varies depending on ionic strength, temperature, and microenvironment (Stryer 1995). Here we show that activity of MgR drops sharply below pH 7.5 (Fig. 2A). This suggests that a histidine residue in MgR may be important for Zn2+-dependent activity. Second, degradation of oligoribonucleotides by MgR yields end products of di- to tetranucleotides, with the majority being trinucleotides. It remains unanswered how these oligoribonucleotides are converted to mononucleotides in Mycoplasma since oligoribonuclease, an essential enzyme carrying out this action in E. coli (Ghosh and Deutscher 1999), has not been identified in Mycoplasma (Zuo and Deutscher 2001). Third, MgR degrades stable RNA efficiently, but is sensitive to 2′-O-methylation. Ribose methylations have been reported to be prominent in the 23S rRNA in Mycoplasma, whereas base methylations are more common in E. coli (Hsuchen and Dubin 1980). It would be interesting to learn if MgR's sensitivity to 2′-O-methylation has any biological significance. In this respect, it is possible that MgR is involved in RNA quality-control mechanisms in Mycoplasmas. For instance, rRNA molecules that fail to be correctly ribose-methylated may be susceptible to degradation by MgR.
MATERIALS AND METHODS
Materials
The vector pET15b and host strains Novablue, BL21(DE3), and Rosetta-gami(DE3)/pLysS were obtained from Novagen Inc. E. coli RNase R and RNase II were purified to apparent homogeneity from a BL21(DE3) strain harboring pETR plasmid as described (Zuo and Deutscher 2002; Zuo et al. 2006). E. coli strain CA244 (lacZ, trp, relA, spoT) was used to prepare ribosomal RNA as described previously (Li et al. 1999). [3H]-poly(A), α-[32P]UTP, and γ-[32P]ATP are the products of GE Healthcare Inc. The ThermoScript RT-PCR System for cDNA synthesis and PCR is the product of Invitrogen. T7 RNA polymerase and RNA ligase are from New England Biolabs. Oligodeoxynucleotides were synthesized by Integrated DNA Technologies. RNA oligonucleotides were synthesized by Dharmacon Research Inc. All other chemicals and reagents are analytical grade. M. genitalium G37 was purchased from ATCC. Total RNA from cultured M. genitalium G37 was isolated by TRI Reagent (Molecular Research Center).
Cloning and overexpression of M. genitalium RNase R (MgR)
The predicted coding sequence of the rnr gene of M. genitalium was amplified from genomic DNA of strain G37 (ATCC Bioproducts Inc.) by the polymerase chain reaction. The following oligonucleotide primers were used in the polymerase chain reaction: mgR5p (5′-AAATCATGAAGGTTTTAACTG-3′) and mgR3p (5′-AAAGGATCCTAACAACCATTGG-3′). The products were cleaved with BspHI and BamHI and ligated into the unique NcoI and BamHI sites of pET15b. In the M. genitalium rnr gene, three tryptophan residues are encoded by TGA codons, which were changed to TGG codons to allow MgR expression in E. coli. This was accomplished by a one-step QuickChange (Stratagene Inc.) reaction using three mutation primers: mgRw54p3 (5′-TTCCTTAAGATGGCCCAATCAAGCAGATCATCAATG-3′), mgRw527p5 (5′-ATAGCTAGCTGGTTAAATGAAAACAAAGATAATCC-3′), and mgRw588p5 (5′-ACCATTCACCGGTTGTTGTGGATGCATCTTTTTACTCC-3′). The resulting plasmid pETmgR was verified by DNA sequencing. Plasmid pETmgR was then transformed into E. coli strain Rosetta-gami(DE3)/pLysS for MgR overexpression. The protein produced from this construct comprises of the full-length 725 residues in the MgR native protein with a calculated molecular weight of 82.9 kDa. Cultures of this strain were grown in LB medium at 37°C to early logarithmic phase and induced with 1 mM isopropyl β-thiogalactopyranoside (IPTG) at room temperature overnight. The cultures were chilled and the cells were harvested by centrifugation, resuspended in buffer A (20 mM Tris-Cl at pH 7.5, 10% glycerol, 1 mM DTT), and then flash-frozen using liquid nitrogen and stored at −20°C.
Purification of MgR from an overexpressing E. coli strain
For MgR purification, the frozen cell suspension (∼6 g of wet cells suspended in 20 mL) was thawed on ice and lysed using a French Press at 12,000 psi in the presence of protease inhibitors (0.1 mM phenylmethylsulfonyl fluoride and 1 tablet of Complete Mini Protease Inhibitor Cocktail; Boehringer Mannheim Inc.) and 10 μg/mL DNase I. The lysate was clarified by centrifugation. This crude extract was first applied to an Affi-Gel Blue column (GE Healthcare Inc.) in Buffer A and eluted stepwise in the presence of increasing amounts of NaCl from 0.15 M to 1 M. Fractions containing MgR were combined and applied directly to a hydroxylapatite column (Bio-Rad Inc.) equilibrated with Buffer A containing 1 M NaCl. The flowthrough containing essentially pure MgR was dialyzed overnight against Buffer A containing 1 mM EDTA and further purified using a MonoQ anion exchange column (GE Healthcare Inc.) and a Superdex S200 column (GE Healthcare Inc.). The purified MgR protein was concentrated by Microcon centrifugation (Millipore Inc.) to 15 mg/mL, as measured by A 280 (using an estimated extinction coefficient of 62,000 M−1 cm−1). The concentrated protein was stored at −80°C after flash-freezing in liquid nitrogen. All purification steps were carried out at 4°C. This procedure yields ∼6 mg of MgR protein from 6 g of cells. For activity assays, a small portion of the protein at 0.5 mg/mL was kept at −20°C in a storage buffer containing 40% glycerol, 10 mM Tris-Cl (pH 7.5), 25 mM NaCl, 0.5 mM DTT, and 12.5 μM EDTA.
Poly(A) degradation assays
Acid-soluble degradation assays were carried out for both EcR and MgR with [3H]-poly(A) as substrate (Cheng and Deutscher 2002). Each reaction was in 50 μL containing 20 mM Tris-Cl (pH 8.5), 100 mM KCl, 0.01 mM ZnCl2, 15 μM [3H]-poly(A) (concentration refers to polymer, same for other substrates), and 0.15 μg of enzyme. Samples were incubated for 15 min at 37°C. The remaining poly(A) substrate was precipitated by the addition of trichloroacetic acid to 10% in the presence of carrier yeast total RNA. Acid-soluble products were counted using a scintillation counter. Other buffer and salt conditions were used when optimal conditions for activity were studied, as described in the Results section.
Oligoribonucleotide degradation assays
5′-32P-labeled oligonucleotides were prepared as described (Zuo and Deutscher 2002). Assay conditions for both EcR and MgR were 20 mM Tris-Cl (pH 8.5), 100 mM KCl, 0.01 mM ZnCl2, and 5 mM DTT. In addition, the reactions included 0.5 mM MgCl2, which was carried over from the 32P-labeling reactions of the oligonucleotide substrates. Each assay contained 25 μM 5′-32P-labeled oligonucleotides and 1.0 μg of EcR or MgR in a 40-μL reaction. Reaction mixtures were incubated at 37°C for the time indicated. Reactions were stopped by adding 2 volumes of loading buffer containing 96% formamide and 1 mM EDTA. Reaction products were resolved on 22.5% denaturing polyacrylamide gels and were analyzed with a PhosphorImager (Molecular Dynamics Inc.).
rRNA degradation assays
Assays were carried out using rRNA prepared from E. coli culture as described previously (Li et al. 1999). Each assay was carried out in 50 μL in the same buffer as for poly(A) degradation, using 0.2 μM rRNA, and MgR or EcR. When Mg2+ was used instead of Zn2+, MgCl2 was added to 0.25 mM. Samples were incubated at 37°C. Aliquots were removed at various time points and placed on ice, and reactions were stopped by adding EDTA to 1 mM. RNAs were separated on a 1.2% agarose gel and were detected under UV light after staining with SYBR Gold (Molecular Probes). rRNA degradation was analyzed by monitoring the reduction of full-length RNA and the formation of any shorter intermediates.
Determination of the 3′-ends of rRNA degradation intermediates by 3′-RACE
Identification of rRNA degradation intermediates was carried out as previously described (Li et al. 1998) with some modifications. In brief, a DNA linker (5′-pTGGTTGGGATACTGCAGGAAp-3′) was ligated to the RNA before or after RNase R treatment. cDNA was synthesized from a primer complementary to the linker sequence (RP, 5′-TTCCTGCAGTATCCCAACCA-3′). To map 3′-ends of 23S products, this primer and another upstream primer (23S-LP1, 5′-TGCGAAAGCAGGTCATAGTG-3′) were used in PCR reactions with the cDNA as template. The expected size of this PCR product is 553 bp from full-length 23S RNA. Sizes of PCR products were determined by agarose gel electrophoresis. PCR products were gel-purified and sequenced using the upstream primer by Davis Sequencing. In addition, another upstream primer (23S-LP2, 5′-GGAGCCGACCTTGAAATACC-3′) was used to generate longer, independent PCR products and validate the results. The expected size of this PCR product is 769 bp from full-length 23S RNA.
Mapping of 3′-ends of 16S rRNA products was carried out using the same procedure, except different primers were used for PCR. Using RP and an upstream primer for 16S (16S-LP1, 5′-CCTTACGACCAGGGCTACAC-3′), the expected size of the PCR product is 354 bp from full-length 16S RNA. Using RP and another upstream primer (16S-LP2, 5′-AGAGCAAGCGGACCTCATAA-3′) would give rise to a PCR product of 294 bp from full-length 16S RNA.
tRNA precursor construction and processing reactions
A DNA fragment containing tRNA1 Glyt precursor sequence was generated by PCR from genomic DNA of M. genitalium. The PCR primer at the 5′-end of pre-tRNA contains an upstream KpnI restriction site, and the primer annealing to the 3′-end of the pre-tRNA contains an EcoRI site. The PCR product was then cloned into pUC19 by double digestion of both the insert DNA and the vector with KpnI and EcoRI, followed by ligation and transformation into E. coli DH10B-competent cells (Invitrogen). Plasmids were prepared and digested using KpnI and EcoRI to identify inserts of expected size. The sequence of the cloned insert was verified by DNA sequencing (Davis Sequencing).
A PCR product was made from the cloned tRNA gene to serve as template for in vitro transcription, using a primer containing a T7 promoter upstream of the pre-tRNA (5′-GTGCGGTACCAAATTAATACGACTCACTATAG-3′), and a primer downstream of the pre-tRNA (5′-ATGGACGGAGGACACACAAGTTGGAGCAGATAACGGG-3′). The tRNA precursor made by in vitro transcription from the T7 promoter is 95 nt in length, starting from the mature 5′-end of the tRNA followed by a 21-nt 3′-trailer sequence (5′-pppGCAGATATAGTTCAATGGTAGAACATAACCTTGCCAAGGTTAAGATGCGGGTTCGATTCCCGTTATCTGCTCCAACTTGTGAGTCCTCCGTCCAT-OH-3′).
tRNA precursor was synthesized by T7 RNA polymerase in the presence of [α-32P]UTP, according to a procedure described previously (Li and Deutscher 1994). The [32P]-labeled RNA products were gel-purified. Reactions with RNase R were carried out in 10 μL containing 0.05 μg of EcR or MgR in the same buffer as used for poly(A) degradation. Reactions were incubated at 37°C followed by the addition of 2 volumes of formamide loading buffer. Samples were run on an 8% polyacrylamide/urea denaturing gel to separate the RNA at single-nucleotide resolution. The products were detected by autoradiography.
Determination of the 3′-ends of tRNA processing products by 3′-RACE
The 3′-termini of the product of MgR-treated pre-tRNA1 Gly or tRNA1 Gly in total RNA from M. genitalium G37 were analyzed by 3′-RACE as described above. The same linker and complementary primer (RP) were used. Other primers internal to the tRNA include tRNA-LP1 (5′-GCAGATATAGTTCAATGGTAGAACATAAC-3′, from 1 to 29 nt of tRNA), tRNA-LP2 (5′-ATGGTAGAACATAACCTTGCCAAG-3′, from 15 to 38 nt of tRNA), and tRNA-LP3 (5′-CAAGGTTAAGATGCGGGTTC-3′, from 35 to 54 nt of tRNA). The sequences of various PCR products were determined by DNA sequencing.
ACKNOWLEDGMENTS
We are grateful to Murray P. Deutscher for sharing the pETR plasmid harboring the gene encoding E. coli RNase R, and to Robin Gutell for the updated 23S rRNA secondary structure plot. We thank Murray P. Deutscher for helpful discussions and critical reading of the manuscript, and Zhe Jiang for carrying out mass spectrometry analysis of purified MgR. This work was supported by a New Project Development Award at Florida Atlantic University and NIH grant S06-GM073621 to Z.L., and by NIH grant R01-GM69972 to A.M. Y.Z. was supported in part by an American Heart Association Florida/Puerto Rico Affiliate Postdoctoral Fellowship (0525530B). M.S.L. is supported by the Graduate Program in Biomedical Sciences at Florida Atlantic University.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.706207.
REFERENCES
- Andrade, J.M., Cairrao, F., Arraiano, C.M. RNase R affects gene expression in stationary phase: Regulation of ompA. Mol. Microbiol. 2006;60:219–228. doi: 10.1111/j.1365-2958.2006.05092.x. [DOI] [PubMed] [Google Scholar]
- Bollenbach, T.J., Lange, H., Gutierrez, R., Erhardt, M., Stern, D.B., Gagliardi, D. RNR1, a 3′–5′ exoribonuclease belonging to the RNR superfamily, catalyzes 3′ maturation of chloroplast ribosomal RNAs in Arabidopsis thaliana . Nucleic Acids Res. 2005;33:2751–2763. doi: 10.1093/nar/gki576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairrao, F., Cruz, A., Mori, H., Arraiano, C.M. Cold shock induction of RNase R and its role in the maturation of the quality control mediator SsrA/tmRNA. Mol. Microbiol. 2003;50:1349–1360. doi: 10.1046/j.1365-2958.2003.03766.x. [DOI] [PubMed] [Google Scholar]
- Cannone, J.J., Subramanian, S., Schnare, M.N., Collett, J.R., D'Souza, L.M., Du, Y., Feng, B., Lin, N., Madabusi, L.V., Muller, K.M., et al. The comparative RNA web (CRW) site: An online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002a;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannone, J.J., Subramanian, S., Schnare, M.N., Collett, J.R., D'Souza, L.M., Du, Y., Feng, B., Lin, N., Madabusi, L.V., Muller, K.M., et al. The comparative RNA web (CRW) site: An online database of comparative sequence and structure information for ribosomal, intron, and other RNAs: Correction. BMC Bioinformatics. 2002b;3:15. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C., Deutscher, M.P. Elevation of RNase R in response to multiple stress conditions. J. Biol. Chem. 2005;280:34393–34396. doi: 10.1074/jbc.C500333200. [DOI] [PubMed] [Google Scholar]
- Cheng, Z.-F. University of Miami Miller School of Medicine; Miami, FL: 2003. Cloning, purification and characterization, and biological function of the Escherichia coli exoribonuclease RNase R. Ph.D. thesis. [Google Scholar]
- Cheng, Z.-F., Deutscher, M.P. Purification and characterization of the Escherichia coli exoribonuclease RNase R. Comparison with RNase II. J. Biol. Chem. 2002;277:21624–21629. doi: 10.1074/jbc.M202942200. [DOI] [PubMed] [Google Scholar]
- Cheng, Z.-F., Deutscher, M.P. Quality control of ribosomal RNA mediated by polynucleotide phosphorylase and RNase R. Proc. Natl. Acad. Sci. 2003;100:6388–6393. doi: 10.1073/pnas.1231041100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, Z.-F., Deutscher, M.P. An important role for RNase R in mRNA decay. Mol. Cell. 2005;2:313–318. doi: 10.1016/j.molcel.2004.11.048. [DOI] [PubMed] [Google Scholar]
- Cheng, Z.-F., Zuo, Y., Li, Z., Rudd, K.E., Deutscher, M.P. The vacB gene required for virulence in Shigella flexneri and Escherichia coli encodes the exoribonuclease RNase R. J. Biol. Chem. 1998;273:14077–14080. doi: 10.1074/jbc.273.23.14077. [DOI] [PubMed] [Google Scholar]
- Deutscher, M.P. Ribonucleases, tRNA nucleotidyltransferase, and the 3′ processing of tRNA. Prog. Nucleic Acid Res. Mol. Biol. 1990;39:209–240. doi: 10.1016/s0079-6603(08)60628-5. [DOI] [PubMed] [Google Scholar]
- Deutscher, M.P. Degradation of RNA in bacteria: Comparison of mRNA and stable RNA. Nucleic Acids Res. 2006;34:659–666. doi: 10.1093/nar/gkj472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donovan, W.P., Kushner, S.R. Polynucleotide phosphorylase and ribonuclease II are required for cell viability and mRNA turnover in Escherichia coli K-12. Proc. Natl. Acad. Sci. 1986;83:120–124. doi: 10.1073/pnas.83.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezraty, B., Dahlgren, B., Deutscher, M.P. The RNase Z homologue encoded by Escherichia coli elaC gene is RNase BN. J. Biol. Chem. 2005;17:16542–16545. doi: 10.1074/jbc.C500098200. [DOI] [PubMed] [Google Scholar]
- Ghosh, S., Deutscher, M.P. Oligoribonuclease is an essential component of the mRNA decay pathway. Proc. Natl. Acad. Sci. 1999;96:4372–4377. doi: 10.1073/pnas.96.8.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta, R.S., Kasai, T., Schlessinger, D. Purification and some novel properties of Escherichia coli RNase II. J. Biol. Chem. 1977;252:8945–8949. [PubMed] [Google Scholar]
- Hopper, A.K., Phizicky, E.M. tRNA transfers to the limelight. Genes & Dev. 2003;17:162–180. doi: 10.1101/gad.1049103. [DOI] [PubMed] [Google Scholar]
- Hsuchen, C.-C., Dubin, D.T. Methylation patterns of mycoplasma transfer and ribosomal ribonucleic acid. J. Bacteriol. 1980;144:991–998. doi: 10.1128/jb.144.3.991-998.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison, C.A., Peterson, S.N., Gill, S.R., Cline, R.T., White, O., Fraser, C.M., Smith, H.O., Venter, J.C. Global transposon mutagenesis and a minimal Mycoplasma genome. Science. 1999;286:2165–2169. doi: 10.1126/science.286.5447.2165. [DOI] [PubMed] [Google Scholar]
- Kasai, T., Gupta, R.S., Schlessinger, D. Exoribonucleases in wild type Escherichia coli and RNase II-deficient mutants. J. Biol. Chem. 1977;252:8950–8956. [PubMed] [Google Scholar]
- Kishine, M., Takabayashi, A., Munekage, Y., Shikanai, T., Endo, T., Sato, F. Ribosomal RNA processing and an RNase R family member in chloroplasts of Arabidopsis . Plant Mol. Biol. 2004;55:595–606. doi: 10.1007/s11103-004-1507-1. [DOI] [PubMed] [Google Scholar]
- Kushner, S.R. mRNA decay in Escherichia coli comes of age. J. Bacteriol. 2002;184:4658–4665. doi: 10.1128/JB.184.17.4658-4665.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushner, S.R. mRNA decay in prokaryotes and eukaryotes: Different approaches to a similar problem. IUBMB Life. 2004;56:585–594. doi: 10.1080/15216540400022441. [DOI] [PubMed] [Google Scholar]
- Li, Z., Deutscher, M.P. The role of individual exoribonucleases in processing at the 3′ end of Escherichia coli tRNA precursors. J. Biol. Chem. 1994;269:6064–6071. [PubMed] [Google Scholar]
- Li, Z., Deutscher, M.P. Maturation pathways for E. coli tRNA precursors: A random multienzyme process in vivo. Cell. 1996;86:503–512. doi: 10.1016/s0092-8674(00)80123-3. [DOI] [PubMed] [Google Scholar]
- Li, Z., Deutscher, M.P. RNase E plays an essential role in the maturation of Escherichia coli tRNA precursors. RNA. 2002;8:97–109. doi: 10.1017/s1355838202014929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z., Deutscher, M.P. Exoribonucleases and endoribonucleases. In: Curtiss R. III, editor. EcoSal—Escherichia coli and Salmonella: Cellular and molecular biology. Chap. 4.6.3. ASM Press; Washington, DC: 2004. [Google Scholar]
- Li, Z., Pandit, S., Deutscher, M.P. Polyadenylation of stable RNA precursors in vivo. Proc. Natl. Acad. Sci. 1998;95:12158–12162. doi: 10.1073/pnas.95.21.12158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z., Pandit, S., Deutscher, M.P. RNase G (CafA protein) and RNase E are both required for the 5′ maturation of 16S ribosomal RNA. EMBO J. 1999;18:2878–2885. doi: 10.1093/emboj/18.10.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z., Gong, X., Joshi, V., Li, M. Coevolution of tRNA 3′ trailer sequences with 3′ processing enzymes in bacteria. RNA. 2005;11:567–577. doi: 10.1261/rna.7287505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limbach, P.A., Crain, P.F., McCloskey, J.A. Summary: The modified nucleosides of RNA. Nucleic Acids Res. 1994;22:2183–2196. doi: 10.1093/nar/22.12.2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCloskey, J.A., Crain, P.F. The RNA modification database–1998. Nucleic Acids Res. 1998;26:196–197. doi: 10.1093/nar/26.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minagawa, A., Takaku, H., Takagi, M., Nashimoto, M. A novel endonucleolytic mechanism to generate the CCA 3′ termini of tRNA molecules in Thermotoga maritime . J. Biol. Chem. 2004;279:15688–15697. doi: 10.1074/jbc.M313951200. [DOI] [PubMed] [Google Scholar]
- Mitchell, P., Tollervey, D. Musing on the structural organization of the exosome complex. Nat. Struct. Biol. 2000;7:843–846. doi: 10.1038/82817. [DOI] [PubMed] [Google Scholar]
- Oussenko, I.A., Abe, T., Ujiie, H., Muto, A., Bechhofer, D.H. Participation of 3′-to-5′ exoribonucleases in the turnover of Bacillus subtilis mRNA. J. Bacteriol. 2005;187:2758–2767. doi: 10.1128/JB.187.8.2758-2767.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini, O., Nezzar, J., Marchfelder, A., Putzer, H., Condon, C. Endonucleolytic processing of CCA-less tRNA precursors by RNase Z in Bacillus subtilis . EMBO J. 2003;22:4534–4543. doi: 10.1093/emboj/cdg435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purusharth, R.I., Madhuri, B., Ray, M.K. Exoribonuclease R in Pseudomonas syringae is essential for growth at low temperature and plays a novel role in the 3′ end processing of 16 and 5 S ribosomal RNA. J. Biol. Chem. 2007;282:16267–16277. doi: 10.1074/jbc.M605588200. [DOI] [PubMed] [Google Scholar]
- Regnier, P., Arraiano, C.M. Degradation of mRNA in bacteria: Emergence of ubiquitous features. Bioessays. 2000;22:235–244. doi: 10.1002/(SICI)1521-1878(200003)22:3<235::AID-BIES5>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Stryer, L. Biochemistry. 4th ed. W.H. Freeman and Company; New York: 1995. p. 23. [Google Scholar]
- Vincent, H.A., Deutscher, M.P. Substrate recognition and catalysis by the exoribonuclease RNase R. J. Biol. Chem. 2006;281:29769–29775. doi: 10.1074/jbc.M606744200. [DOI] [PubMed] [Google Scholar]
- Wen, T., Oussenko, I.A., Pellegrini, O., Bechhofer, D.H., Condon, C. Ribonuclease PH plays a major role in the exonucleolytic maturation of CCA-containing tRNA precursors in Bacillus subtilis . Nucleic Acids Res. 2005;33:3636–3643. doi: 10.1093/nar/gki675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo, Y., Deutscher, M.P. Exoribonuclease superfamilies: Structural analysis and phylogenetic distribution. Nucleic Acids Res. 2001;5:1017–1026. doi: 10.1093/nar/29.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo, Y., Deutscher, M.P. The physiological role of RNase T can be explained by its unusual substrate specificity. J. Biol. Chem. 2002;277:29654–29661. doi: 10.1074/jbc.M204252200. [DOI] [PubMed] [Google Scholar]
- Zuo, Y., Vincent, H.A., Zhang, J., Wang, Y., Deutscher, M.P., Malhotra, A. Structural basis for processivity and single-strand specificity of RNase II. Mol. Cell. 2006;24:149–156. doi: 10.1016/j.molcel.2006.09.004. [DOI] [PubMed] [Google Scholar]