Abstract
We previously cloned RRP14/YKL082c, whose product exhibits two-hybrid interaction with Ebp2p, a regulatory factor of assembly of 60S ribosomal subunits. Depletion of Rrp14p results in shortage of 60S ribosomal subunits and retardation of processing from 27S pre-rRNA to 25S rRNA. Furthermore, 35S pre-rRNA synthesis appears to decline in Rrp14p-depleted cells. Rrp14p interacts with regulatory factors of 60S subunit assembly and also with Utp11p and Faf1p, which are regulatory factors required for assembly of 40S ribosomal subunits. We propose that Rrp14p is involved in ribosome synthesis from the beginning of 35S pre-rRNA synthesis to assembly of the 60S ribosomal subunit. Disruption of RRP14 causes an extremely slow growth rate of the cell, a severe defect in ribosome synthesis, and a depolarized localization of cortical actin patches throughout the cell cycle. These results suggest that Rrp14p has dual functions in ribosome synthesis and polarized cell growth.
Keywords: ribosome assembly, pre-rRNA processing, cell polarity, RRP14, EBP2
INTRODUCTION
Yeast ribosomes are composed of four rRNAs and 79 ribosomal proteins. As yeast cells consume a great deal of energy to synthesize ribosomes, production of the components is highly regulated in response to environmental changes and intracellular insults (see, for example, Warner and Udem 1972; Mizuta and Warner 1994; for review, see Warner 1999). Three mature (25S, 18S, and 5.8S) rRNAs are synthesized as a long precursor, 35S pre-rRNA, in the nucleolus (for review, see Woolford and Warner 1991). The 90S preribosome particle contains 35S pre-rRNA, ribosomal proteins, small subunit (SSU) processome, and other nonribosomal proteins, most of which are required for 40S subunit assembly (Grandi et al. 2002). The 35S pre-rRNA is cleaved at sites A0, A1, and A2, and the 90S complex is converted to precursors of 40S and 60S subunits (see Fig. 3A, below) (for review, see Kressler et al. 1999; Venema and Tollervey 1999). A combination of TAP purification and proteome analysis has revealed that about 200 nonribosomal proteins are involved for pre-rRNA processing and assembly of the 60S and 40S subunits (e.g., Hampichamchai et al. 2001; Bassler et al. 2001; for review, see Warner 2001; Tschochner and Hurt 2003). However, it remains to be elucidated how these regulatory factors regulate the assembly. We previously demonstrated that Ebp2p is one of the regulatory factors essential for maturation of 25S rRNA and assembly of 60S ribosomal subunits (Tsujii et al. 2000). We cloned RRP14/YKL082c in a two-hybrid screen by using EBP2 as bait (Shirai et al. 2004). Rrp14p shares an amino acid sequence with the mouse SURF-6 gene product, and both Rrp14p and the mouse homolog are localized to the nucleolus (Magoulas and Fried 1996; Drees et al. 2001; Polzikov et al. 2005). The mammalian SURF-6 gene was discovered as the sixth member of the tightly clustered mammalian Surfeit locus (Magoulas and Fried 1996).
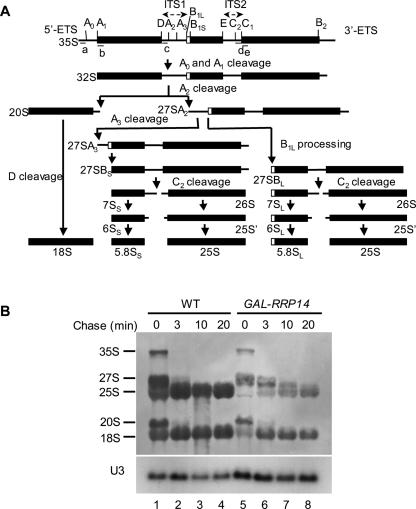
FIGURE 3.
Rrp14p depletion causes a defect in 25S rRNA maturation. (A) The processing pathway of 35S pre-rRNA to the mature rRNAs in S. cerevisiae. Northern probes (a), (b), (c), (d), and (e) used in Figure 4 are shown. (B) Pulse–chase analysis of rRNA synthesis in Rrp14p-depleted cells. W303–1A transformed with pRS414 (WT, lanes 1–4) and KM1411 (GAL1–RRP14 lanes 5–8) were cultured at 30°C in SCGal–Met and Trp, shifted to SC–Met and Trp, and cultured for 18 h. Each culture was pulsed with [methyl-3H] methionine for 3 min and chased with nonradioactive methionine for the indicated times. Total RNA prepared from each sample was analyzed by electrophoresis and blotted to a membrane. The lower part of the membrane was probed for U3 snoRNA as a loading marker.
Yeast Rrp14p was found in the fractions that were copurified with Ssf1p–TAP (Fatica et al. 2002) and Rrp1p–TAP (Horsey et al. 2004), both of which are regulatory proteins of 60S ribosomal subunit biogenesis. However, it remains to be elucidated whether Rrp14p is a bona fide factor for ribosome synthesis. Furthermore, it was demonstrated that Rrp14p has two-hybrid interactions with Zds2p, Gic1p, Gic2p, and Bud8p, which are related to cell polarity (Drees et al. 2001).
It was reported that RRP14 is essential for growth in systematic analysis (Winzeler et al. 1999). In this paper, however, we find that the rrp14Δ strain is viable with slow growth rate. We report that the rrp14Δ strain has a severe defect in ribosome synthesis and exhibits aberrant cell polarity and cell morphology. This is the first report showing that a regulatory factor of ribosome synthesis has an important role in the cell polarity derived from actin localization.
RESULTS
Rrp14p is enriched in the nucleolus
In order to ascertain the subcellular localization of Rrp14p, we constructed a strain KM1419 expressing Rrp14p fused with Yellow fluorescent protein (YFP) at the N terminus. The integrated YFP–RRP14 could complement the growth defect of the rrp14 null mutation (see below), indicating that the construct is biologically functional. The subcellular localization of YFP–Rrp14p was analyzed by fluorescence microscopy. Nop1p–Cyan fluorescent protein (CFP), used as a marker for the nucleolus, was detected in the region adjacent to the DAPI-stained nucleoplasm. The signal of YFP–Rrp14p was seen in the same region as Nop1p–CFP (data not shown), which is consistent with a previous observation that Rrp14p is primarily localized in the nucleolus (Drees et al. 2001). Furthermore, a weak signal of YFP–Rrp14p was also seen in the nucleoplasm. These results suggest that Rrp14p functions mainly in the nucleolus and also in the nucleoplasm.
Depletion of Rrp14p results in shortage of 60S ribosomal subunits
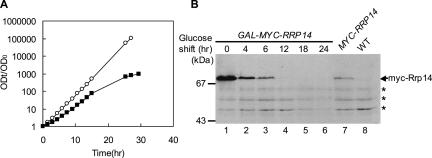
In order to achieve conditional expression of RRP14, we constructed the KM1411 strain in which the chromosomal RRP14 gene was disrupted and expression of myc–Rrp14p was driven by the galactose-inducible and glucose-repressible GAL1 promoter. When KM1411 was shifted from galactose to glucose medium, the growth rate of the cell was temporarily increased followed by decline; the growth was severely impeded 24 h after the shift (Fig. 1A). Western blot analysis using anti-myc antibodies revealed that myc–Rrp14p was overproduced when the KM1411 strain was cultured in galactose medium compared to the level in the KM1412 strain in which expression of MYC–RRP14 was controlled by the RRP14 promoter (Fig. 1B, lanes 1,7). The expression level of myc–Rrp14p was decreased after the shift of the cells from galactose to glucose medium; 6 h after the shift, the expression level of myc–Rrp14p was similar to the wild-type level and significantly decreased 12 h after the shift (Fig. 1B).
FIGURE 1.
Rrp14p depletion causes a growth defect. (A) Growth curves of W303–1A transformed with pRS414 (WT; open circle) and KM1411 (GAL1–MYC–RRP14; closed square) cultured at 30°C in SCGal medium and shifted to SC medium. The changes in optical density at 600 nm were followed after the shift. The cell cultures were diluted to keep the optical density lower than 1.0. Data are represented as ODt/OD0 on a logarithmic scale, where t is the time in hours after shifting medium. (B) Western blotting of KM1411 (GAL1–MYC–RRP14) cultured at 30°C in SCGal medium, shifted to SC medium, and cultured for the indicated time. Cell extracts from KM1411 (GAL1–MYC–RRP14, lanes 1–6), KM1412 (MYC–RRP14, lane 7) and W303–1A transformed with pRS414 (WT, negative control; lane 8) were subjected to SDS-PAGE. Western blotting using anti-myc antibodies is shown. The positions of size markers are shown on the left. Asterisks indicate nonspecific bands.
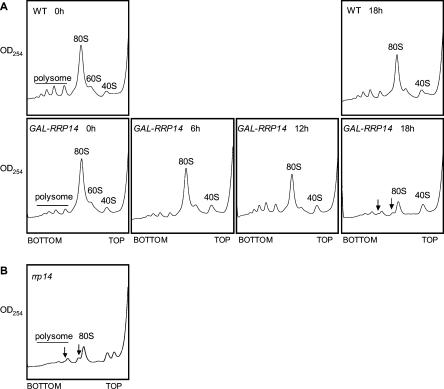
The effect of depletion of Rrp14p on ribosome biogenesis was examined by using the KM1411 strain. We performed sucrose density gradient ultracentrifugation using cell extracts with equal A260 units. The polysome profile of the strain KM1411 cultured in glucose medium for 18 h clearly revealed that the level of 60S subunits was reduced in the Rrp14p-depleted cells; 60S subunits, 80S monosomes, and polysomes decreased, and half-mer polysomes that contain 43S initiation complexes stalled at the AUG start codon appeared (Fig. 2A). These results indicate that Rrp14p is required for 60S ribosomal subunit assembly. Nevertheless, it is noteworthy that only a small amount of 40S subunits accumulated, suggesting that Rrp14p is also required for assembly and/or maintenance of 40S ribosomal subunits.
FIGURE 2.
Rrp14p depletion causes a defect in assembly of 60S ribosomal subunits. (A) The polysome profiles from W303–1A transformed with pRS414 (WT) and KM1411 (GAL1–RRP14) strains cultured at 30°C in SCGal, shifted to SC, and cultured for the indicated time. (B) The polysome profiles from the KM1421 (rrp14Δ) strain cells cultured at 30°C in SC. The positions of 40S, 60S, and 80S ribosomal particles, and polysomes are indicated. Arrows indicate half-mer polysomes.
RRP14 is required for production of 25S rRNA and also 18S rRNA
We performed [methyl-3H] methionine pulse–chase analysis to investigate whether the depletion of Rrp14p caused a defect in pre-rRNA processing. The 35S pre-rRNA, the longest detectable precursor, is cleaved to 27S and the 20S pre-rRNAs, which are further processed to the mature 25S and 18S rRNAs, respectively (Fig. 3A). The cells were pulsed with [methyl-3H] methionine for 3 min. In wild-type cells, most precursor rRNAs were processed to 25S and 18S after a 3-min chase (Fig. 3B, lanes 1–4). On the other hand, when the KM1411 (GAL–RRP14) strain was cultured in glucose medium for 18 h, the processing rate of the 27S pre-rRNA to 25S rRNA appeared to be slower and less amount of 25S rRNA was produced than that of the wild-type cells (Fig. 3B, lanes 5–8). Moreover, although the maturation from the 20S pre-rRNA to 18S rRNA was not significantly slower, a lesser amount of the mature 18S rRNA was produced in the Rrp14p-depleted cells compared to the wild-type cells (Fig. 3B). It is noteworthy that the incorporation of radioactivity of [methyl-3H] methionine into RNA reduced when Rrp14p was depleted; the relative incorporations were 26%, 25%, 22%, and 31% after chase for 0, 3, 10, and 20 min, respectively, in the Rrp14p-depleted cells compared to wild-type cells (data not shown). These data indicate that newly synthesized pre-rRNA and processed rRNA appear rather stable for at least 20 min in the Rrp14p-depleted cells. These results suggest that when Rrp14p was depleted, transcription of pre-rRNA declined, and maturation of 25S rRNA was delayed.
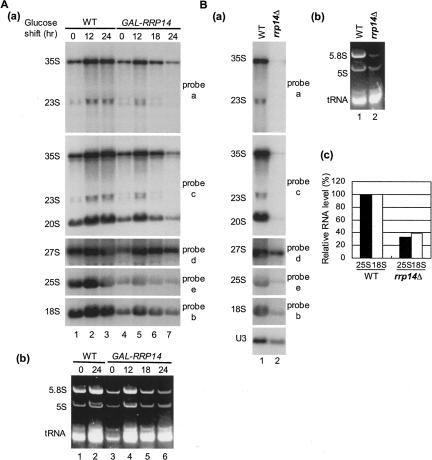
We next performed Northern analysis showing the steady-state levels of mature rRNAs and pre-rRNAs in the wild-type and KM1411(GAL-RRP14) strains cultured in galactose medium and after the shift to glucose medium for various periods of time (Fig. 4A). As the level of total RNA in the cell declined when Rrp14p was depleted, RNA samples corresponding to the equal OD600 units of cells were compared. When the wild-type cells were shifted from galactose medium to glucose medium, both 35S pre-rRNA and mature rRNAs increased and the levels were maintained 24 h after shifting to glucose medium, consistent with our previous reports (Tsuno et al. 2000; Tsujii et al. 2000). Following the transfer of the GAL1–RRP14 strain to glucose medium, the steady-state level of both 25S rRNA and 18S rRNA temporarily increased and then reduced with time of incubation in glucose medium [Fig. 4A(a)]. Compared to the level of RNAs in the wild-type cells 24 h after shifting to glucose medium, the Rrp14p-depleted cells have smaller amounts of 35S pre-rRNA and mature 25S, 18S, and 5.8S rRNAs [Fig. 4A(a), lanes 3,7, 4A(b), lanes 2,6], whereas the levels of 5S rRNA and tRNA are not significantly declined [Fig. 4A(b), lanes 2,6]. 23S RNA, which is produced by the cleavage of 35S at a site A3 without the cleavages at sites A0, A1, or A2, was not accumulated in the Rrp14p-depleted cells [Fig. 4A(a)]. This suggests that processing of 35S at A0, A1, or A2 is not delayed. These results suggest that the synthesis of 35S pre-rRNA declines when Rrp14p is depleted, consistent with the results of the [methyl-3H] methionine pulse–chase analysis.
FIGURE 4.
Northern analysis for steady-state level of rRNAs. (A) W303–1A transformed with pRS414 (WT) and KM1411 (GAL1–RRP14) cultured at 30°C in SCGal were shifted to SC and cultured for the indicated times. Total RNA corresponding to 0.5 OD600 of cells was used for Northern blot analysis. [A(a)] Northern blot analysis was carried out using 32P-labeled DNA probes a, b, c, d, and e, which are shown in Figure 3A. [A(b)] Ethidium bromide staining of a denaturing 6% polyacrylamide/8M urea gel is shown. (B) W303–1A (WT) and KM1422 (rrp14Δ) cultured at 30°C in SC medium. Total RNA corresponding to 0.5 OD600 of cells was used for Northern blot analysis. [B(a)] Northern blot analysis was carried out using 32P-labeled DNA probes a, b, c, d, and e, and a probe for U3 snoRNA. [B(b)] Ethidium bromide staining of a denaturing 6% polyacrylamide/8M urea gel is shown. [A(c)] Mature 25S and 18S rRNA levels shown in B(a) were quantified using BAS-2000 and BAS-1800 (Fuji Photo Film Co.), normalized with the U3 level, and the ratio of the radioactivity value of the rrp14Δ strain per that of wild-type strain serves as the percent.
The rrp14Δ cells grow extremely slowly with a defect in ribosome synthesis
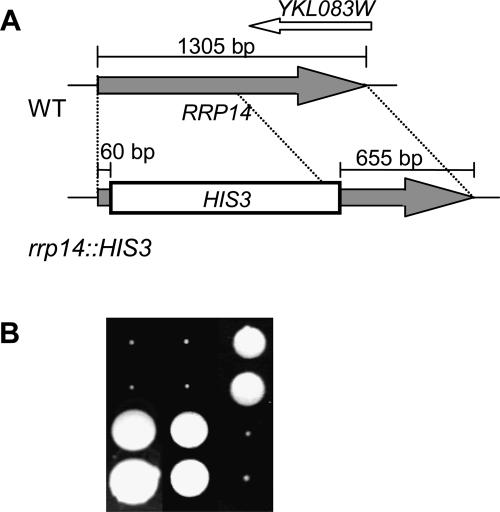
An RRP14-null allele was created by replacing one of the RRP14 ORF of the diploid strain, W303, with the HIS3 gene followed by tetrad analysis. As the 3′-end region of the RRP14 ORF overlaps the YKL083W ORF, we deleted 590 base-pairs (bp) from the 60 bp downstream initiation codon of the RRP14 ORF to avoid deleting the YKL083W ORF (Fig. 5A). PCR analysis of chromosomal DNA isolated from a candidate of W303 rrp14Δ/+ demonstrated that the resultant diploid cell carried one intact RRP14 gene and one disrupted by insertion of HIS3 (data not shown). Twenty-eight tetrads from the rrp14Δ/+ diploid were dissected. After growing for 3 d at 25°C, only two colonies were observed from each set of tetrad, and after longer growing, the tetrads yielded two large colonies and two small colonies (Fig. 5B). The cells from the small colonies were His+, indicating that the rrp14Δ cells grow extremely slowly. The rrp14Δ cells grow with doubling times of ∼15 h and ∼11 h in liquid SC medium at 25°C and 30°C, respectively. As RRP14 was reported to be essential for growth in systematic analysis (Winzeler et al. 1999), we constructed the rrp14Δ strain in another background. The rrp14Δ strain derived from BY4741 was also viable with a similar doubling time to rrp14Δ derived from W303 (data not shown).
FIGURE 5.
The rrp14Δ cells grow extremely slowly. (A) Construction of rrp14Δ with the HIS3 gene. (B) Tetrad analysis of the RRP14/rrp14Δ ∷HIS3 strain. Three sets of tetrads were cultured on a YPD plate at 25°C for 12 d.
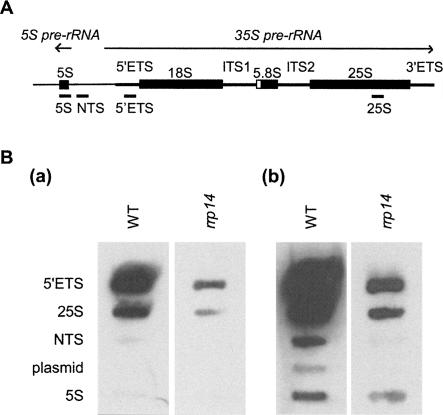
In order to examine if rrp14Δ caused a defect in ribosome synthesis, we performed sucrose density gradient ultracentrifugation and Northern analysis. The polysome profile of rrp14Δ cells shows the appearance of half-mer peaks, indicating that rrp14Δ results in the shortage of 60S subunits (Fig. 2B). Northern analysis revealed that the rrp14Δ cells have only a small amount of mature 25S, 18S, and 5.8S rRNAs, and the level of 35S pre-rRNA was also significantly declined. The rrp14Δ cells have similar levels of both 5S rRNA and tRNA to that in wild-type cells (Fig. 4B).
To examine whether the transcriptional activity of rDNA is declined in the rrp14Δ strain, we performed run-on transcription analysis (Fig. 6). Yeast cells were chilled rapidly, permeabilized, and incubated with α-32P-UTP to elongate previously initiated transcripts. The amount of RNA synthesized was measured by hybridization of the radiolabeled RNA to slot blots containing plasmids with the DNA sequences (Fig. 6A) representing transcribed portions of the 35S rRNA (5′ETS, 25S) and 5S rRNA genes. Disruption of RRP14 resulted in greatly reduced transcription of 35S rRNA genes (Fig. 6B). In contrast, 5S rRNA genes appeared to be transcribed in rrp14Δ cells at a similar rate to that in wild-type cells (Fig. 6B).
FIGURE 6.
Run-on transcription assays. (A) The rDNA transcription unit. RNA polymerase I transcribes the 35S primary rRNA transcript, whereas RNA polymerase III transcribes the 5S rRNA in the opposite direction. PCR products corresponding to the indicated segments of the 5′ external transcribed spacer (5′ETS), the 25S rRNA, NTS, and the 5S rRNA were cloned into the pTOPO plasmid. (B) Transcription run-on analysis. The wild-type and rrp14Δ strains were cultured, permeabilized, and exposed to α-32P-UTP for 10 min. RNA was extracted and hybridized to the plasmids slot-blotted onto Nitran membrane. Autoradiograph [B(b)] is exposed for longer than autoradiograph [B(a)].
Taken together, it is suggested that Rrp14p is required for both 35S pre-rRNA synthesis and 25S rRNA maturation/60S subunit assembly.
Two-hybrid interactions of Rrp14p with a ribosomal protein and regulatory factors of ribosome synthesis
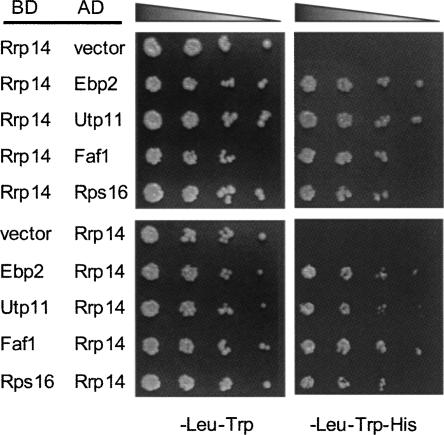
RRP14 was previously isolated in two-hybrid screening by using EBP2 as bait. In the screen, we found that Ebp2p interacts with Utp11p and Faf1p that are required for 40S subunit assembly, and ribosomal protein Rps16p besides several factors required for 60S subunit assembly (Shirai et al. 2004; for the nomenclature of ribosomal proteins, see Mager et al. 1997). Each combination of Utp11p, Faf1p, and Rps16p exhibited a positive two-hybrid interaction (Shirai et al. 2004). We proposed that Ebp2p associates with Utp11p, Faf1p, and Rps16p in the 90S particle and remains in the 60S preribosomal subunit after the 90S particle is converted into two preribosomal subunits (Shirai et al. 2004). Thus, in this study, we tested to see if Rrp14p interacts with these factors. Interestingly, Rrp14p exhibited a two-hybrid interaction with Utp11p, Faf1p, and Rps16p (Fig. 7) as well as the factors that are required for 60S subunit assembly such as Loc1p, Nop12p, and Dbp9 (data not shown). These results suggest that Rrp14p, like Ebp2p, associates with the 90S preribosomal particle and functions in pre-60S ribosomal subunits after the 90S particle is divided into two preribosomal subunits.
FIGURE 7.
Rrp14p interacts with Utp11p, Faf1p, and Rps16p. Protein–protein interactions were analyzed by the yeast two-hybrid system. Fivefold serial dilutions of cultures of L40 strain cells transformed with lexA binding domain-fused (BD) and Gal4p activation domain-fused (AD) genes were plated on SC–Trp, Leu, and SC–Trp, Leu, His. As the empty vector containing lexA binding domain (pBTM116) exhibits a background level of expression, 1 mM 3-aminotriazole was added to a SC–Trp, Leu, His plate for the lower five combinations.
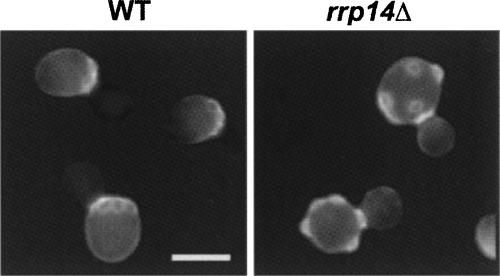
The rrp14Δ cells exhibit aberrant morphology
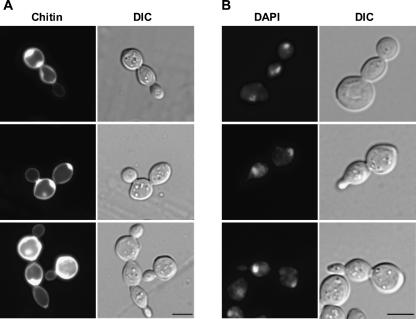
Microscopic observation revealed that rrp14Δ resulted in aberrant morphological cells. Chained cells and multibudded cells were observed in several percentages of the rrp14Δ cells even after brief sonication (Fig. 8A). Staining with 4′,6-diamidino-2-phenylindole (DAPI) showed that nuclear division had occurred without cell division (Fig. 8B). These data suggest that some cells initiate a new round of cell cycle in the absence of cell separation. Similar phenotypes were observed in the rrp14Δ cells in the background of BY4741 (data not shown).
FIGURE 8.
The rrp14Δ cells exhibit aberrant cell morphology. (A) Calcofluor staining and cell morphology (DIC) of the KM1422 (rrp14Δ) cells. (B) DAPI staining and cell morphology of the KM1422 (rrp14Δ) cells. Bars indicate 5 μm.
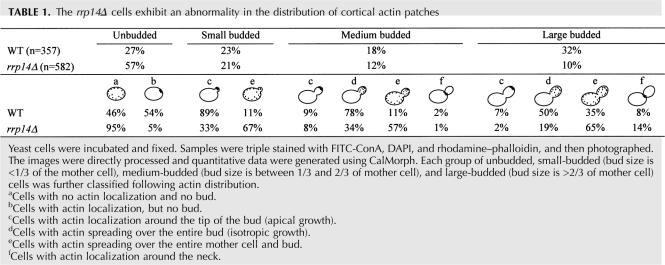
The rrp14Δ cells has a defect in cell polarity
As cell separation is intimately related to cell polarity, we examined the distribution of cortical actin patches in the rrp14Δ cells by using CalMorph, an image processing program that automatically recognizes budding yeast cell morphology (Ohtani et al. 2004; Saito et al. 2004, 2005). CalMorph can extract morphological data directly from fluorescence microscopic images of the budding yeast cell wall, actin, and nucleus stained with fluorescein isothiocyanate–Concanavalin A (FITC-ConA), rhodamine–phalloidin, and DAPI, and can generate quantitative data about yeast cell shape, nuclear shape and location, and actin distribution. Logarithmically growing cells of the wild-type and rrp14Δ strains were collected, stained, and photographed, and the images were processed with CalMorph. Bud index and actin distribution are shown in Table 1. The rrp14Δ strain shows an increased proportion of unbudded cells. The wild-type cells exhibited a polarized localization of cortical actin patches; 89% of small-budded, 87% of medium-budded, and more than a half unbudded and large-budded wild-type cells showed a staining pattern of cortical actin patches toward the bud tips (pattern c in Table 1) or throughout bud (Table 1, pattern d). In contrast, a large percentage of the rrp14Δ cells exhibited a depolarized pattern throughout the cell cycle; 95%, 67%, 57%, and 65% of unbudded, small-, medium-, and large-budded rrp14Δ cells, respectively, showed a nonpolarized localization of cortical actin patches spreading over the entire mother cell and bud (Table 1, patterns a and e). The result indicates that loss of Rrp14p function results in depolarized cell growth.
TABLE 1.
The rrp14Δ cells exhibit an abnormality in the distribution of cortical actin patches
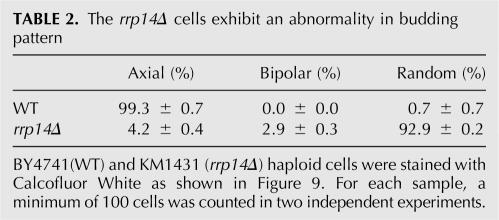
During division by budding, yeast cells polarize toward division sites that are chosen based on cell type (for review, see Chant 1999). Haploid cells exhibit an axial budding pattern in which mother and daughter cells bud immediately adjacent to their previous sites of cell division. Cells that are defective in actin-based polarized morphogenesis often display errors in bud site selection. Furthermore, Gic1p and Gic2p that interact with Rrp14p were suggested to be required for axial bud site selection (Chen et al. 1997). Thus, we examined the budding patterns of haploid wild-type and rrp14Δ cells that contained three or more calcofluor-stained bud scars. For wild-type haploids, 99.3% of the cells displayed an axial budding pattern, whereas only 4.2% of haploid rrp14Δ cells showed an axial pattern (Fig. 9; Table 2); 92.9% and 2.9% of rrp14Δ cells exhibited random and bipolar patterns, respectively. These results indicates that polarity for bud site selection is completely lost in the rrp14Δ cells.
FIGURE 9.
The rrp14Δ cells lose polarity for bud site selection. BY4741(WT) and KM1431 (rrp14Δ) haploid cells were grown to a midlogarithmic phase, sonicated for cell separation, and stained with Calcofluor White to visualize bud scars.
TABLE 2.
The rrp14Δ cells exhibit an abnormality in budding pattern
DISCUSSION
We demonstrated in this paper that Rrp14p has dual functions in ribosome synthesis and cell division. We obtained viable rrp14Δ cells with a slow growth rate in the background of both W303 and BY4741 strains. It was reported that RRP14 is necessary for viability from knockout yeast mutants (Winzeler et al. 1999). There are two possibilities for the discrepancy. In the genome-wide analysis, the positions of the deletions were not adjusted, even though 10% of ORFs in Saccharomyces cerevisiae overlap one another. As the RRP14 ORF overlaps the YKL083W ORF, which is also categorized as an essential gene in the same analysis, deletion of YKL083W might affect the viability of the rrp14Δ strain. A second possibility is that it could be considered to be inviable because of its extremely slow growth. As we delete 590 bp from the 60 bp downstream initiation codon of the RRP14 ORF and inserted HIS3, it is unlikely that functional proteins are expressed from the remaining region of the RRP14 ORF. Using the rrp14Δ strain, in this paper, we demonstrate that the function of Rrp14p is necessary for cell polarity derived from polarity of the actin cytoskeleton.
Rrp14p has a role in ribosome biogenesis
RRP14 was previously cloned in two-hybrid screening by using EBP2 as bait (Shirai et al. 2004). Ebp2p is essential for 25S rRNA maturation and 60S subunit assembly. Previous reports demonstrated that Rrp14p was copurified with Ssf1p–TAP (Fatica et al. 2002) and with Rrp1p–TAP (Horsey et al. 2004). As expected from these results, we confirmed that Rrp14p has an important role in ribosome synthesis. In the polysome profile, it appears that Rrp14p is required for the 60S subunit assembly. [Methyl-3H] methionine pulse–chase analysis reveals delay of processing from 27S to 25S rRNA in the Rrp14p-depleted strain, indicating that Rrp14p also has an important function in 25S rRNA maturation, consistent with the polysome profile and suggesting the shortage of 60S subunits. The defect of the Rrp14p depletion, however, is not restricted in 25S rRNA maturation/60S subunit assembly. [Methyl-3H] methionine pulse–chase analysis also suggests that pre-rRNA transcription declines and consequently lesser amounts of 25S and 18S rRNAs are produced in the Rrp14p-depleted strain. Run-on transcription analysis confirms that Rrp14p is required for rDNA transcription.
Rrp14p has a SURF-6 domain that is evolutionarily conserved from human to yeast. The domain is localized in the carboxyl terminus of all eukaryotic proteins, and has an average length of 191 amino acids with an average residue identity of 36% between different species. It was demonstrated that SURF-6 binds to both DNA and RNA in vitro (Magoulas et al. 1998). It is possible that Rrp14p binds to the promoter region of rDNA, affects its transcription, and subsequently binds to the 90S preribosomal particle to function in pre-rRNA processing and ribosomal subunit assembly. Ribosomal RNA transcription and pre-RNA processing are coordinated via specific components of the small subunit processome that associates with 35S pre-rRNA (Gallagher et al. 2004). In this study, we find that Rrp14p interacts with Utp11p and Faf1p. It was demonstrated that Utp11p is a component of the SSU processome and Faf1p associates with a large number of components of SSU (Dragon et al. 2002; Rempola et al. 2006). Interestingly, Faf1p was categorized as a middle-strength transcription activator in systematic analysis using the yeast one-hybrid system (Titz et al. 2006). We speculate that Rrp14p is specifically involved in transcription of rDNA, processing of pre rRNA, and 60S subunit assembly.
Rrp14p has a role in cell polarity
We demonstrate in this paper that Rrp14p has an important role in cell polarity. The data were automatically and quantitatively analyzed with the CalMorph program, which was used for the Saccharomyces cerevisiae Morphological Database (SCMD; http://scmd.gi.k.u-tokyo.ac.jp/) (Ohtani et al. 2004; Saito et al. 2004, 2005). In S. cerevisiae, following the selection of a bud site, all growth was directed into the bud. This polarized growth of the budding yeast derives from the polarity of the actin cytoskeleton; at the last step of the cell cycle, cytokinesis is accomplished by the concerted action of actomyosin ring contraction and septum formation, followed by cell separation of the daughter cell from the mother cell by the degradation of the septum. Chitinases and glucanases were transported to the bud neck and degrade the septum (Cabib et al. 2001; Baladron et al. 2002). The establishment and maintenance of a polarized actin cytoskeleton is necessary for polarized secretion. It is possible that a defect of rrp14Δ in cell polarity causes mislocalization of the enzymes, which causes a defect in cell separation. Our results are consistent with large-scale two-hybrid data showing that Rrp14p interacts with Gic1p, Gic2p, and Zds2p (Drees et al. 2001). Gic1p and Gic2p, which share sequence similarity, are suggested to be involved in initiation of budding and cellular polarization via interaction with the Rho family GTPase Cdc42p (Chen et al. 1997). It was suggested that Zds2p is a negative regulator of Cdc42p, while it appears to be involved in multiple cellular events (Bi and Pringle 1996).
Connection between ribosome synthesis and cell cycle
While we were preparing the manuscript, Oeffinger et al. (2007) reported that Rrp14p has dual functions, ribosome synthesis and positioning of the mitotic spindle. We find independently that Rrp14p has dual functions in ribosome synthesis and cell polarity. It is important to note that in our hands pre-rRNA synthesis declined when Rrp14p was depleted. This suggests that Rrp14p is involved in not only 60S subunit assembly but also in transcription of rDNA. We also found that cortical actin patches were depolarized throughout the cell cycle and the establishment of bud sites was compromised in the rrp14Δ cells. The cell morphology phenotype of our rrp14Δ cells was different from that of Rrp14p depleted cells found by Oeffinger et al. (2007); we observed that nuclei divide in the rrp14Δ strain, whereas they demonstrated that upon depletion of Rrp14, nuclei do not divide but cells form buds. The difference might be due to strain differences and/or different experimental conditions such as deletion or depletion, and asynchronicity or synchronicity. Cell polarity is related to various cell cycle events including mitotic exit and cytokinesis. It is possible that a defect of cell polarity may result in various phenotypes. As polarized growth and intracellular polarity in the budding yeast are derived from actin polarization, depolarization of actin might be a fundamental cause of various phenotypes such as the abnormal cell morphology of the rrp14Δ cells (Fig. 8) and incorrect positioning of the mitotic spindle in the Rrp14p-depleted cells (Oeffinger et al. 2007).
Recent studies suggest that several bona fide regulatory factors of ribosome synthesis have functions in cell cycle (Shafaatian et al. 1996; Du and Stillman 2002; Zhang et al. 2002; Oeffinger and Tollervey 2003; Dosil and Bustelo 2004). Moreover, it was suggested that ribosome synthesis is related to cell size homeostasis (Jorgensen and Tyers 2004; Bernstein et al. 2007). We found for the first time that a regulatory factor of ribosome synthesis has an important role in the cell polarity derived from actin localization. Further work is necessary regarding how Rrp14p connects ribosome synthesis and the cell polarity. This will provide us with useful insights into the molecular mechanisms underlying the linkage between ribosome synthesis and cell division.
MATERIALS AND METHODS
Plasmid construction
pRS316-RRP14 was constructed as follows. A PCR fragment produced from yeast genomic DNA using the primers 5′-ATAACGTCGGAACCTAATAAGC-3′ and 5′-GGATGTAATTGCCATCAGGTTT-3′ was digested with SacI and ScaI, and inserted into the SacI and SmaI sites of pRS316. pRS314–GAL1–MYC–BS, a pRS314-based expression vector that contains the GAL1 promoter, the three myc epitopes-encoding region just downstream of the initiation codon, a multicloning site, and the TDH3 terminator, was kindly provided by K. Tanaka (Hokkaido University). pRS314–GAL1–MYC–RRP14 was constructed by inserting into pRS314–GAL1–MYC–BS with the BamHI–SmaI PCR fragment of the RRP14 ORF produced from yeast genomic DNA with the primers 5′-GGGGATCCAGTAATTCACTTGAGGAG-3′, and 5′-GGCCCGGGTTATTTAGGTCCTCCCTT-3′. A plasmid expressing an myc-tagged Rrp14p by its own promoter, pRS314–MYC–RRP14, was constructed as follows. The SacI–EcoRI (followed by blunt-ending) region of the GAL1 promoter of pRS314–GAL1–MYC–RRP14 was replaced with the SacI–SmaI PCR fragment of the RRP14 promoter produced from yeast genomic DNA with the primers 5′- GGGAGCTCGAAGAAAAATGACGTGCCAA-3′ and 5′- GGCCCGGGCTCAGTCTCCTATATCTTTC-3′. For tagging N-terminal YFP, pRS304–YFP–RRP14 was created as follows. The SacI–EcoRI (followed by blunt-ending) region of the EBP2 promoter of pRS304–YFP–EBP2 was replaced with the SacI–SmaI PCR fragment of the RRP14 promoter produced as above, and the BamHI–SmaI region of the EBP2 ORF was replaced with the BamHI–SmaI PCR fragment of the RRP14 ORF as described above. pRS304–YFP–EBP2 was created by inserting it into pRS314–GAL1–MYC–BS with the BamHI–SmaI PCR fragment of the EBP2 ORF, replacing the GAL1 promoter and MYC region with the EBP2 promoter and YFP produced from pDH5 (provided from The Yeast Resource Center, Seattle), respectively, and replacing the insert into pRS304. pRS316–NOP1–CFP expressing C-terminal CFP-tagged Nop1p was constructed as follows. The HindIII–SpeI fragment cleaved from YCplac33–NOP1–GFP (kindly provided by Y. Kikuchi, University of Tokyo) was inserted into the same sites of pRS315 to create pRS315–NOP1–GFP. pRS315–NOP1–CFP was created by replacing the PstI–SpeI GFP region of pRS315–NOP1–GFP with the EcoT22I–SpeI CFP fragment amplified from a plasmid pDH3 (provided from The Yeast Resource Center, Seattle) with the primers 5′-TTTATGCATATGAAAGCAGAAGAACTTTTC-3′ and 5′-TCCACTAGTCTATTTGTATAGTTCATCCATG-3′. The HindIII–SpeI fragment of pRS315–NOP1–CFP was inserted into pRS316 to create pRS316–NOP1–CFP.
Gene disruption
A deletion-insertion mutation of RRP14 was constructed in the diploid W303 and BY4741. A primer pair, 5′-ATGAGTAATTCACTTGAGGAGCGCCTTCGTGCTAATTCAAGCGCCTTCGATGGTTTATTGTTAAGAGCTTGGTGAGCGCTAGGA-3′ and 5′-ATGTCTTCCATATCGGAATCAGACGCAATTTCATCCTGATCCTGTTCCTGTTCACTTTCTCCTCGTTCAGAATGACACGTATAG-3′, which included sequences complementary to the RRP14 ORF, was used to PCR amplify the HIS3 sequence from plasmid pRS313. The resulting PCR product (containing the HIS3 ORF) was used to transform the W303 and BY4741 diploid cells. His+ transformants were obtained and correct integration of the rrp14∷HIS3 gene at the homologous locus was confirmed by PCR. A correct integrant was sporulated and tetrads were dissected to obtain the rrp14Δ strain.
Yeast strains and media
The yeast strains used were W303–1A (MATa ade2–1 can1–100 ura3–1 leu2–3,112 trp1–1 his3–11,15), W303–1B (MATα ade2–1 can1–100 ura3–1 leu2–3,112 trp1–1 his3–11,15), KM1421 (W303–1B rrp14∷HIS3), KM1422 (W303–1A rrp14∷HIS3), KM1410 (W303–1A rrp14∷HIS3 pRS316–RRP14), KM1411 (W303–1A rrp14∷HIS3 pRS314–GAL1–MYC–RRP14), KM1412 (W303–1A rrp14∷HIS3 pRS314–MYC–RRP14), KM1419 (W303–1A rrp14∷HIS3 YFP–RRP14–TRP1 integrated at RRP14 pRS316–NOP1–CFP), BY4741(MATa his3 leu2 met15 ura3), and KM1431 (BY4741 rrp14∷HIS3). KM1419 was constructed as follows. KM1410 was transformed with the EcoRI-digested DNA fragment of pRS304–YFP–RRP14, and transformants without pRS316–RRP14 were selected on a plate containing 0.87 mg/mL 5-fluoro-orotic acid. Homologous recombination was checked by colony PCR and pRS316–NOP1–CFP was transformed. Yeast cells were grown in YPD-rich medium, synthetic complete (SC) medium containing 2% glucose (SC), or 2% galactose (SCGal), or SC dropout medium, depending on the plasmid markers.
Polysome analysis
Yeast crude cell extracts were overlaid on top of 11 mL of a 7%–47% (wt/vol) sucrose gradient and centrifuged for 3.4 h at 35,000 rpm at 4°C in an Hitachi RPS40T rotor as previously described (Shirai et al. 2004). Gradients were collected by pumping up using a peristaltic pump and monitored at 254 nm.
[Methyl-3H] methionine pulse–chase and Northern blot analyses
Processing of pre-rRNA was analyzed by [methyl-3H] methionine pulse chase as previously described (Miyoshi et al. 2001, 2003). Briefly, yeast cell cultures in SC–Met were pulsed with [methyl-3H] methionine (10 μCi/mL) for 3 min and were chased with nonradioactive methionine (500 μg/mL). Samples were taken by pouring cultures onto crushed sterile ice for preparing total RNA. Eight micrograms of total RNA were analyzed by electrophoresis and blotted to a Nytran membrane. The upper part of the membrane was sprayed with En3Hance (NEN) and exposed to film for 14 d. The lower part of the membrane was probed for U3 snoRNA as a loading marker. For Northern blot analysis, the total RNA equivalent of 0.5 OD600 or 1.5 OD600 of cells was separated on an agarose gel or a denaturing 6% polyacrylamide/8 M urea gel, respectively, and transferred to a Nytran membrane by capillary or stained with ethidium bromide. Northern hybridization was carried out using 32P-labeled oligonucleotide probes.
Run-on transcription analysis
Yeast strains were grown in liquid YPD to an OD600 of 0.4–0.5. Cells corresponding to 3.0 OD600 units were harvested. Run-on transcription was performed according to the protocol of Elion and Warner (1986) with the following change: the cells were incubated with α-32P-UTP for 10 min. Slot-blot hybridization was performed according to the protocol of Gallagher et al. (2004) with the following changes: PCR fragments were cloned into the pTOPO plasmid and spotted onto a Nitran membrane (Schleicher & Schuell). Plasmids containing 5′ETS, 25S, and NTS of rDNA were kindly provided by Baserga and Gallagher (Yale University) (Gallagher et al. 2004). The sequences of the primers for the 5S rRNA gene are 5′-CGGGATCCACCCATAACACCTCTCACTCCCAC-3′ and 5′-CGCTCGAGAGATTGCAGCACCTGAGTTTCGCG-3′.
Two-hybrid system
Two kinds of plasmid for production of lexA binding domain-fusion proteins and Gal4p activation domain-fusion proteins were cotransformed into yeast L40 strain cells as previously described (Tsujii et al. 2000). Leu+ Trp+ transformants were selected and fivefold serial dilutions of the cell cultures were stamped on SC–Leu, Trp, and His plates containing 3-amino-1, 2, and 4-triazole and incubated at 30°C for 3 d unless indicated.
Western blotting
Western blotting was performed following standard techniques, and signals were visualized by Enhanced Chemiluminescence (Amersham), as instructed by the manufacturer. Mouse anti-myc monoclonal antibodies (9E10; BAbCO) and horseradish peroxidase-conjugated sheep antimouse IgG antibodies (NA931; Amersham) were used at 1:500 and 1:1000, respectively.
Analysis of cell morphology
The cells were cultured and stained with fluorescein isothiocyanate–Concanavalin A (FITC-ConA) for cell wall identification, with 4,6-diamidino-2-phenylindole (DAPI) to localize nuclei and with rhodamine-conjugated phalloidin to visualize the actin distribution following the CalMorph user manual with minor modification. Yeast cells were grown to the exponential phase (4–6 × 106 cells/mL) at 30°C. The cells were fixed for 30 min at 30°C in growth medium with formaldehyde (3.7% final concentration) and potassium phosphate buffer (100 mM, pH 6.8). The cells were collected, resuspended in formaldehyde solution (3.7% formaldehyde and 100 mM potassium phosphate, pH 6.8), and incubated for 45 min at 30°C. The cells were washed with PBS (137 mM NaCl, 2.68 mM KCl, 8.03 mM Na2HPO4, and 1.47 mM KH2PO4) twice and stained with 20 U/mL rhodamine–phalloidin in PBS with 0.1% Triton X-100 at 4°C overnight. The cells were washed with PBS and then with P buffer (10 mM sodium phosphate and 150 mM NaCl, pH 7.2), and stained with 24 μg/mL FITC-ConA in P buffer for 5 min at room temperature. The cells were washed with P buffer and resuspended in P buffer. After a brief sonication, the cells were placed on slides, mounted with mounting solution (1 mg/mL p-phenylenediamine, 0.1× PBS, 90% glycerol) containing 1 μg/mL DAPI, and observed under a fluorescence microscope (Olympus BX51) equipped with a 100× objective. All images were captured using a black-and-white charge-coupled device-coupled camera (Nippon Roper CoolSNAP HQ) and Slide Book 4 Digital Microscopy software (Intelligent Imaging Innovations). The data were processed with Adobe Photoshop and automatically analyzed with the CalMorph program.
Analysis of budding patterns
Yeast cells were cultured and sonicated for cell separation, and Calcofluor White was added to 2 μg/mL concentration. Bud scars were visualized by fluorescence microscopy and bud patterns were defined as described previously (Chant and Pringle 1995; Nelson et al. 2003). Only cells containing three or more bud scars were counted. Bud patterns were considered axial only if scars resided immediately adjacent to one another. Bud scars residing on opposite thirds of the cells were considered to be bipolar. Cells were considered to have random patterns if bud scars resided in the middle third of the cell.
ACKNOWLEDGMENTS
We thank R. Sternglanz, K. Tanaka, Y. Kikuchi, B. Ono, J.E.G. Gallagher, and S.J. Baserga, for yeast strains and/or plasmids. The CalMorph program has been provided freely by the University of Tokyo for use in this publication. This research was supported by Grants-in-Aid for Scientific research from the Japan Society for the Promotion of Science and from the Ministry of Education, Culture, Sports, Science, and Technology of Japan to K.M. C.H. and C.S. were supported by JSPS Research Fellowships for Young Scientists.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.553807.
REFERENCES
- Baladron, V., Ufano, S., Duenas, E., Martin-Cuadrado, A.B., del Rey, F., Vazquez de Aldana, C.R. Eng1p, an endo-1,3-β-glucanase localized at the daughter side of the septum, is involved in cell separation in Saccharomyces cerevisiae . Eukaryot. Cell. 2002;1:774–786. doi: 10.1128/EC.1.5.774-786.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassler, J., Grandi, P., Gadal, O., Lebmann, T., Petfalski, E., Tollervey, D., Lechner, J., Hurt, E. Identification of a 60S preribosomal particle that is closely linked to nuclear export. Mol. Cell. 2001;8:517–529. doi: 10.1016/s1097-2765(01)00342-2. [DOI] [PubMed] [Google Scholar]
- Bernstein, K.A., Bleichert, F., Bean, J.M., Cross, F.R., Baserga, S.J. Ribosome biogenesis is sensed at the start cell cycle checkpoint. Mol. Biol. Cell. 2007;18:953–964. doi: 10.1091/mbc.E06-06-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi, E., Pringle, J.R. ZDS1 and ZDS2, genes whose products may regulate Cdc42p in Saccharomyces cerevisiae . Mol. Cell. Biol. 1996;16:5264–5275. doi: 10.1128/mcb.16.10.5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabib, E., Roh, D.H., Schmidt, M., Crotti, L.B., Varma, A. The yeast cell wall and septum as paradigms of cell growth and morphogenesis. J. Biol. Chem. 2001;276:19679–19682. doi: 10.1074/jbc.R000031200. [DOI] [PubMed] [Google Scholar]
- Chant, J. Cell polarity in yeast. Annu. Rev. Cell Dev. Biol. 1999;15:365–391. doi: 10.1146/annurev.cellbio.15.1.365. [DOI] [PubMed] [Google Scholar]
- Chant, J., Pringle, J.R. Patterns of bud-site selection in the yeast Saccharomyces cerevisiae . J. Cell Biol. 1995;129:751–765. doi: 10.1083/jcb.129.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, G.C., Kim, Y.J., Chan, C.S. The Cdc42 GTPase-associated proteins Gic1 and Gic2 are required for polarized cell growth in Saccharomyces cerevisiae . Genes & Dev. 1997;11:2958–2971. doi: 10.1101/gad.11.22.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosil, M., Bustelo, X.R. Functional characterization of Pwp2, a WD family protein essential for the assembly of the 90 S pre-ribosomal particle. J. Biol. Chem. 2004;279:37385–37397. doi: 10.1074/jbc.M404909200. [DOI] [PubMed] [Google Scholar]
- Dragon, F., Gallagher, J.E.G., Compagnone-Post, P.A., Mitchell, B.M., Porwancher, K.A., Wehner, K.A., Wormsley, S., Settlage, R.E., Shabanowitz, J., Osheim, Y., et al. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature. 2002;417:967–970. doi: 10.1038/nature00769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drees, B.L., Sundin, B., Brazeau, E., Caviston, J.P., Chen, G.C., Guo, W., Kozminski, K.G., Lau, M.W., Moskow, J.J., Tong, A., et al. A protein interaction map for cell polarity development. J. Cell Biol. 2001;154:549–571. doi: 10.1083/jcb.200104057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, Y.C., Stillman, B. Yph1p, an ORC-interacting protein: Potential links between cell proliferation control, DNA replication, and ribosome biogenesis. Cell. 2002;109:835–848. doi: 10.1016/s0092-8674(02)00773-0. [DOI] [PubMed] [Google Scholar]
- Elion, E.A., Warner, J.R. An RNA polymerase I enhancer in Saccharomyces cerevisiae . Mol. Cell. Biol. 1986;6:2089–2097. doi: 10.1128/mcb.6.6.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatica, A., Cronshaw, A.D., Dlakic, M., Tollervey, D. Ssf1p prevents premature processing of an early pre-60S ribosomal particle. Mol. Cell. 2002;9:341–351. doi: 10.1016/s1097-2765(02)00458-6. [DOI] [PubMed] [Google Scholar]
- Gallagher, J.E.G., Dunbar, D.A., Granneman, S., Mitchell, B.M., Osheim, Y., Beyer, A.L., Baserga, S.J. RNA polymerase I transcription and pre-rRNA processing are linked by specific SSU processome components. Genes & Dev. 2004;18:2506–2517. doi: 10.1101/gad.1226604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi, P., Rybin, V., Baßler, J., Petfalski, E., Strauß, D., Marzioch, M., Schäfer, T., Kuster, B., Tschochner, H., Tollervey, D., et al. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol. Cell. 2002;10:105–115. doi: 10.1016/s1097-2765(02)00579-8. [DOI] [PubMed] [Google Scholar]
- Hampichamchai, P., Jakovljevic, J., Horsey, E., Miles, T., Roman, J., Rout, M., Meagher, D., Imai, B., Guo, Y., Brame, C.J., et al. Composition and functional characterization of yeast 66S ribosome assembly intermediates. Mol. Cell. 2001;8:505–515. doi: 10.1016/s1097-2765(01)00344-6. [DOI] [PubMed] [Google Scholar]
- Horsey, E.W., Jakovljevic, J., Miles, T.D., Harnpicharnchai, P., Woolford J.L., Jr Role of the yeast Rrp1 protein in the dynamics of pre-ribosome maturation. RNA. 2004;10:813–827. doi: 10.1261/rna.5255804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen, P., Tyers, M. How cells coordinate growth and division. Curr. Biol. 2004;14:1014–1027. doi: 10.1016/j.cub.2004.11.027. [DOI] [PubMed] [Google Scholar]
- Kressler, D., Linder, P., de la Cruz, J. Protein trans-acting factors involved in ribosome biogenesis in Saccharomyces cerevisiae . Mol. Cell. Biol. 1999;19:7897–7912. doi: 10.1128/mcb.19.12.7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mager, W.H., Planta, R.J., Ballesta, J.-P.G., Lee, J.C., Mizuta, K., Suzuki, K., Warner, J.R., Woolford J.L., Jr A new nomenclature for the cytoplasmic ribosomal proteins of Saccharomyces cerevisiae . Nucleic Acids Res. 1997;25:4872–4875. doi: 10.1093/nar/25.24.4872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoulas, C., Fried, M. The Surf-6 gene of the mouse surfeit locus encodes a novel nucleolar protein. DNA Cell Biol. 1996;15:305–316. doi: 10.1089/dna.1996.15.305. [DOI] [PubMed] [Google Scholar]
- Magoulas, C., Zatsepina, O.V., Jordan, P.W., Jordan, E.G., Fried, M. The SURF-6 protein is a component of the nucleolar matrix and has a high binding capacity for nucleic acids in vitro. Eur. J. Cell Biol. 1998;75:174–183. doi: 10.1016/S0171-9335(98)80059-9. [DOI] [PubMed] [Google Scholar]
- Miyoshi, K., Miyakawa, T., Mizuta, K. Repression of rRNA synthesis due to a secretory defect requires the C-terminal silencing domain of Rap1p in Saccharomyces cerevisiae . Nucleic Acids Res. 2001;29:3297–3303. doi: 10.1093/nar/29.16.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi, K., Shirai, C., Mizuta, K. Transcription of genes encoding trans-acting factors required for rRNA maturation/ribosomal subunit assembly is coordinately regulated with ribosomal protein genes and involves Rap1 in Saccharomyces cerevisiae . Nucleic Acids Res. 2003;31:1969–1973. doi: 10.1093/nar/gkg278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuta, K., Warner, J.R. Continued functioning of the secretory pathway is essential for ribosome synthesis. Mol. Cell. Biol. 1994;14:2493–2502. doi: 10.1128/mcb.14.4.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, B., Kurischko, C., Horecka, J., Mody, M., Nair, P., Pratt, L., Zougman, A., McBroom, L.D., Hughes, T.R., Boone, C., et al. RAM: A conserved signaling network that regulates Ace2p transcriptional activity and polarized morphogenesis. Mol. Biol. Cell. 2003;14:3782–3803. doi: 10.1091/mbc.E03-01-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeffinger, M., Tollervey, D. Yeast Nop15p is an RNA-binding protein required for pre-rRNA processing and cytokinesis. EMBO J. 2003;22:6573–6583. doi: 10.1093/emboj/cdg616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeffinger, M., Fatica, A., Rout, M.P., Tollervey, D. Yeast Rrp14p is required for ribosomal subunit synthesis and for correct positioning of the mitotic spindle during mitosis. Nucleic Acids Res. 2007;35:1354–1366. doi: 10.1093/nar/gkl824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtani, M., Saka, A., Sano, F., Ohya, Y., Morishita, S. Development of image processing program for yeast cell morphology. J. Bioinform. Comput. Biol. 2004;1:695–709. doi: 10.1142/s0219720004000363. [DOI] [PubMed] [Google Scholar]
- Polzikov, M., Zatsepina, O., Magoulas, C. Identification of an evolutionary conserved SURF-6 domain in a family of nucleolar proteins extending from human to yeast. Biochem. Biophys. Res. Commun. 2005;327:143–149. doi: 10.1016/j.bbrc.2004.11.148. [DOI] [PubMed] [Google Scholar]
- Rempola, B., Karkusiewicz, I., Piekarska, I., Rytka, J. Fcf1p and Fcf2p are novel nucleolar Saccharomyces cerevisiae proteins involved in pre-rRNA processing. Biochem. Biophys. Res. Commun. 2006;346:546–554. doi: 10.1016/j.bbrc.2006.05.140. [DOI] [PubMed] [Google Scholar]
- Saito, T.L., Ohtani, M., Sawai, H., Sano, F., Saka, A., Watanabe, D., Yukawa, M., Ohya, Y., Morishita, S. SCMD: Saccharomyces cerevisiae morphological database. Nucleic Acids Res. 2004;32:D319–D322. doi: 10.1093/nar/gkh113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito, T., Sese, J., Nakatani, Y., Sano, F., Yukawa, M., Ohya, Y., Morishita, S. Data mining tools for Saccharomyces cerevisiae morphological database. Nucleic Acids Res. 2005;33:W753–W757. doi: 10.1093/nar/gki451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafaatian, R., Payton, M.A., Reid, J.D. PWP2, a member of the WD-repeat family of proteins, is an essential Saccharomyces cerevisiae gene involved in cell separation. Mol. Gen. Genet. 1996;252:101–114. doi: 10.1007/BF02173210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirai, C., Takai, T., Nariai, M., Horigome, C., Mizuta, K. Ebp2p, the yeast homolog of Epstein-Barr virus nuclear antigen 1-binding protein 2, interacts with factors of both the 60S and 40S ribosomal subunit assembly. J. Biol. Chem. 2004;279:25353–25358. doi: 10.1074/jbc.M403338200. [DOI] [PubMed] [Google Scholar]
- Titz, B., Thomas, S., Rajagopala, S.V., Chiba, T., Ito, T., Uetz, P. Transcriptional activators in yeast. Nucleic Acids Res. 2006;34:955–967. doi: 10.1093/nar/gkj493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschochner, H., Hurt, E. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 2003;13:255–263. doi: 10.1016/s0962-8924(03)00054-0. [DOI] [PubMed] [Google Scholar]
- Tsujii, R., Miyoshi, K., Tsuno, A., Matsui, Y., Toh-e, A., Miyakawa, T., Mizuta, K. Ebp2p, yeast homologue of a human protein that interacts with Epstein-Barr virus Nuclear Antigen 1, is required for pre-rRNA processing and ribosomal subunit assembly. Genes Cells. 2000;5:543–553. doi: 10.1046/j.1365-2443.2000.00346.x. [DOI] [PubMed] [Google Scholar]
- Tsuno, A., Miyoshi, K., Tsujii, R., Miyakawa, T., Mizuta, K. RRS1, a conserved essential gene, encodes a novel regulatory protein required for ribosome biogenesis in Saccharomyces cerevisiae . Mol. Cell. Biol. 2000;20:2066–2074. doi: 10.1128/mcb.20.6.2066-2074.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venema, J., Tollervey, D. Ribosome synthesis in Saccharomyces cerevisiae . Annu. Rev. Genet. 1999;33:261–311. doi: 10.1146/annurev.genet.33.1.261. [DOI] [PubMed] [Google Scholar]
- Warner, J.R. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999;24:437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- Warner, J.R. Nascent ribosomes. Cell. 2001;107:133–136. doi: 10.1016/s0092-8674(01)00531-1. [DOI] [PubMed] [Google Scholar]
- Warner, J.R., Udem, S.A. Temperature sensitive mutations affecting ribosome synthesis in Saccharomyces cerevisiae . J. Mol. Biol. 1972;65:243–257. doi: 10.1016/0022-2836(72)90280-x. [DOI] [PubMed] [Google Scholar]
- Winzeler, E.A., Shoemaker, D.D., Astromoff, A., Liang, H., Anderson, K., Andre, B., Bangham, R., Benito, R., Boeke, J.D., Bussey, H., et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Woolford J.L., Jr, Warner, J.R. The ribosome and its synthesis. In The molecular and cellular biology of the yeast Saccharomyces: Genome dynamics, protein synthesis, and energetics . In: Broach J.R., et al., editors. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1991. pp. 587–626. [Google Scholar]
- Zhang, Y., Yu, Z., Fu, X., Liang, C. Noc3p, a bHLH protein, plays an integral role in the initiation of DNA replication in budding yeast. Cell. 2002;109:849–860. doi: 10.1016/s0092-8674(02)00805-x. [DOI] [PubMed] [Google Scholar]