Abstract
Though the “RNA world” hypothesis has gained a central role in ideas concerning the origin of life, the scenario concerning its emergence remains uncertain. It has been speculated that the first scene may have been the emergence of a template-dependent RNA synthetase ribozyme, which catalyzed its own replication: thus, “RNA replicase.” However, the speculation remains uncertain, primarily because of the large sequence length requirement of such a replicase and the lack of a convincing mechanism to ensure its self-favoring features. Instead, we propose a nucleotide synthetase ribozyme as an alternative candidate, especially considering recent experimental evidence suggesting the possibility of effective nonenzymatic template-directed synthesis of RNA. A computer simulation was conducted to support our proposal. The conditions for the emergence of the nucleotide synthetase ribozyme are discussed, based on dynamic analysis on a computer. We suggest the template-dependent RNA synthetase ribozyme emerged later, perhaps after the emergence of protocells.
Keywords: nucleotide pool, self-replication, RNA catalysis, Darwinian evolution, Monte Carlo method
INTRODUCTION
A major difficulty in the field concerning the origin of life, which aims to study the rise of primary life on the Earth from early, nonlife backgrounds, comes from the fact that we cannot define well enough what the primary life was like. An essential interdependence, as we know from modern life, of the dependence of DNA replication on the protein enzyme catalysis and the dependence of protein synthesis on the DNA-based coding breeds the “RNA world” (Gilbert 1986) hypothesis, which proposed that the genetic and catalytic functions were both RNA based in the primary life forms, thus avoiding the paradox “which came first, DNA or protein?” The hypothesis has gained quite a lot of supporting evidence and has become the central hypothesis in the field (Joyce 2002; Joyce and Orgel 2006).
The RNA world hypothesis presents a model for the molecular nature of early life. However, we are still puzzled about the detailed form of this primary life. Was it cellular? Or could it have been just naked RNA molecules? Because a cellular form would seem to be very complex to arise de novo, it is reasonable to assume that it arose from a naked molecular form via Darwinian evolution. It is then important to identify naked RNA molecules capable of self-replication, which may emerge from a nonlife background and have the potential to evolve into a cellular form. This is the so-called problem of the origin of the RNA world (Orgel 2004; Joyce and Orgel 2006).
Though the prebiotic synthesis of nucleotides still faces some problems, the experiments seeking to mimic it have made many achievements (Miller 1998; Schwartz 1998; Orgel 2004; Joyce and Orgel 2006). It is reasonable to imagine that the nonlife background from which the naked RNA molecules arose is some nucleotide pools that existed in the early Earth (Joyce and Orgel 2006). Typically, it is speculated that, from the nucleotide pools, the naked RNA species emerging first was an RNA replicase, a template-dependent RNA synthetase ribozyme capable of catalyzing the copying of its own sequence, via an intermediate of its complement (thus, self-replicating) (Cech 1989; Joyce 2002; Joyce and Orgel 2006). A major reason for such speculation is that the mechanism of nonenzymatic template-directed copying (Joyce 1987; Orgel 1992; Kozlov and Orgel 2000) seems not to be sufficiently effective to support the replication of any RNA species as a potential ribozyme (with a certain sequence length). Without replication, Darwinian evolution cannot occur.
However, there remain two significantly problematic aspects of this speculation. First, the template-dependent RNA synthetase ribozyme itself seems to be too long (typically ∼160–200 nucleotides [nt]) (Johnston et al. 2001) to have appeared de novo (e.g., based on mineral catalysis, up to ∼50 nt is reasonable) (Ferris 2002) and to be too long to be copied by its own catalysis (e.g., with a successive extension of 14 nt as a “good” result) (Johnston et al. 2001). A recent effort improved the successive extension to around or a little longer than 20 nt, but the ribozyme itself is still ∼200 nt long (Zaher and Unrau 2007). Second, without a cellular context, the replicase should be self-favoring enough (i.e., overcoming the parasite problem) to ensure that it could “grow out” from the nucleotide pool. They may recognize themselves by “tags” (Wein and Maizels 1987) or rely on limited dispersal (perhaps on mineral surfaces) (Szabo et al. 2002). However, these mechanisms seem insufficiently reliable, as discussed previously (Ma and Yu 2006), leading to the proposal of “intramolecular replicase.” However, this intramolecular replicase hypothesis still faces the first problem.
Recently, it was revealed that the nonenzymatic template-directed synthesis of RNA could be much more efficient (1000-fold rate enhancement) with the aid of intercalating molecules (so-called molecular midwives) (Jain et al. 2004). These intercalating molecules were suggested to have existed on the early Earth (Hud and Anet 2000). Such evidence shook the belief that RNA replicase was the first ribozyme to emerge in the RNA world. Indeed, if all RNA species could self-replicate without the help of an RNA synthetase ribozyme, why would we insist that such a ribozyme should be the first to emerge? RNA species with other advantages may also grow out from the nucleotide pool.
In a nucleotide pool, if all RNA species compete for nucleotides to replicate themselves, based on the mechanism of nonenzymatic template-directed synthesis, nucleotides should be scarce. Then, if an RNA species could catalyze the synthesis of nucleotides (a “nucleotide synthetase ribozyme”), nucleotides may be more abundant near the species and that species may replicate more efficiently than others. Actually, the synthesis of a nucleotide could be a chain process of a number of steps, which may involve a series of different ribozymes (Joyce 2002, Muller 2006; Joyce and Orgel 2006). The so-called nucleotide synthetase ribozyme, which may have emerged first, would tend to be a ribozyme enhancing the rate-limiting step in the chain (see Comments and Perspectives, below, for a more detailed discussion).
When the population of the nucleotide synthetase ribozyme spreads in the pool, Darwinian evolution may occur, perhaps moving toward improving its efficiency and the emergence of ribozymes participating in other steps of the chain process. Meanwhile, if a ribozyme favoring the formation of a membrane, such as an amphiphilic molecule synthetase ribozyme, could appear, it could cooperate with the nucleotide synthetase ribozyme, and a protocell may emerge. In the protocell, the amphiphilic molecule synthetase ribozyme could replicate efficiently utilizing the nucleotides synthesized by the nucleotide synthetase ribozyme, and the nucleotide synthetase ribozymes could replicate more efficiently than their naked analogs, because the nucleotides would not diffuse away due to the limitation of the “cell membrane.”
Thus, a nucleotide synthetase ribozyme is a good candidate as the first (naked) RNA species emerging in the RNA world and has the potential to evolve into a cellular form. The template-dependent RNA synthetase ribozyme, RNA replicase, may emerge subsequently, in the cellular form.
In the present study, we conducted a computer simulation, using the Monte Carlo method, to explore the possibility that a nucleotide synthetase ribozyme emerged in a nucleotide pool and to analyze the dynamics involved (the conditions for its successful emergence). We use “emerge” to mean “appear and spread” (grow into a population).
COMPUTER SIMULATION
We conducted a computer simulation using a Monte Carlo method similar to the cellular automata approach. The principle of the method can be found in a previous report (Ma et al. 2007). Generally, we assumed a two-dimensional surface for the system, with an N×N square grid. The raw materials, nucleotides, and RNA molecules in a grid cell may interact with each other and may move to adjacent cells. An RNA molecule containing a special domain (arbitrarily presumed to be an active domain) may act as a nucleotide synthetase ribozyme. Each action in the system is assumed with some probability (Table 1).
TABLE 1.
Parameters used in the Monte Carlo simulations
Setting values of parameters based on realistic conditions may be an effective method for simulation. However, the parameters in the model (e.g., the probability of nucleotide decay and the probability of breaking a phosphodiester bond) could be affected by a number of factors (e.g., temperature, pH value, concentrations of metal ions, etc.), which are yet not quite certain according to our current knowledge regarding the conditions prevalent on the primordial Earth.
A practical method is to estimate first a relative magnitude of the parameters (especially for the probabilities), based on their fundamental relationships, and then determine it quantitatively, but still approximately, via test simulations, with a goal of achieving the successful emergence of the nucleotide synthetase ribozyme. Then, with a definite set of parameters according with the magnitude, we can explore how a certain parameter would affect the emergence of the nucleotide synthetase ribozyme by varying it over a much broader range. The conditions for the emergence of the ribozyme could then be inferred, and the realistic possibility of the emergence of the ribozyme may be evaluated when we have more knowledge regarding the conditions on the primordial Earth in the future.
For the estimation of the magnitude, basically, PNFR should be much greater than PNF, PLT should be much greater than PRL, PND may be a little greater than PNF, PBB may be of the same order as PRL, and PAT may be of the same order as PSP. Additionally, PFP is a parameter related to the replicating fidelity [in the model, Fidelity=1/(3×PFP+1)] (Ma et al. 2007). Because nonenzymatic replication tends not to have a very high fidelity, PFP should not be too small (e.g., PFP=0.05, corresponding to Fidelity=0.87). By subsequent test simulations with a goal of achieving the successful emergence of the nucleotide synthetase ribozyme, we determined the relative magnitude of the parameters (Table 1). It should be noted that there is no absolute significance to the values (e.g., 0.001–0.01 for PND), and the determination of the magnitude was just to make the setting of parameters basically reasonable and to facilitate subsequent analysis by narrowing the scope. The setting of the common parameter lists in Figure 2 and Figure 4, below, is just according to the magnitude.
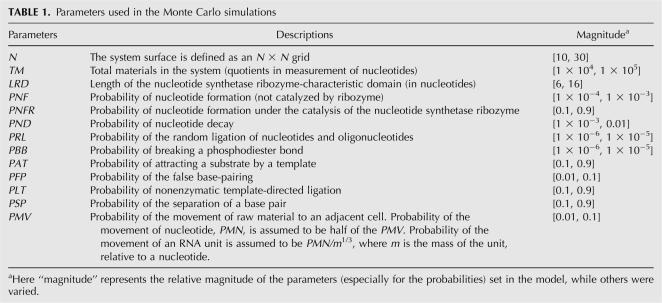
FIGURE 2.
Emergence of a nucleotide synthetase ribozyme in a nucleotide pool. Circles represent the nucleotide synthetase ribozyme. Pluses, stars, and triangles represent three control RNA species with a presumed random sequence domain as long as the characteristic domain of the nucleotide synthetase ribozyme. (A) LRD=8 nt (AGCAUGCU); the first sequence is inoculated at step 1×104; (B) LRD=8 nt (UGAUGCAG); the first sequence is inoculated at step 1×104; (C) LRD=16 nt (AGUCAGCAUGCUGACU); the first sequence is inoculated at step 1×104; (D) LRD=6 nt (UCGCGA); the first sequence appears inside the pool (emerging de novo). The common parameter list: N=20, TM = 4×104, PNF=2×10−4, PNFR=0.9, PND=0.01, PRL= 2×10−6, PBB=1×10−6, PAT=0.1, PFP=0.01, PLT=0.9, PSP=0.9, and PMV=0.01. (Note: the Y-axes of the subgraphs are not on the same scale.)
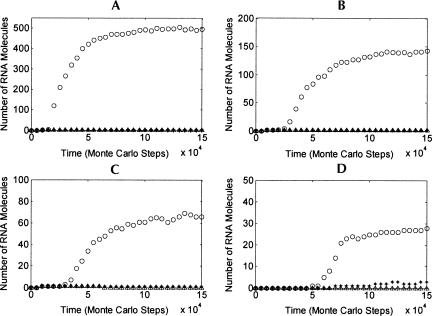
FIGURE 4.
The dynamics of the spread of the nucleotide synthetase ribozyme. The X-axes tick labels are values of the corresponding parameters. The Y-axes represent the molecular number of the nucleotide synthetase ribozyme (the first bar in a four-bar group) and the control RNA species (the following three bars in the four-bar group) after 1.5×105 Monte Carlo steps, which are the means of the results of the program running with three different random seeds. Ten sequences of each species were inoculated into the system at step 1×104. The common parameter list: N=20, TM=4×104, LRD=10, PNF=2×10−4, PNFR=0.2, PND=0.01, PRL=2×10−6, PBB=1×10−6, PAT=0.1, PFP=0.01, PLT=0.9, PSP=0.9, and PMV=0.01. For a subgraph specializing on a parameter, values of other parameters can be found in the common list.
The system can be imagined as a shallow pool with a rocky bottom or even a thin fluid layer spreading on the surface of a rock. The events happening in the system can be described as follows. Raw materials may form nucleotides at a low level without the aid of a ribozyme (Fig. 1). After a period of time, some random RNA molecules may be synthesized by random ligation inside the system or adjacently outside the system (perhaps at the rocky bottom of the pool by mineral catalysis) (Ferris et al. 1996; Ferris 2002) and introduced into the system. The RNA molecules may replicate via nonenzymatic template-directed synthesis at some level (Fig. 1B). They compete for nucleotides for their replication, and nucleotides become scarce. If one of the random RNA species can catalyze the synthesis of nucleotides, it may have more building blocks around itself and replicate more efficiently (Fig. 1C). The aim of the simulation is to see whether the nucleotide synthetase ribozyme could spread in the system and become a constant population on a significant scale and how the parameters in the model affect such an outcome. The constant population of the nucleotide synthetase ribozyme may then supply a base for further Darwinian evolution that might take place in the system (Fig. 1D,E; see Comments and Perspectives, below, for a detailed discussion).
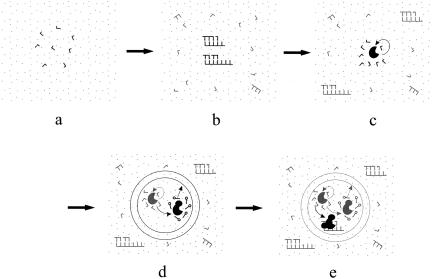
FIGURE 1.
A scheme describing the events that happen successively in the modeled system (A–C) and that might happen thereafter (D,E), which as a whole could be read as a proposed process for the origin of the RNA world. In each subgraph, new events are drawn in black, while old events are in gray. (A) Formation of nucleotides (L-shaped) from raw materials (dots) without ribozyme catalysis. (B) Nonenzymatic template-directed synthesis of RNA. The template molecules are synthesized by random ligations. (C) The emergence of a nucleotide synthetase ribozyme. The synthesized nucleotides around the ribozyme may be priorly used by itself in its replication (arrow). (D) The emergence of an amphiphilic molecule synthetase ribozyme, which promotes the formation of the protocell's membrane (arrow), thus limiting the diffusion of the synthesized nucleotides. The synthesized nucleotides may also be used by the amphiphilic molecule synthetase ribozyme (arrow). (E) The emergence of a template-directed RNA synthetase ribozyme, which speeds the replication of the RNA species in the protocell. The three ribozymes cooperate to facilitate the replication (reproduction) of the whole protocell.
RESULTS
The nucleotide synthetase ribozyme can spread in the system if such a molecule could appear (Fig. 2). The presumed active domain of the ribozyme could be palindromic (Fig. 2A) or nonpalindromic (Fig. 2B), with the former being easier to spread, because each turn of template copying produces a copy of the ribozyme at once. The domain could be relatively long (Fig. 2C), but a longer domain would mean a lower efficiency of spreading. If the ribozyme is not introduced into the system from the outside, it may even emerge de novo in the system (Fig. 2D), but with much lower efficiency.
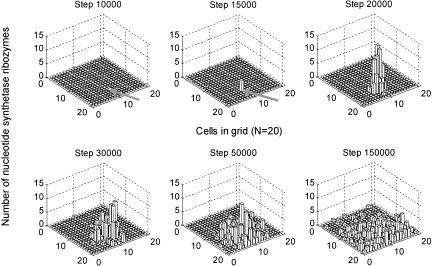
Figure 3 shows the spatial distribution of the nucleotide synthetase ribozyme during its spread. Apparently, the spread originated from the replication of the first molecule of the nucleotide synthetase ribozyme, which then gains more “nutrients” (nucleotides) around it. Via diffusion and replication, the ribozyme molecules radiate into other grid cells of the system and become a constant population.
FIGURE 3.
The spatial distribution of the nucleotide synthetase ribozyme during its spread. The case corresponds to that in Figure 2A. The arrows point to the grid cell where the first molecule of the nucleotide synthetase ribozyme is introduced (inoculated).
The dynamics of the spread of the nucleotide synthetase ribozyme were studied, and the relationship between the parameters and outcomes is shown in Figure 4. To avoid the effect of randomness on the outcome, we inoculated 10 (instead of 1) molecules of the nucleotide synthetase ribozyme and each control RNA species and did the simulations with three different random seeds for each parameter case and then used the means of the results in the bar figures.
DISCUSSION
Analysis of the results
The results show that the spread of a nucleotide synthetase ribozyme in the pool is quite robust (Figs. 2, 4). The appearance of the first molecule of the nucleotide synthetase ribozyme is a key event. Though it is possible that the first molecule may appear de novo in the pool by random ligation of nucleotides and oligonucleotides (Fig. 2D), this is less efficient. To achieve higher efficiency of the de novo appearance in the pool, the rate of random ligation in the pool should be increased. However, a higher rate of random ligation in the pool also means a lower efficiency of the spread (Fig. 4, PRL). This may be interpreted as the higher the rate of random ligation in the pool, the more random RNA species in the pool would appear, competing for limited resources in their replication. Though the template-directed synthesis of the nucleotide synthetase ribozyme may be favored by exploiting short oligonucleotides (especially dimers and trimers) forming in random ligations, other unrelated RNA species may also exploit them. Thus, the key limiting factor is still the scarcity of the resources. The emergence of the ribozyme means its appearance and spread. If its appearance is difficult in the condition favoring its spread, and its spread is difficult in the condition favoring its appearance, it seems that the appearance and the spread should occur in different backgrounds. A possible scenario in the context of our model is that the first ribozyme is synthesized at the surface of “the rocky bottom of the pool” by mineral catalysis (Ferris et al. 1996; Ferris 2002), which favors its appearance, and is then released (introduced) as a “seed” into the pool, in which conditions with a relatively low rate of random ligation favor its spread.
A spatial limitation is important to assure the advantage of the nucleotide synthetase ribozymes for themselves—synthesizing “nutrients” (nucleotides) around, thus favoring their own replication (Figs. 3, 4, PMV). However, too serious a limitation on moving is also not advantageous (Fig. 4, PMV). The reason is that raw materials elsewhere are less available for the nucleotide synthetase ribozymes. A good solution is the formation of a protocell, in which the diffusion of synthesized nucleotides is limited, while the cell can itself move around to “catch” raw materials elsewhere. The solution provides an immediate direction for evolution: the emergence of a ribozyme favoring the formation of a cell membrane, perhaps an amphiphilic molecule synthetase ribozyme (Fig. 1D).
Without the help of a template-dependent RNA synthetase ribozyme (replicase), replication of the nucleotide synthetase ribozyme would have to rely on nonenzymatic template-directed copying. The low efficiency of such nonenzymatic template-directed copying may lead to unsuccessful spread (Fig. 4, PLT=0.01, 0.05, 0.1). Meanwhile, it seems that high efficiency of nonenzymatic template-directed copying does not always favor the spread of the nucleotide synthetase ribozyme (Fig. 4, PLT=0.9), which may be because nonenzymatic template-directed replication of other random RNA species becomes an issue, especially of shorter RNAs that would more readily replicate, thus making them competitive.
It is not surprising that the resistance of RNA molecules to degradation is important for the successful spread of a nucleotide synthetase ribozyme (Fig. 4, PBB). It has been suggested that the RNA world originated in a cold or even frozen environment (Bada and Lazcano 2002; Vlassov et al. 2004; Trinks et al. 2005). Such an environment may favor the RNA molecules' resistance to degradation. It is also not surprising that there is an optimal middle value for the rate of the attraction of a template to substrates (Fig. 4, PAT) and the separation of templates and substrates (Fig. 4, PSP), because successful template-directed replication requires a balance between the attraction of substrates and the dissociation of the synthesized chains. It is somewhat surprising that there was actually an optimal middle value for the decay rate of nucleotides (Fig. 4, PND). This may be because too high a decay rate of nucleotides makes the complete replication of the nucleotide synthetase ribozyme difficult, while too low a decay rate renders the raw materials for synthesizing nucleotide scarce and the nucleotide synthetase ribozyme could not benefit from its advantage. It is not surprising that a higher fidelity of replication (lower probability of false base-pairing) results in more successful spreading (Fig. 4, PFP).
It is interesting that there was an optimal concentration for the solution in the pool (Fig. 4, N). For a total materials of 4×104 in this case, the optimal number of cells in the grid is 40×40. Both higher concentrations (e.g., corresponding to grid cells of 10×10) and lower concentrations (e.g., corresponding to grid cells of 100×100) lead to unsuccessful spread of the nucleotide synthetase ribozyme. This can be a practical guide to related experiments in the future.
To spread successfully, the demands on the efficiency of the nucleotide synthetase ribozyme seem not to be too high. As shown in Figure 4, PNFR, 0.05–0.1 for PNFR is sufficient, while the PNF is 2×10−4, meaning that the efficiency of the ribozyme-catalyzed reaction should be 250–500 times higher than that of a corresponding nonenzymatic reaction. As shown in Figure 4, PNF, PNF should be <4×10−4 to 2×10−4, while the PNFR is 0.2, meaning that the efficiency of the ribozyme-catalyzed reaction should be 500–1000 times higher than that of a corresponding nonenzymatic reaction. In total, a rate enhancement of ∼1000-fold is plenty enough for the nucleotide synthetase ribozyme to spread successfully.
Without the help of an RNA synthetase ribozyme (replicase), the nucleotide synthetase ribozyme could not be very long (Fig. 4, LRD) to spread successfully. The longer the nucleotide synthetase ribozyme is, the more difficult its complete replication becomes. A domain of 14–16 nt is possible at the least (see also Fig. 2C) in the system we assumed. It is possible that, by adjusting parameters, the nucleotide synthetase ribozyme with such an active domain of 20–30 nt (perhaps as a two stem–loop structure) could spread successfully in the system. It is unclear whether in practice an RNA molecule with a core active domain of only ∼20–30 nt could act as a nucleotide synthetase ribozyme. However, it should not be forgotten that a famously simple ribozyme, the hammerhead ribozyme, which catalyzes the cleavage/ligation of RNA chains, contains a core sequence only ∼20–30 nt, and the ribozyme itself can be just ∼30–40 nt long (Hammann and Lilley 2002). To date, ribozymes constructed via in vitro molecular evolution similar or related to the nucleotide synthetase ribozyme are ∼80–140 nt long, with rate enhancements of ∼105–107-fold (Lorsch and Szostak 1994; Unrau and Bartel 1998; Huang 1998; Curtis and Bartel 2005). A shorter nucleotide synthetase ribozyme with a much lower level of efficiency (e.g., 1000-fold as above) does not seem unreasonable.
Comments and perspectives
The synthesis of a nucleotide could be a chain process of many steps, like the synthesis of ribose, bases, nucleosides, nucleoside phosphates, and nucleoside polyphosphates, which may involve a series of quite different ribozymes (Joyce 2002; Joyce and Orgel 2006; Muller 2006). The nucleotide synthetase ribozyme mentioned here tends to refer to a ribozyme participating in the rate-limiting step in the chain (e.g., the ribozyme catalyzing the synthesis of ribose if the sugar is the scarcest intermediate product). Other ribozymes in the chain may also emerge when the rate-limiting step changes because of the emergence of the first ribozyme, under the general selective pressure of a need for nucleotides. Then, a “naked” RNA-based biochemical pathway for the synthesis of nucleotides may finally emerge; in such a scenario the ribozymes involved cooperate to synthesize nucleotides. It is not clear how plausible it is that the naked pathway could emerge, without a cellular context. Perhaps further computer simulation would help address this, but that is beyond the scope of this study.
As mentioned in the analysis of the results, for the spread of a nucleotide synthetase ribozyme, the formation of a protocell is advantageous, because the diffusion of the synthesized nucleotides is limited, while the protocell can itself move around to catch raw materials elsewhere for the synthesis of nucleotides. Though the amphiphilic molecules forming the protocell's membrane may be synthesized nonenzymatically and the membrane vesicle may self-replicate automatically, coupling the self-replication of the ribozymes inside, there is apparently a selective pressure for the emergence of a ribozyme favoring the synthesis of the amphiphilic molecules (Fig. 1D; Szostak et al. 2001). The membrane should be permeable to raw materials but impermeable to the synthesized nucleotides and should withstand conditions favoring ribozyme catalysis, such as divalent cations in millimolar concentration. It has been proposed that instead of the more popular phospholipids of modern cells, the membrane of protocells in the RNA world would be composed of fatty acids/fatty acid alcohols (Muller 2006). Interestingly, such membranes may withstand millimolar concentrations of Mg2+ and show different permeability to NMPs, NDPs, and NTPs, at least in certain membrane compositions and at certain temperatures. In the context of this article, it would be interesting to investigate further the permeability of such membranes to ribose, bases, and nucleosides. No matter how, it is yet not quite sure what kind(s) of amphiphilic molecules the protocell's membrane should be composed of, and more experiments designed to explore the permeability of those possible membranes to intermediate products in the process of nucleotide synthesis are needed.
Compared with the template-dependent RNA synthetase ribozyme (replicase), the advantage of the nucleotide synthetase ribozyme being the first ribozyme to emerge in the RNA world can be seen at four points at least. First, it may be quite short, which is important for both the appearance of the first molecule and its subsequent spread (replication). Second, the parasite problem is not as serious, because it is the products (nucleotides, as “nutrients”), instead of the ribozyme itself, that are occupied by molecular parasites; thus its spread would not be seriously blocked. Third, its emergence naturally brings a need for the emergence of the protocell (to prevent the diffusion of nucleotides, but also to be able to move around to search for raw materials elsewhere). Finally, its emergence at the very beginning, especially considering the possible establishment of a naked RNA-based biochemical pathway for nucleotide synthesis, as mentioned above, may greatly alleviate the dependence of the early RNA world on the prebiotic nonenzymatic synthesis of nucleotides, which seems not to be very efficient (Schwartz 1998; Orgel 2004; Joyce and Orgel 2006). The template-dependent RNA synthetase ribozyme, a more complex ribozyme, may have emerged after the emergence of protocells (Fig. 1E), in which context the parasite problem has been already solved and the special supply of nucleotides is assured.
In fact, a recent work has proposed a scenario in the early stages of the RNA world consistent with our suggestion here (Manrubia and Briones 2007). First, “short, replicating RNA sequences are selected to perform a simple function.” Apparently, the short RNA sequences could only undergo nonenzymatic template-directed replication. The “simple function,” as suggested in the present article, should be “catalyzing the synthesis of nucleotides.” Second, “the combination of independently evolved populations may have facilitated the emergence of larger molecules able to perform more complex function (including RNA replication).” The function of “RNA replication” mentioned here referred to “catalyzing the template-directed RNA synthesis.” Though we cannot comment on the evolution mode (“modular evolution,” as it was called by the authors) in the context of our work here, we are in agreement that the function of RNA replication, carried by the template-directed RNA synthetase ribozymes, is a more complex function that should emerge later, rather than first.
Actually, the scenario described here provides a solution to the so-called another chicken-and-egg paradox in the context of the RNA world hypothesis (Joyce and Orgel 2006): “Without evolution, it appears unlikely that a self-replicating ribozyme could arise, but without some form of self-replication, there is no way to conduct an evolutionary search for the first, primitive self-replicating ribozyme.” Our answer is that without evolution, a simple nucleotide synthetase ribozyme may self-replicate by nonenzymatic template-directed synthesis and arise, and in this form of self-replication, an evolutionary search for the template-dependent RNA synthetase ribozyme, which may speed the self-replication, could be conducted.
Key related experiments in the future would be the search for nucleotide synthetase ribozymes of sufficiently short (e.g., about or a little longer than 30 nt), even those with a relatively low efficiency (e.g., about or even less than 1000-fold rate enhancement), and to accumulate more evidence on efficient, nonenzymatic template-directed RNA synthesis, perhaps with the aid of so-called midwife molecules (Hud and Anet 2000; Jain et al. 2004). If the idea raised here is plausible, correspondingly, efforts in “recreating an RNA world” (Muller 2006) to understand the arising of the RNA world, including those of constructing an RNA-based protocell (Bartel and Unrau 1999; Szostak et al. 2001; Chen et al. 2006), should pay more attention to nucleotide synthetase ribozymes than to template-dependent RNA synthetase ribozymes (replicases).
The model we have described here provides a possible route by which a functional polymer establishes its predominance in a pool of random polymers capable of undergoing nonenzymatic template-directed replication, offering the possibility of further Darwinian evolution. We emphasize that the function emerging first is related to catalyzing the synthesis of nutrients (thus, related to metabolism), instead of catalyzing template-directed replication itself. Then, even if the nonenzymatic template-directed replication of RNA was not efficient enough to support the replication of a functional ribozyme, the idea is still significant. We may expect that, for example, some RNA-like polymer in a “pre-RNA world” (Joyce 2002; Orgel 2004; Joyce and Orgel 2006) may undergo nonenzymatic template-directed replication efficiently. If so, the model still describes a scenario for the emergence of living species, with the feature of self-replication and Darwinian evolution (Luisi 1998) from a nonlife background. Then, of course, the RNA world may be an evolving result of such a pre-RNA world.
ACKNOWLEDGMENTS
Financial support by the National Natural Science Foundation of China (No.90409013) and the National 973 Fundamental Research Program (No.2005CB523308), Hubei Natural Science Foundation of China (No.2003S2113), and State Key Laboratory of Software Engineering in Wuhan University is gratefully acknowledged.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.658507.
REFERENCES
- Bada, J.L., Lazcano, A. Origin of life—Some like it hot, but not the first biomolecules. Science. 2002;296:1982–1983. doi: 10.1126/science.1069487. [DOI] [PubMed] [Google Scholar]
- Bartel, D.P., Unrau, P.J. Constructing an RNA world. Trends Biochem. Sci. 1999;24:M9–M13. [PubMed] [Google Scholar]
- Cech, T.R. RNA chemistry—ribozyme self-replication. Nature. 1989;339:507–508. doi: 10.1038/339507a0. [DOI] [PubMed] [Google Scholar]
- Chen, I.A., Hanczyc, M.M., Sazani, P.L., Szostak, J.W. Protocells: Genetic polymers inside membrane vesicles. In: Gesteland R.F., et al., editors. The RNA world. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2006. pp. 57–88. [Google Scholar]
- Curtis, E.A., Bartel, D.P. New catalytic structures from an existing ribozyme. Nat. Struct. Mol. Biol. 2005;12:994–1000. doi: 10.1038/nsmb1003. [DOI] [PubMed] [Google Scholar]
- Ferris, J.P. Montmorillonite catalysis of 30–50 mer oligonucleotides: Laboratory demonstration of potential steps in the origin of the RNA world. Orig. Life Evol. Biosph. 2002;32:311–332. doi: 10.1023/a:1020543312109. [DOI] [PubMed] [Google Scholar]
- Ferris, J.P., Hill, A.R., Liu, R., Orgel, L.E. Synthesis of long prebiotic oligomers on mineral surfaces. Nature. 1996;381:59–61. doi: 10.1038/381059a0. [DOI] [PubMed] [Google Scholar]
- Gilbert, W. The RNA world. Nature. 1986;319:618. [Google Scholar]
- Hammann, C., Lilley, D.M.J. Folding and activity of the hammerhead ribozyme. ChemBioChem. 2002;3:691–700. doi: 10.1002/1439-7633(20020802)3:8<690::AID-CBIC690>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Huang, F.Q. RNA enzymes with two small-molecule substrates. Chem. Biol. 1998;5:669–678. doi: 10.1016/s1074-5521(98)90294-0. [DOI] [PubMed] [Google Scholar]
- Hud, N.V., Anet, F.A.L. Intercalation-mediated synthesis and replication: A new approach to the origin of life. J. Theor. Biol. 2000;205:543–562. doi: 10.1006/jtbi.2000.2084. [DOI] [PubMed] [Google Scholar]
- Jain, S.S., Anet, F.A.L., Stahle, C.J., Hud, N.V. Enzymatic behavior by intercalating molecules in a template-directed ligation reaction. Angew. Chem. Int. Ed. Engl. 2004;43:2004–2008. doi: 10.1002/anie.200353155. [DOI] [PubMed] [Google Scholar]
- Johnston, W.K., Unrau, P.J., Lawrence, M.S., Glasner, M.E., Bartel, D.P. RNA-catalyzed RNA polymerization: Accurate and general RNA-templated primer extension. Science. 2001;292:1319–1325. doi: 10.1126/science.1060786. [DOI] [PubMed] [Google Scholar]
- Joyce, G.F. Nonenzymatic template-directed synthesis of informational macromolecules. Cold Spring Harb. Symp. Quant. Biol. 1987;52:41–51. doi: 10.1101/sqb.1987.052.01.008. [DOI] [PubMed] [Google Scholar]
- Joyce, G.F. The antiquity of RNA-based evolution. Nature. 2002;418:214–221. doi: 10.1038/418214a. [DOI] [PubMed] [Google Scholar]
- Joyce, G.F., Orgel, L.E. Progress toward understanding the origin of the RNA world. In: Gesteland R.F., et al., editors. The RNA world. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 2006. pp. 23–568. [Google Scholar]
- Kozlov, I.A., Orgel, L.E. Nonenzymatic template-directed synthesis of RNA from monomers. Mol. Biol. 2000;34:781–789. [PubMed] [Google Scholar]
- Lorsch, J.R., Szostak, J.W. In vitro evolution of new ribozymes with polynucleotide kinase activity. Nature. 1994;371:31–36. doi: 10.1038/371031a0. [DOI] [PubMed] [Google Scholar]
- Luisi, P.L. About various definitions of life. Orig. Life Evol. Biosph. 1998;28:613–622. doi: 10.1023/a:1006517315105. [DOI] [PubMed] [Google Scholar]
- Ma, W.T., Yu, C.W. Intramolecular RNA replicase: Possibly the first self-replicating molecule in the RNA world. Orig. Life Evol. Biosph. 2006;36:413–420. doi: 10.1007/s11084-005-9006-1. [DOI] [PubMed] [Google Scholar]
- Ma, W.T., Yu, C.W., Zhang, W.T. Monte Carlo simulation of early molecular evolution in the RNA World. Biosystems. 2007;90:28–39. doi: 10.1016/j.biosystems.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Manrubia, S.C., Briones, C. Modular evolution and increase of functional complexity in replicating RNA molecules. RNA. 2007;13:97–107. doi: 10.1261/rna.203006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, S.L. The endogenous synthesis of organic compound. In: Brack A., editor. The molecular origins of life: Assembling pieces of the puzzle. Cambridge University Press; Cambridge: 1998. pp. 59–85. [Google Scholar]
- Muller, U.F. Re-creating an RNA world. Cell. Mol. Life Sci. 2006;63:1278–1293. doi: 10.1007/s00018-006-6047-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orgel, L.E. Molecular replication. Nature. 1992;358:203–209. doi: 10.1038/358203a0. [DOI] [PubMed] [Google Scholar]
- Orgel, L.E. Prebiotic chemistry and the origin of the RNA world. Crit. Rev. Biochem. Mol. Biol. 2004;39:99–123. doi: 10.1080/10409230490460765. [DOI] [PubMed] [Google Scholar]
- Schwartz, A.W. Origins of the RNA world. In: Brack A., editor. The molecular origins of life: Assembling pieces of the puzzle. Cambridge University Press; Cambridge: 1998. pp. 237–254. [Google Scholar]
- Szabo, P., Scheuring, I., Czaran, T., Szathmary, E. In silico simulations reveal that replicators with limited dispersal evolve towards higher efficiency and fidelity. Nature. 2002;420:340–343. doi: 10.1038/nature01187. [DOI] [PubMed] [Google Scholar]
- Szostak, J.W., Bartel, D.P., Luisi, P.L. Synthesizing life. Nature. 2001;409:387–390. doi: 10.1038/35053176. [DOI] [PubMed] [Google Scholar]
- Trinks, H., Schroder, W., Biebricher, C.K. Ice and the origin of life. Orig. Life Evol. Biosph. 2005;35:429–445. doi: 10.1007/s11084-005-5009-1. [DOI] [PubMed] [Google Scholar]
- Unrau, P.J., Bartel, D.P. RNA-catalyzed nucleotide synthesis. Nature. 1998;395:260–263. doi: 10.1038/26193. [DOI] [PubMed] [Google Scholar]
- Vlassov, A.V., Johnston, B.H., Landweber, L.F., Kazakov, S.A. Ligation activity of fragmented ribozymes in frozen solution: Implications for the RNA world. Nucleic Acids Res. 2004;32:2966–2974. doi: 10.1093/nar/gkh601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wein, A.M., Maizels, N. 3′ Terminal tRNA-like structures tag genomic RNA molecules for replication: Implications for the origin of protein synthesis. Proc. Natl. Acad. Sci. 1987;84:7383–7387. doi: 10.1073/pnas.84.21.7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaher, H.S., Unrau, P.J. Selection of an improved RNA polymerase ribozyme with superior extension and fidelity. RNA. 2007;13:1017–1026. doi: 10.1261/rna.548807. [DOI] [PMC free article] [PubMed] [Google Scholar]