Abstract
We report that MOP3 is a general dimerization partner for a subset of the basic-helix–loop–helix (bHLH)-PER–ARNT–SIM (PAS) superfamily of transcriptional regulators. We demonstrated that MOP3 interacts with MOP4, CLOCK, hypoxia-inducible factor 1α (HIF1α), and HIF2α. A DNA selection protocol revealed that the MOP3-MOP4 heterodimer bound a CACGTGA-containing DNA element. Transient transfection experiments demonstrated that the MOP3-MOP4 and MOP3-CLOCK complexes bound this element in COS-1 cells and drove transcription from a linked luciferase reporter gene. We also deduced the high-affinity DNA binding sites for MOP3-HIF1α complex (TACGTGA) and used transient transfection experiments to demonstrate that the MOP3-HIF1α and MOP3-HIF2α heterodimers bound this element, drove transcription, and responded to cellular hypoxia. Finally, we found that MOP3 mRNA expression overlaps in a number of tissues with each of its four potential partner molecules in vivo.
The PAS domain is found in a variety of proteins that play roles in development and adaptation to the environment (1–6). The PAS domain is also commonly found in proteins that harbor basic-helix–loop–helix (bHLH) domains, and that act in pairs as heterodimeric transcription factors (6–8). For example, the aryl hydrocarbon receptor nuclear translocator (ARNT) protein has been shown to act as a general partner for a number structurally related proteins that appear to act as sensors for environmental stimuli. ARNT dimerizes with the aryl hydrocarbon receptor (AHR) and mediates metabolic responses to 2,3,7,8-tetrachlorodibenzo-p-dioxin and related environmental contaminants. ARNT also dimerizes with hypoxia-inducible factor 1α (HIF1α) and mediates cellular responses to low oxygen and glucose (2, 6).
Recently, a number of “orphan” bHLH-PAS proteins have emerged from searches of expressed sequence tag databases and low stringency hybridization screens (9–14). For newly discovered bHLH-PAS proteins that have close homologs (e.g., HIF1α and HIF2α§ or ARNT and ARNT2), partnering and DNA binding specificity can often be predicted from amino acid sequence similarities in their bHLH-PAS domains (9, 12, 14). For divergent orphans like MOP3, MOP4, and MOP5, amino acid sequence does not provide the information necessary for similar predictions. In an attempt to characterize this class of orphans, we have employed a series of assays that allow us to: (i) identify heterodimeric partnerships, (ii) determine the DNA response element bound by these heterodimers, (iii) verify that these complexes drive transcription in mammalian cells, and (iv) identify those tissues where these partnerships may occur. This report describes application of this approach to two bHLH-PAS orphans, MOP3 and MOP4.
MATERIALS AND METHODS
Reagents.
Oligonucleotides were supplied by GIBCO/BRL and designated OL522 5′-GACAGTATCACGCCTCTCCTT-3′, OL579 5′-AGCGGCGTCGGGATAAAATGA-3′, OL595 5′-ATGCTGAACTGTGCCGAAAACTGT-3′, OL656 5′-GAACAGTGGGGTGGGTCCTCTTT-3′, OL990 5′-GGAATTCTGAGTCTGAAC-3′, OL991 5′-GGAATTCCACGCTCAGG-3′, OL992 5′-GGAATTCTGAGTCTGAAC(N)13CCTGAGCGTGGATTCC-3′, OL1116 5′-GATCGGAC- ACGTGACCATTGGTCACGTGTCCATTGGACACG- TGACC-3′, OL1117 5′-GATCGGTCACGTGTCCAATGGACACGTGACCAATGGTCACGTGTCC-3′, OL1155 5′-GATCGGATACGTGACCATTGGTTACGTGTCCAT-TGGATACGTGACC-3′, and OL1156 5′-GATCGGTCACGTATCCAATGGACACGTAACCAATGGTCACGTATCC-3′. The yeast LexA fusion plasmid pBTM116 was provided by P. Bartel and S. Fields (State University of New York, Stony Brook). The yeast strain L40 was a kind gift of S. Hollenberg (Fred Hutchinson Cancer Research Center, Seattle, WA). The yeast strain AMR70 was constructed by Rolf Sternglanz, and was a kind gift of S. Hollenberg. LexA antiserum was a kind gift of J. W. Little (University of Arizona). pSGBCU11 was a kind gift of Stephen Goff (CIBA–Geigy, Research Triangle Park, NC). Human CLOCK was a kind gift of T. Nagase (Kazusa DNA Research Institute, Chiba, Japan). Mammalian expression vectors were purchased from GIBCO/BRL (pSVSport) and Promega (pTarget). Antibodies specific for MOP3 and MOP4 were prepared against peptides specific for each protein as described (15). The MOP3 peptide sequence was DNDQGSSSPSNDEAAC, and the MOP4 peptide sequence was KDKGSSLEPRQHFNALDVGC.
Expression Plasmid Construction.
Yeast expression plasmids harboring the LexA DNA binding domain fused to the bHLH-PAS domains of HIF1α (PL856), HIF2α (PL857), MOP4 (PL859), AHR (PL739), and ARNT (PL701) have been described (14). LexAbHLH-PAS fusion plasmids for MOP3 (PL831) and CLOCK (PL828) were constructed in pBTM116 by an identical approach. Plasmids harboring the full-length ORFs of MOP3, MOP4, and CLOCK were constructed by PCR amplification of the ORF of each cDNA and cloned into the appropriate vectors for expression in yeast or mammalian systems. For yeast expression of full-length proteins, PCR products were cloned into the appropriate sites of pSGBCU11. For mammalian expression, PCR products were cloned into pSVSport and pTarget. The yeast expression vector for full-length ARNT has been described (PL574) (14). The yeast expression vector for full-length MOP3 was designated PL694. Mammalian expression vectors for ARNT (PL87), HIF1α (PL611), and HIF2α (PL447) have been described (14, 16). Mammalian expression vectors were constructed for MOP3 (PL691 and PL861), MOP4 (PL695 and PL871), and CLOCK (PL941).
Two-Hybrid cDNA Library Screen.
The yeast interaction trap was performed using the yeast strain L40 (MATa, his3Δ200, trp1-901, leu2-3,112, ade2, LYS∷lexAop4HIS, URA3∷lexAop8lacZ) or AMR70 (MATα, his3, lys2, trp1, leu2, URA∷lexAop8-lacZ) as described (14, 17, 18). The bait plasmid (PL859) was a fusion of the bHLH-PAS domain of MOP4 to the DNA binding domain of LexA (14). The MOP4 bait construct was used to screen a human fetal brain cDNA library fused to the transactivation domain of Gal4 (CLONTECH) and transformants were plated on selective media (minus tryptophan, uracil, histidine, and leucine). The cDNAs from surviving colonies, positive for lacZ activity were sequenced by the chain termination method (19). These sequences were analyzed using the blast algorithm in May of 1997 (20).
Interaction Screen Against Known bHLH-PAS Proteins.
LexAbHLH-PAS fusion proteins (“baits”) of HIF1α, HIF2α, MOP3, MOP4, AHR, ARNT, and CLOCK were transformed into the L40 strain of yeast. The full-length (“fish”) MOP3 and ARNT plasmids were transformed into the AMR70 yeast strain, and these transformants were plated onto yeast complete media plates (21). The L40 yeast harboring the bait constructs were replica plated onto these yeast complete media plates and mated for 8 hr at 30°C. The plates were then replica plated onto selective media and grown for an additional day at 30°C. 5-Bromo-4-chloro-3-indolyl β-d-galactoside (X-Gal) overlay assays were performed to determine the relative expression of the lacZ reporter gene (22). Western blot analysis, using LexA-specific sera, was performed on extract from each transformant to confirm expression of the fusion protein (14).
DNA Binding Specificity.
To determine high-affinity DNA binding sites for MOP3-MOP4 heterodimers, site selection and amplification was performed as described (23). Briefly, reticulocyte lysate expressed MOP3 and MOP4 proteins (≈0.5 fmol each) were incubated with DNA oligonucleotide randomers corresponding to ≈7 × 107 different nucleotide sequences. Randomers were generated and amplified by PCR using oligonucleotides OL990 and OL991 as primers and OL992 as template. After incubating the complexes with the randomers for 30 min at 30°C, samples were loaded directly on 4% polyacrylamide-TBE (90 mM Tris/64.6 mM boric acid/2.5 mM EDTA, pH 8.0) gels to separate MOP3-MOP4 bound DNA from free DNA (23). Gel slices corresponding to the migration of bound DNA were excised, incubated overnight in TE (10 mM Tris/1 mM EDTA, pH 8.0), and the eluate subjected to additional PCR using oligonucleotides OL990 and OL991.
Cell Culture and Transient Transfection.
Transient transfections of COS-1 cells were performed by the Lipofectamine protocol (GIBCO) as described (14). To mimic hypoxia, 100 μM of cobalt chloride was included in the cell growth media and incubated at 37°C until harvest. To monitor the transcriptional activity of the MOP3-MOP4 or MOP3-CLOCK heterodimers, a synthetic reporter was constructed by annealing phosphorylated oligonucleotides OL1116 and OL1117 and cloning them into the BglII site in the reporter plasmid pGL3p (Promega). To measure the transcriptional activity of the MOP3-HIF1α or MOP3-HIF2α heterodimers, a synthetic reporter was constructed by annealing phosphorylated oligonucleotides OL1155 and OL1156 and then cloning them as above. Luciferase levels were reported in relation to β-galactosidase activity as described (14).
mRNA Expression Analysis.
To generate antisense riboprobes, partial cDNAs of the mouse MOP3 and MOP4 were cloned into plasmid vectors harboring bacteriophage promoters. A partial 1.2-kb mouse fragment of MOP3 was obtained by PCR of a mouse kidney cDNA library using oligonucleotides OL579 and OL656, and cloned into pGEM-T in the T7 orientation. For MOP4, reverse transcription–PCR was performed on 3 μg of E17.5d placenta total RNA with oligonucleotides OL522 and OL595. The resultant fragment was subcloned in pGEM-T in the SP6 orientation. Total RNA from various mouse tissues was prepared using the Trizol reagent (GIBCO/BRL) according to manufacturer’s protocols. Ribonuclease protection assay (RPA) was performed as described for both MOP3 and MOP4 (24). For in situ analysis, sense and antisense MOP3 and MOP4 riboprobes were generated with [α-[35S]thio]UTP, 80 μCi (Amersham, >1,000 Ci/mmol; 1 Ci = 37 GBq) as the radioactive ribonucleotide and subjected to alkaline hydrolysis for 13 min at 60°C as described (25). Tissue sections (5 μm) were processed and hybridized with the specific riboprobes (25).
RESULTS
MOP4 Two-Hybrid Library Screen.
The MOP4 bait plasmid was used to screen a human fetal brain cDNA library fused to the transactivation domain of Gal4. After screening ≈7 × 105 colonies, 21 survived selection and were blue in the presence of 5-bromo-4-chloro-3-indolyl-β-d-galactoside. blast searches revealed that seven of these clones represented four independent MOP3 cDNA fragments. These cDNAs differed in their first 57 codons from the MOP3 cDNA we have described previously (GenBank accession no. U60415). These 57 amino acids are identical to that reported by a second group, and appear to be derived from a second promoter (13). All subsequent functional studies were done using constructs derived from the MOP3 cDNAs identified by the yeast interaction trap.
MOP3 and MOP4 Screened Against Known bHLH-PAS Proteins.
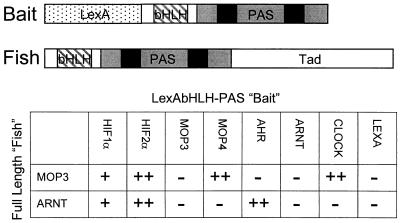
To confirm the specificity of the MOP3-MOP4 interaction, we reversed the interaction trap strategy and screened full-length MOP3 against all bHLH-PAS proteins available in this laboratory. As a positive control we compared these results to a parallel screen using full-length ARNT. Western blot analysis using anti-LexA sera indicated approximately equal expression levels for all fusions (data not shown). The full-length MOP3 protein interacted strongly with LexAbHLH-PAS fusions of MOP4, CLOCK, and HIF2α and weakly with HIF1α (Fig. 1). No interaction of full-length MOP3 could be detected with LexA fusions of MOP3, AHR, ARNT, or the LexA control. Full-length ARNT demonstrated robust interactions with HIF2α and the AHR, and weaker interactions with HIF1α. We did not detect full-length ARNT interactions with LexAbHLH-PAS fusions of MOP3, MOP4, CLOCK, ARNT, or the LexA control (Fig. 1).
Figure 1.
Interaction panel of LexAbHLH-PAS fusion proteins with full-length MOP3 and ARNT. (Upper) Schematic representation of the LexAbHLHPAS “bait” and the full-length “fish.” The bHLH and PAS domains are boxed. The “A” and “B” repeats of the PAS domains are indicated. The transactivation domain of the full-length “fish” is indicated. (Lower) LexA fusion protein plasmids containing the bHLH-PAS domains of HIF1α, HIF2α, MOP3, MOP4, AHR, ARNT, and CLOCK were coexpressed with plasmids harboring full-length MOP3 and ARNT (see Materials and Methods). LexAAHR interactions were assayed on plates containing 1 μM β-naphthoflavone (14). After incubation, an 5-bromo-4-chloro-3-indolyl β-d-galactoside overlay assay was performed. ++, A strong interaction, turning blue within 2 hr; +, a weaker interaction, turning blue between 8 hr and overnight; and −, a negative interaction after overnight incubation. The experiment was performed three times with identical results.
DNA Binding Specificity of the MOP3-MOP4 Heterodimer.
We performed a selection and amplification protocol to identify the DNA sequence bound with high-affinity by the MOP3-MOP4 complex. After three rounds of selection and amplification, a gel shift assay was performed using radiolabeled selected randomers to identify the migration of the complex. We identified a species dependent on the presence of both proteins (data not shown). A band corresponding to this migration was excised from the polyacrylamide gel, and used as template for a fourth round of amplification before cloning the pool. Analysis of the sequencing data from 10 clones revealed that the MOP3-MOP4 heterodimeric pair bound the sequence G/TGA/GACACGTGACCC (Fig. 2A). This sequence is an imperfect palindrome containing a core E-box element (underlined) and specificity for nucleotides in the flanking region (e.g., +4 “A”). We refer to this response element bound by the MOP3-MOP4 as M34. To demonstrate sequence binding specificity and to confirm the selectivity for the +4 nucleotide, we performed competition experiment varying the +4 position to A, C, G, or T (Fig. 2A). In agreement with our selection results, we observed a strong preference for the flanking +4 “A” nucleotide by the MOP3-MOP4 complex (Fig. 2B).
Figure 2.
DNA binding specificity of the MOP3-MOP4 heterodimer. (A) The consensus DNA binding site for MOP3-MOP4 heterodimer in vitro. Ten selected DNA sequences bound by the MOP3-MOP4 complex are indicated with the E-box core boxed. Underneath, the M34 consensus is indicated. Nucleotide positions relative to the E-box core are shown. Bases in uppercase are randomer derived, while bases in lower case are primer derived. (B) Demonstration of flanking region specificity by competition analysis. A gel shift analysis was performed in which the double-stranded radiolabeled consensus oligonucleotide, GGGACACGTGACCC, was incubated in the presence of MOP3, MOP4, and either 10, 30, or 100 ng of double-stranded unlabeled competitor oligonucleotides. The unlabeled competitors were designed identical to the labeled oligonucleotide except the most conserved +4 “A” was changed to A, C, G, or T.
MOP3 Forms Transcriptionally Active Complexes with MOP4 and CLOCK.
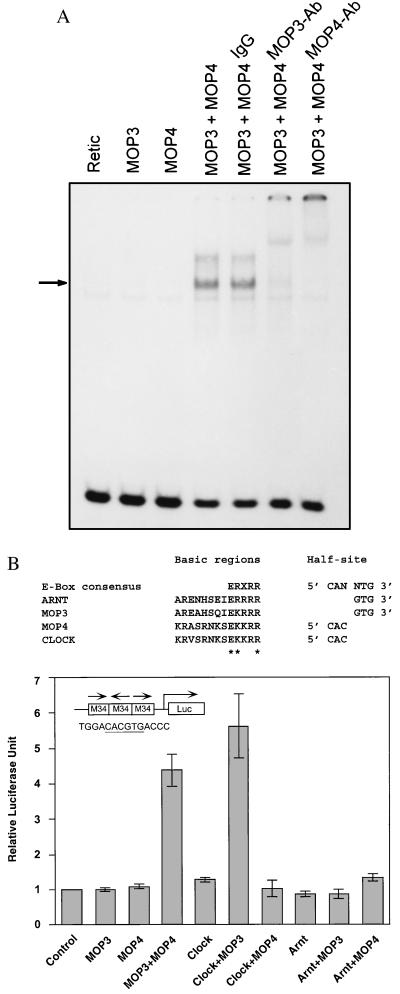
To demonstrate that both MOP3 and MOP4 are required for binding to the M34 element, we performed additional gel shift experiments. A specific band was present only with the combination of MOP3 and MOP4, and was not present with either protein alone (Fig. 3A). As an additional specificity control, affinity-purified anti-MOP3 or anti-MOP4-specific Igs were used in gel shift experiments. Fig. 3A shows that both MOP3-specific and MOP4-specific IgG were capable of retarding the mobility of the MOP3-MOP4 complex, while purified preimmune IgG alone was not.
Figure 3.
Interaction of MOP3 with MOP4 or CLOCK. (A) In vitro interaction of MOP3 and MOP4. Gel shift analysis was performed using the radiolabeled consensus response element, M34, in the presence of unprogrammed reticulocyte lysate, MOP3, MOP4, and both proteins. One microgram of purified IgG, purified MOP3-specific antibodies (MOP3Ab), or purified MOP4-specific antibodies (MOP4Ab) was used to demonstrate specificity of the complex. (B) In vivo interaction of MOP3 with MOP4 or CLOCK. (Upper) Basic region consensus and DNA half-site specificity for ARNT, MOP3, MOP4, and CLOCK are shown. Residues thought to contact DNA are denoted with an asterisk. (Lower) COS-1 cells were transfected with the M34 luciferase reporter with each expression plasmid as indicated. Cells were harvested 20 hr posttransfection. Luciferase activities were normalized with that of β-galactosidase as described (14). (Inset) The M34 responsive element is illustrated and the core sequence is underlined.
To determine whether the MOP3-MOP4 complex could drive transcription in vivo, we constructed a vector with three copies of the M34 element upstream of a minimal simian virus 40 promoter-luciferase reporter (Fig. 3B). Upon cotransfection of the reporter plasmid into COS-1 cells with MOP3 and MOP4, we observed that this combination enhanced transcription 3.3-fold, while neither protein alone was capable of driving transcription over control (Fig. 3B). The observations that CLOCK also interacted with MOP3 in the yeast interaction trap (Fig. 1 Lower) and that CLOCK shares extensive homology with MOP4 prompted us determine if MOP3-CLOCK complex could also drive transcription in vivo from an M34 element. Cotransfection of MOP3 and CLOCK revealed that this complex was also active, driving transcription 5.6-fold over control (Fig. 3B). Transfections with MOP3, MOP4, CLOCK, and ARNT alone, as well as combinations of ARNT and MOP3 or MOP4 failed to drive transcription over control (Fig. 3B).
MOP3 Forms Functional DNA Binding Complexes with HIF1α and HIF2α.
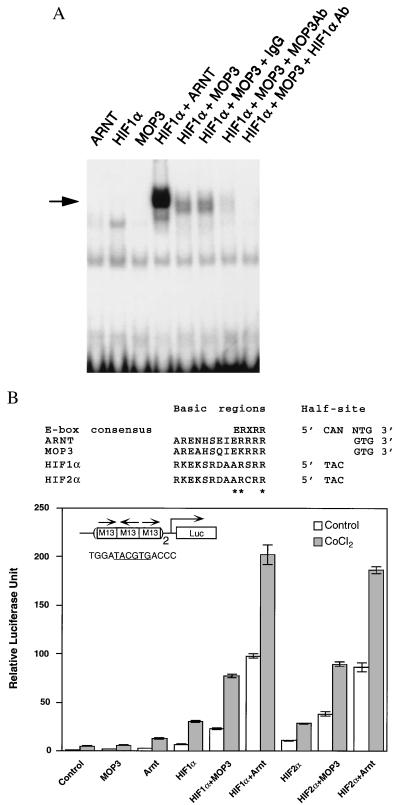
Prompted by our yeast interaction results, we set out to determine the ability of MOP3 to form DNA binding complexes with HIF1α in vitro. Because of the asymmetry at the +4 position of the M34 element, we were uncertain which half-site was bound by MOP3. Therefore, we synthesized enhancer elements with the HIF1α 5′ half site (TAC) fused to both of the potential MOP3 3′ half-sites described above (GCCCTACGTGACCC or GCCCTACGTGTTCC) (14, 26). We found that the HIF1α/MOP3 complex preferred the GCCCTACGTGACCC element in vitro (data not shown), suggesting that MOP3 preferred an “A” at the +4 position. Therefore the corresponding response element bound by the HIF1α-MOP3 complex, which we refer to as M13, was used in subsequent experiments. Fig. 4A demonstrates that the M13 element is bound in the presence of the MOP3-HIF1α combination, but not by either protein alone. MOP3-specific and HIF1α-specific antisera abolished this complex while preimmune IgG did not. For comparison we included ARNT in these experiments, and found that ARNT-HIF1α band was more intense than the MOP3-HIFα complex when all proteins were used at equimolar concentrations (Fig. 4A).
Figure 4.
Interaction of MOP3 with HIF1α or HIF2α. (A) In vitro analysis of MOP3 and HIF1α. Gel shift analysis was performed using a deduced radiolabeled response element, M13. Normalized quantities of reticulocyte expressed MOP3, HIF1α, and ARNT proteins were used for all gel shift studies. To demonstrate specificity, 1 μg of purified IgG, MOP3-specific (MOP3Ab), or HIF1α-specific antibodies (HIF1αAb) were used. (B) In vivo interaction of MOP3 with HIF1α or HIF2α (Upper) Basic region consensus and DNA half-site specificity for ARNT, MOP3, HIF1α, and HIF2α are shown. (Lower) COS-1 cells were transfected with the M13 luciferase reporter with each expression plasmid as indicated. Two oligonucleotides, containing three M13 elements were cloned upstream of the simian virus 40 promoter (denoted by a 2 beside the element). Hypoxia studies were performed with the addition of 100 μM CoCl2 at the time of transfection. Cells were harvested 20 hr posttransfection. □, Relative luciferase levels; ▪, relative luciferase levels in the presence of CoCl2. (Inset) The M13 responsive element is illustrated and the core sequence is underlined.
To determine if MOP3 formed a transcriptionally active complex with either HIF1α or HIF2α in vivo, we constructed a synthetic reporter using six copies of the M13 element described above. The M13 reporter was up-regulated when cotransfected with combinations of MOP3-HIF1α and MOP3-HIF2α (3.3-fold and 3.6-fold, respectively). ARNT formed more active complexes with both HIF1α and HIF2α (14.1-fold and 8.1-fold, respectively), consistent with our in vitro results. Like ARNT, upon exposure of these transfected cells to cobalt chloride to simulate cellular hypoxia, MOP3 interacted and drove transcription in complexes with both HIF1α and with HIF2α (Fig. 4B).
Coexpression of MOP3, MOP4, and HIF1α in Neonatal and Adult Murine Tissues.
To determine if MOP3 was coexpressed with MOP4 in any murine tissue, RPA and in situ hybridization analysis were performed. Parallel RPA analysis of neonatal and adult tissues indicated that MOP3 was most highly expressed in brain, thymus, and muscle (Fig. 5). MOP4 showed highest expression in the brain. We performed in situ hybridization analysis on tissues where RPA data indicated overlap between MOP3 and MOP4, or MOP3 and HIF1α. Sense controls were negative in all tissues except eye, where the pigment of the retina gave a nonspecific signal. In transverse sections of E15.5 brain, we observed that both MOP3 and MOP4 showed their highest expression levels in the thalamus. In E15.5 eye, we were able to detect colocalization of MOP3 and HIF1α in the retina, but were unable to detect specific labeling of MOP4. Fig. 5B shows that both MOP3 and HIF1α are colocalized in the thymic cortex of postnatal animals. Prompted by the observation of others that the MOP4 mRNA is expressed at low levels in the colon, we assayed that target tissue and observed that MOP4 and HIF1α were coexpressed in postnatal colonic mucosa, while MOP3 was undetectable there (Fig. 5B and data not shown) (11).
Figure 5.
Expression of MOP3 and MOP4 in the mouse. (A) RPA analysis of MOP3 and MOP4 in various mouse tissues. Total RNA (10 μg) from whole embryo (embryonic day 9.5 (E9.5), E13.5, E17.5), placenta, neonatal liver, kidney, heart, lung, thymus, skeletal muscle, brain, and adult liver, kidney, heart, lung, thymus, skeletal muscle, and brain was analyzed for MOP3 and MOP4 expression. The results were quantitated and expressed as relative units. (Inset) The MOP3 and MOP4 probes (lane 1), tRNA control (lane 2), and protected fragments (lane 3) are indicated. (B) In situ analysis of the MOP3 mRNA. Whole mount in situ hybridization was performed on transverse brain section from an E15.5 embryo, adult eye, thymus, and colon. Slides were probed with antisense probes derived from HIF1α, MOP3, and MOP4 and photographed under identical conditions. Hematoxylin and eosin stains of parallel slides of E15.5 transverse embryos and adult eye are shown. Transverse section: ca, cerebral aqueduct; T, thalamus; lv, lateral ventricle; 3v, third ventricle; st, striatum. Eye: pr, pigment of the retina; or, outer layer of the retina; ir, inner layer of the retina; le, lens epithelium; co, cornea.
DISCUSSION
In an effort to determine the pairing rules of MOP3 and MOP4, we employed the yeast interaction trap to identify the bHLH-PAS partners of these orphans. Our initial experiment using a MOP4 bait construct to screen a brain cDNA library identified MOP3 as a partner. In further experiments, we reversed this approach and used full-length MOP3 to detect interactions with other bHLH-PAS members. This analysis confirmed the MOP3-MOP4 interaction and also demonstrated that CLOCK, HIF1α and HIF2α were additional partners of MOP3 (Fig. 1). As demonstrated previously, ARNT interacted with the AHR, HIF1α, and HIF2α, but not with MOP4 or CLOCK (14). The fact that both MOP4 and CLOCK interacted with MOP3 was not surprising given their 75% amino acid sequence identity in their bHLH-PAS domains. The observation that MOP3 was a partner of both HIF1α and HIF2α, but that it did not dimerize with the AHR in the yeast interaction trap was an unexpected result. Due to lack of expression in our yeast system, we were unable to examine the interactions of MOP3 or MOP4 with a number of additional bHLH-PAS proteins, including mSIM1, mSIM2, hARNT2, and hSRC1. Thus, we do not exclude the possibility that additional MOP3 and MOP4 interactions with these proteins may be important. Nevertheless, our data lead us to suggest that MOP3 is a general partner of a number of bHLH-PAS factors, with a distinct interaction profile from that of the more well characterized general partner ARNT.
Analysis of MOP3 and MOP4 revealed that these proteins did not share perfect identity with any other known bHLH proteins in their basic residues thought to contact DNA (see Fig. 3B Top). Therefore, we could not readily predict the response elements that the MOP3-MOP4 heterodimer would bind. To overcome this limitation, we employed a DNA selection and amplification protocol and determined that the MOP3-MOP4 complex bound an E-box, with flanking region specificity for an “A” at +4 (i.e., CACGTGA, M34 element) (Fig. 2). In keeping with our prediction that MOP4 and CLOCK are functional homologs, transfection experiments demonstrated that the combination of either MOP3-MOP4 or MOP3-CLOCK was capable of driving transcription from M34 elements, while neither MOP3, MOP4, or CLOCK alone displayed this activity. In support of our argument that MOP3 harbors a distinct partnering specificity from that of ARNT, we observed that neither MOP3 nor MOP4 was capable of interacting with ARNT and driving transcription from the M34 element in its presence (Fig. 3B).
What could be the consequence of these interactions? Experiments from a number of laboratories indicate that circadian behavior may be regulated at the transcriptional level by complex interactions between multiple PAS domain containing proteins. Strong genetic evidence supports a role for CLOCK in the maintenance of circadian behavior in mice and the product of the period gene (PER) for control of circadian rhythms in Drosophila (27, 28). The fact that MOP4 is a brain specific homologue of CLOCK and that these factors share MOP3 as a common dimeric partner suggests that both MOP3 and MOP4 may play a role in this process as well. In addition to the mammalian MOP3, MOP4 and CLOCK proteins, murine and human homologs of Drosophila PER have recently been characterized (29–32). Like Drosophila PER, the mRNA levels of these mammalian homologs respond to light and display circadian rhythmicity (30, 32). Sequence analysis of PER proteins indicates that they contain PAS domains, but do not contain consensus bHLH domains. Coupled with additional biochemical evidence from other laboratories, these data suggest that PER proteins may affect their own transcription through interactions mediated by their PAS domains (1, 33). Thus, these PERs may impact transcriptional activity of other bHLH-PAS dimers by acting as either dominant negative inhibitors that block pairing of trancriptionally active complexes, or they may act in a positive manner as coactivators of these complexes.
In addition to defining the pairing rules and DNA binding specificities of MOP3 and MOP4, our data lead us to a testable model that describes circadian oscillation of transcription. We speculate that MOP3-CLOCK or MOP3-MOP4 complexes regulate PER transcription through CACGTGA-containing enhancers. The transcriptional activity of these promoters could in turn be modified by feedback inhibition/activation by the PER protein products themselves. In support of this idea, an E-box element in the Drosophila PER promoter, required for normal cycling of the PER mRNA, bears striking resemblance to the M34 element we have identified (i.e., 5′-CACGTGAGC-3′ compared with 5′-CACGTGACC-3′ from Fig. 2) (34). Given that we are borrowing from both Drosophila and mammalian systems, our model assumes that these signal transduction pathways have been largely conserved throughout evolution. In keeping with this idea, a search of Drosophila expressed sequence tags revealed the existence of an uncharacterized MOP4/CLOCK homolog (GenBank accession no. AA698290) and an uncharacterized MOP3 homolog (GenBank accession no. AA695336).
What could be the consequences of MOP3-HIF interactions? Transient transfection experiments showed that MOP3 formed transcriptionally active complexes with HIF1α and HIF2α and that these complexes responded to cellular hypoxia (Fig. 4). MOP3 may play a specialized role in hypoxia signaling. The different tissue specific expression profiles of MOP3 and ARNT suggests that MOP3 may regulate cellular responses to hypoxia at distinct sites, such as the retina, thymic cortex, and thalamus. Moreover, the observation that MOP3 binds a GTG half-site with flanking region specificity for an “A” at +4, may indicate that MOP3/HIF complexes may have greater affinity for a distinct subset of hypoxia response elements (i.e., TACGTGA vs. the more commonly observed TACGTGG elements observed in known hypoxia responsive enhancers). Finally, the observation that MOP4 is expressed at a site where MOP3 expression appears low, i.e., colonic mucosa, suggests that additional partners may exist for MOP4 and CLOCK and that all bHLH-PAS signaling pathways may involve complex equilibria between multiple PAS protein partnerships.
Acknowledgments
This work was supported by The Burroughs Wellcome Foundation and by National Institutes of Health Grants P30-CA07175, ES05703, and GM08061.
ABBREVIATIONS
- PAS
PER–ARNT–SIM homology domain
- bHLH
basic-helix–loop–helix domain
- AHR
aryl hydrocarbon receptor
- ARNT
aryl hydrocarbon receptor nuclear translocator
- HIF1α
hypoxia-inducible factor 1α
- PER
product of the Drosophila period gene
- RPA
ribonuclease protection assay
- M34
the response element bound by the MOP3-MOP4 complex
- M13
the response element bound by the HIF1α-MOP3 complex
- CLOCK
product of the Clock locus in mice
- E-box
an enhancer element defined as CANNTG
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF044288).
References
- 1.Huang Z J, Edery I, Rosbash M. Nature (London) 1993;364:259–262. doi: 10.1038/364259a0. [DOI] [PubMed] [Google Scholar]
- 2.Burbach K M, Poland A, Bradfield C A. Proc Natl Acad Sci USA. 1992;89:8185–8189. doi: 10.1073/pnas.89.17.8185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoffman E C, Reyes H, Chu F F, Sander F, Conley L H, Brooks B A, Hankinson O. Science. 1991;252:954–958. doi: 10.1126/science.1852076. [DOI] [PubMed] [Google Scholar]
- 4.Nambu J R, Lewis J O, Wharton K A, Jr, Crews S T. Cell. 1991;67:1157–1167. doi: 10.1016/0092-8674(91)90292-7. [DOI] [PubMed] [Google Scholar]
- 5.Jackson F R, Bargiello T A, Yun S H, Young M W. Nature (London) 1986;320:185–188. doi: 10.1038/320185a0. [DOI] [PubMed] [Google Scholar]
- 6.Wang G L, Jiang B H, Rue E A, Semenza G L. Proc Natl Acad Sci USA. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reyes H, Reisz-Porszasz S, Hankinson O. Science. 1992;256:1193–1195. doi: 10.1126/science.256.5060.1193. [DOI] [PubMed] [Google Scholar]
- 8.Dolwick K M, Swanson H I, Bradfield C A. Proc Natl Acad Sci USA. 1993;90:8566–8570. doi: 10.1073/pnas.90.18.8566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirose K, Morita M, Ema M, Mimura J, Hamada H, Fujii H, Saijo Y, Gotoh O, Sogawa K, Fujii-Kuriyama Y. Mol Cell Biol. 1996;16:1706–1713. doi: 10.1128/mcb.16.4.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan C M, Kuwana E, Bulfone A, Fletcher C F, Copeland N G, Jenkins N A, Crews S, Martinez S, Puelles L, Rubenstein J L R, Tessier-Lavigne M. Mol Cell Neurosci. 1996;7:1–16. doi: 10.1006/mcne.1996.0001. [DOI] [PubMed] [Google Scholar]
- 11.Zhou Y D, Barnard M, Tian H, Li X, Ring H Z, Francke U, Shelton J, Richardson J, Russell D W, McKnight S L. Proc Natl Acad Sci USA. 1997;94:713–718. doi: 10.1073/pnas.94.2.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tian H, McKnight S L, Russell D W. Genes Dev. 1997;11:72–82. doi: 10.1101/gad.11.1.72. [DOI] [PubMed] [Google Scholar]
- 13.Ikeda M, Nomura M. Biochem Biophys Res Commun. 1997;233:258–264. doi: 10.1006/bbrc.1997.6371. [DOI] [PubMed] [Google Scholar]
- 14.Hogenesch J B, Chan W C, Jackiw V H, Brown R C, Gu Y Z, Pray-Grant M, Perdew G H, Bradfield C A. J Biol Chem. 1997;272:8581–8593. doi: 10.1074/jbc.272.13.8581. [DOI] [PubMed] [Google Scholar]
- 15.Poland A, Glover E, Bradfield C A. Mol Pharmacol. 1991;39:20–26. [PubMed] [Google Scholar]
- 16.Jain S, Dolwick K M, Schmidt J V, Bradfield C A. J Biol Chem. 1994;269:31518–31524. [PubMed] [Google Scholar]
- 17.Carver L A, Bradfield C A. J Biol Chem. 1997;272:11452–11456. doi: 10.1074/jbc.272.17.11452. [DOI] [PubMed] [Google Scholar]
- 18.Vojtek A B, Hollenberg S M, Cooper J A. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 19.Sanger F, Nicklen S, Coulson A R. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 21.Kaiser C, Michaelis S, Mitchell A. Methods in Yeast Genetics. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. [Google Scholar]
- 22.Bohen S P, Yamamoto K R. Proc Natl Acad Sci USA. 1993;90:11424–11428. doi: 10.1073/pnas.90.23.11424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swanson H I, Chan W K, Bradfield C A. J Biol Chem. 1995;270:26292–26302. doi: 10.1074/jbc.270.44.26292. [DOI] [PubMed] [Google Scholar]
- 24.Luo G, Gu Y-Z, Jain S, Chan W K, Carr K M, Hogenesch J B, Bradfield C A. Gene Expression. 1997;6:287–299. [PMC free article] [PubMed] [Google Scholar]
- 25.Jain S, Maltepe E, Lu M M, Simon C, Bradfield C A. Mech Dev. 1998;73:117–123. doi: 10.1016/s0925-4773(98)00038-0. [DOI] [PubMed] [Google Scholar]
- 26.Semenza G L, Wang G L. Mol Cell Biol. 1992;12:5447–5454. doi: 10.1128/mcb.12.12.5447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.King D P, Zhao Y, Sangoram A M, Wilsbacher L D, Tanaka M, Antoch M P, Steeves T D, Vitaterna M H, Kornhauser J M, Lowrey P L, et al. Cell. 1997;89:641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hall J C, Rosbash M. BioEssays. 1987;7:108–112. doi: 10.1002/bies.950070304. [DOI] [PubMed] [Google Scholar]
- 29.Sun Z S, Albrecht U, Zhuchenko O, Bailey J, Eichele G, Lee C C. Cell. 1997;90:1003–1011. doi: 10.1016/s0092-8674(00)80366-9. [DOI] [PubMed] [Google Scholar]
- 30.Albrecht U, Sun Z S, Eichele G, Lee C C. Cell. 1997;91:1055–1064. doi: 10.1016/s0092-8674(00)80495-x. [DOI] [PubMed] [Google Scholar]
- 31.Tei H, Okamura H, Shigeyoshi Y, Fukuhara C, Ozawa R, Hirose M, Sakaki Y. Nature (London) 1997;389:512–516. doi: 10.1038/39086. [DOI] [PubMed] [Google Scholar]
- 32.Shearman L P, Zylka M J, Weaver D R, Kolakowski L F J, Reppert S M. Neuron. 1997;19:1261–1269. doi: 10.1016/s0896-6273(00)80417-1. [DOI] [PubMed] [Google Scholar]
- 33.Gekakis N, Saez L, Delahaye-Brown A M, Myers M P, Sehgal A, Young M W, Weitz C J. Science. 1995;270:811–815. doi: 10.1126/science.270.5237.811. [DOI] [PubMed] [Google Scholar]
- 34.Hao H, Allen D L, Hardin P E. Mol Cell Biol. 1997;17:3687–3693. doi: 10.1128/mcb.17.7.3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ema M, Taya S, Yokotani N, Sogawa K, Matsuda Y, Fujii-Kuriyama Y. Proc Natl Acad Sci USA. 1997;94:4273–4278. doi: 10.1073/pnas.94.9.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]