Abstract
RNA-specific nucleotidyl transferases (rNTrs) are a diverse family of template-independent polymerases that add ribonucleotides to the 3′-ends of RNA molecules. All rNTrs share a related active-site architecture first described for DNA polymerase β and a catalytic mechanism conserved among DNA and RNA polymerases. The best known examples are the nuclear poly(A) polymerases involved in the 3′-end processing of eukaryotic messenger RNA precursors and the ubiquitous CCA-adding enzymes that complete the 3′-ends of tRNA molecules. In recent years, a growing number of new enzymes have been added to the list that now includes the “noncanonical” poly(A) polymerases involved in RNA quality control or in the readenylation of dormant messenger RNAs in the cytoplasm. Other members of the group are terminal uridylyl transferases adding single or multiple UMP residues in RNA-editing reactions or upon the maturation of small RNAs and poly(U) polymerases, the substrates of which are still not known. 2′-5′Oligo(A) synthetases differ from the other rNTrs by synthesizing oligonucleotides with 2′-5′-phosphodiester bonds de novo.
Keywords: CCA-adding enzyme, poly(A) polymerase, RNA modification, terminal uridylyl transferase, 2′-5′-oligo(A) synthetase
INTRODUCTION
Poly(A) polymerizing activity in eukaryotic cells was discovered around 1960 when an enzyme that generated poly(A) from ATP was identified from extracts of calf thymus (Edmonds and Abrams 1960). Around the same time, it was found that stretches of poly(A) at the 3′-ends of mRNAs were common in prokaryotes and eukaryotes, and the enzyme responsible for poly(A) synthesis was purified and named poly(A) polymerase (PAP) (Edmonds 2002). After cDNA clones and recombinant PAPs became available (Lingner et al. 1991; Raabe et al. 1991; Wahle et al. 1991), it was shown that PAPs have sequence homology with DNA polymerase β (Pol β) and other nucleotidyl transferases (NTrs) and that all these enzymes share structural homology in their catalytic domain (Holm and Sander 1995; Martin and Keller 1996). Much of the initial work, including X-ray crystallography, was done with Pol β, a template-dependent DNA repair enzyme (Pelletier et al. 1994; Sawaya et al. 1994; for review, see Ramadan et al. 2004) and the related terminal deoxynucleotidyl transferase (TdT), an enzyme involved in the generation of antibody diversity by random nucleotide addition in V(D)J recombination (Delarue et al. 2002; Thai and Kearney 2005). Classification of related nucleotidyltransferases revealed the large superfamily of Pol β-like nucleotidyltransferases (Polβ-NTrs) (Yue et al. 1996; Aravind and Koonin 1999). More recently, the knowledge about these enzymes has expanded dramatically. Not only were the crystal structures and catalytic mechanisms of poly(A) polymerases, terminal uridylyl transferases (TUTases), and CCA-adding enzymes (CCAtrs) determined, but it has also been shown that different eukaryotic poly(A) polymerases have specialized functions in different cell compartments and during the cell cycle (Wang et al. 2002; Kadaba et al. 2004; LaCava et al. 2005; Vanacova et al. 2005; Wyers et al. 2005).
Here, we present an overview of those nucleotidyl transferases that act on RNA (rNTrs) by covalently adding nucleotides to the 3′-end, and we describe a few of the most studied members of this subfamily. It has become customary to divide eukaryotic poly(A) polymerases, TUTases, and poly(U) polymerases into canonical and noncanonical rNTrs. Canonical rNTrs include the nuclear PAPs α, β, and γ that were first isolated and characterized from mammals and yeast (Lingner et al. 1991; Raabe et al. 1991; Wahle et al. 1991), whereas the recently discovered GLD-2 and Trf4-type PAPs are termed noncanonical rNTrs. A representative set of known and predicted human rNTrs is shown in Figure 1. In a somewhat simplified classification, the canonical rNTrs form one group of enzymes with similar catalytic, central and RNA-binding domains and only contain PAPs. The second group includes the Gld-2-, Trf4/5-, and Cid1-type poly(A) or poly(U) polymerases, 2′-5′-oligo(A) synthetases, and the TUTases (see below for abbreviations). These enzymes share the catalytic domain with canonical rNTrs but contain a different nucleotide base-recognition motif. CCA-adding enzymes with a similar catalytic domain but an even more diverged nucleotide recognition system represent the third group. All these enzymes share a common catalytic domain signature.
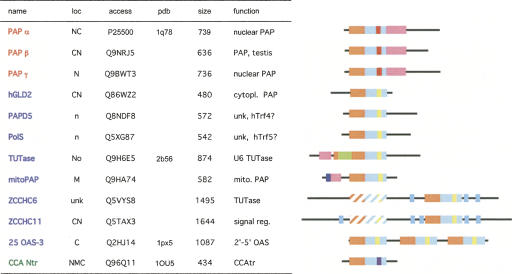
FIGURE 1.
Human ribonucleotidyl transferases. (Names in red) Canonical and (blue) noncanonical rNTrs. Cellular localizations (loc) are abbreviated as C (cytoplasmic), N (nuclear), M (mitochondrial), and No (nucleolar). (Uppercase letters) Experimentally demonstrated and (lowercase letters) predicted localization; (unk) unknown; (signal reg.) signal regulator. The SWISS-PROT accession code is indicated in the “access” column. PDB structure database codes (pdb) of structures from the most similar organism are indicated with lowercase letters or with uppercase letters if the structure of the human protein is available. Color code for domains: (orange) catalytic domain; (light blue) central domain; (violet) RBD; (red) nucleotide recognition motif (NRM type 1) in canonical rNTrs; (yellow) NRM type 2 in noncanonical rNTrs; (purple) NRM in CCAtrs; (cyan) zinc finger motifs; (olive) insert in U6 TUTase; (hatched bars) predicted inactive NTr motifs.
STRUCTURAL FEATURES OF POL β-TYPE NUCLEOTIDYL TRANSFERASES
Pol β-NTrs have sequence and structural homology with the catalytic domain of DNA Pol β. This structure consists of a five-stranded β-sheet backed by two α-helixes, forming an αβ two-layer sandwich. A hallmark in Pol β-NTrs is a single or double helical turn between strands 1 and 2 (the numbering refers to Pol β), depending on the organism. The β-strand 2 features two catalytic Asp or Glu residues separated by one mostly hydrophobic amino acid, resulting in a DxD or DxE motif. A third catalytic residue (Asp or Glu) of the triad is located on strand 3, which runs parallel to strand 2. The general signature of this highly conserved catalytic-site motif is hG[GS]x(7–13)Dh[DE]h, with h indicating hydrophobic, uppercase letters invariant, and x any amino acid.
The domain organization of rNTrs typically consists of a catalytic domain followed by a central domain (Fig. 2). All canonical and a few noncanonical rNTrs contain, in addition, an RNA-binding domain (RBD) at the N or C terminus or inserted into the catalytic domain, which has structural homology with the RNA-recognition motif (RRM) protein family. RBDs in canonical-type PAPs were found to be involved in sequence-nonspecific RNA substrate binding, and deletion of the RBD led to complete loss of activity (Zhelkovsky et al. 1995; Martin and Keller 1996). The function of the RBD structures in TUTases and mitochondrial PAP is most likely RNA binding, but this has not been investigated systematically (see below). Some proteins also feature Zn-fingers (Fig. 1).
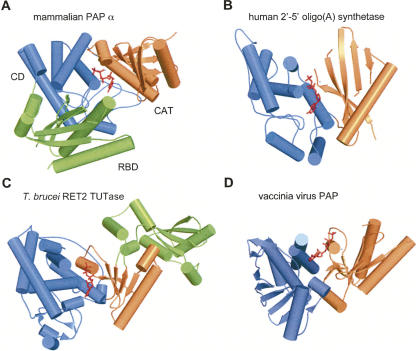
FIGURE 2.
Catalytic and central domains and RNA-binding domains (RBDs) of canonical and noncanonical rNTrs. (A) Bovine PAP pdb:1Q78 (Martin et al. 2004) with bound 3′-dATP. Color code for domains: (orange) catalytic domain (CAT); (blue) central domain (CD); and (green) RBD. Color codes are the same for B–D. The domain nomenclature used by the authors of these structures may differ. (B) 2′-5′-Oligo(A) synthetase (pdb:1PX5) (Hartmann et al. 2003) with ATP modeled from A. (C) Trypanosoma brucei RET2 (pdb:2B56) (Deng et al. 2005) with UTP. (D) Vaccinia virus PAP (pdb:2GA9) (Moure et al. 2006) with bound ATP-γ-S.
Comparative analysis of nucleotidyl transferases revealed a conserved motif within the central domain in PAPs and in archaeal CCAtrs (Rogozin et al. 2003). This motif has structural homology with the ATP-cone, an ancient nucleotide-binding and regulatory motif also found in ribonucleotide reductase, an enzyme that converts ribonucleotides to deoxynucleotides. In the structure of bovine PAP crystallized in a complex with Mg-ATP (Martin et al. 2004), the adenine base contacted a loop with the sequence TPAYP (Fig. 3), a motif that is part of the ATP-cone. Moreover, in the structure of the trypanosomal RET2 TUTase, the conserved Asp421 in the homologous loop motif DPADP is responsible for interacting with the uracil base of UTP via water molecules (see below). This nucleotide-recognition motif (NRM) (Fig. 3) occurs in two different versions, one characteristic for canonical rNTrs (NRM type 1) and the second for noncanonical rNTrs (NRM type 2) (Fig. 3).
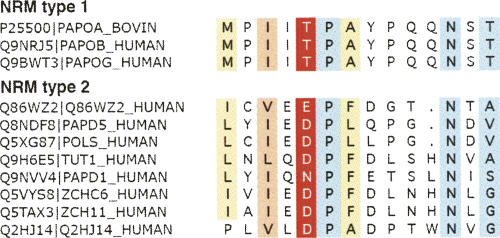
FIGURE 3.
Nucleotide base recognition motifs in human rNTrs. (Red shading) The main nucleotide-contacting residue, (blue) small, (yellow) hydrophobic, and (orange) aliphatic residues. The sequences included in the alignment correspond to the proteins in Figure 1 and are named by SWISS-PROT accession.
ACTIVE-SITE ARCHITECTURE AND CATALYTIC MECHANISM
All members of the Pol β-NTrs, with the exception of 2′-5′-oligo(A) synthetase, act via a two-metal-ion catalytic mechanism and add ribo- or deoxynucleoside monophosphates to the 3′-hydroxyl group of the recipient substrate (Steitz 1998). One divalent metal ion binds as a cosubstrate (as MgATP2−) and after catalysis leaves with one of the products as MgPPi 2−. The other metal coordinates the primer 3′-OH and the α-phosphate of the incoming nucleotide and functions catalytically to stabilize the negative charge that develops at the transition state.
It has been proposed that an “induced fit” mechanism is responsible for the selection of the correct nucleotide in Pol β (Sawaya et al. 1997) and PAP (Balbo et al. 2005). According to this model, ATP specificity is mediated by stabilization of both the transition state and the ground state by use of free energy derived from the recognition of the correct substrate (MgATP2−). This binding energy is subsequently used to increase the free energy of the ground state (ground-state destabilization) and to accelerate the velocity for the incorporation of the correct substrate (Balbo et al. 2005, 2007).
Because rNTrs do not use a DNA or RNA template for the insertion of the nucleotide, a protein template was suggested to be responsible for selection. This is supported by crystal structures of CCA-adding enzymes, where the CCA tail grows into a dynamic protein pocket, which reshapes after each nucleotide addition and defines a new template at every step of the reaction (Xiong and Steitz 2004; Tomita et al. 2006). In contrast, it is not clear how the high specificity for AMP or UMP incorporation is accomplished in rNTrs that add homopolymeric tracts of As or Us-in particular, in processive canonical and noncanonical rNTrs. From the crystal structures of protein–nucleotide complexes of yeast and bovine PAP, only partial specificity for ATP could be predicted (Fig. 4A; Bard et al. 2000; Martin et al. 2004). The PAP structure of Martin et al. (2004) displays two residues (N202 and T317) that form at most weak hydrogen bonds with the adenine because the H-bond distances are larger than ideal (3.5–3.6 Å; indicated in Fig. 4A). It is conceivable that these H-bond distances become shorter after closure of the catalytic cleft with motion of the N-terminal domain, as suggested for yeast PAP (Balbo et al. 2007). In contrast, the ATP-γ-S nucleotide in vaccinia PAP (VP55; see below) was found to make specific interactions at the adenine base (Moure et al. 2006). It is possible that base stacking between the adenine of the incoming nucleotide with the 3′-adenylate residue of the RNA primer and also water molecules buried in the active site contribute to nucleotide selection when there is a lack of specific interactions with the nucleotide. As canonical-type nuclear PAPs are highly processive enzymes in vivo, a mechanism with few ligand contacts could allow a faster synthesis rate compared to a mechanism whereby each nucleotide has to be recognized by many ligands at the active site. Specificity for the recognition of UTP was demonstrated in recent crystal structures of the RNA-editing TUTase RET2 and the minimal catalytically active TUTase TUT4 from Trypanosoma (Fig. 4B; Deng et al. 2005; Stagno et al. 2007). RET2, which will be described in detail below, is only found in kinetoplastid mitochondria, where it adds single UMP residues to the 3′-ends of RNA-editing intermediates.
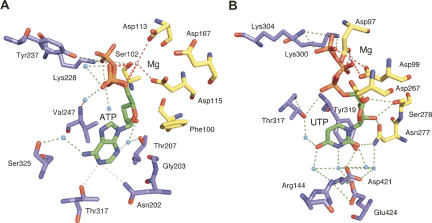
FIGURE 4.
Comparison of catalytic sites of canonical and noncanonical rNTrs. (A) Bovine PAP (pdb:1Q78) (Martin et al. 2004), apparent nonspecific nucleotide recognition. (Red dotted lines) Metal coordination; (green dotted lines) hydrogen bonds (light pink dotted lines indicate weak hydrogen bonds; see text); (cyan spheres) water; (gray spheres) Mg ions. Only waters relevant for substrate binding are displayed. (B) RET2 (pdb:2B56) (Deng et al. 2005) suggesting specific recognition of UTP via water molecules. The same color code applies as for A. Molecular graphics were done with PyMOL (DeLano 2002).
In the absence of other factors, mammalian nuclear PAP displays distributive addition of AMP to an RNA primer, where after each addition the protein dissociates from the RNA. In contrast, if PAP is part of the specific 3′-end processing complex, polyadenylation is processive most of the time, i.e., PAP does not dissociate from the primer, but the RNA translocates away from the active site by one nucleotide and the enzyme can add another AMP. This is made possible by several proteins that act as processivity factors for PAP within the pre-mRNA 3′-processing complex. These proteins, which include the 160-kDa subunit of the cleavage and polyadenylation specificity factor (CPSF-160), the factor interacting with PAP (Fip1) (Kaufmann et al. 2004) and the nuclear poly(A) binding protein 1 (PABPN1) (Kühn et al. 2003; Kühn and Wahle 2004) prevent the RNA from dissociating from the polymerase active site. PABPN1 is special because it binds in multiple copies to the emerging poly(A) tail, measures the length of the tail, and promotes dissociation of the polyadenylation complex when ∼200 AMP residues have been synthesized (Wahle 1991).
PHYLETIC DISTRIBUTION AND CLASSIFICATION OF RNA-SPECIFIC RIBONUCLEOTIDYL TRANSFERASES
Phylogenetic comparisons suggest that the families of Pol β-NTrs have evolved rapidly according to specific needs, although their number per genome does not necessarily correlate with the complexity of the organism. For example, Saccharomyces cerevisiae and Drosophila melanogaster can live with a single canonical nuclear PAP, whereas this number increases to three or more PAP genes in Caenorhabditis elegans and vertebrates. Furthermore, the noncanonical rNTrs are present in two copies in S. cerevisiae, in six copies in the fission yeast Schizosaccharomyces pombe, seven in Drosophila and mammals, and 12 in the nematode C. elegans. A model for the evolution of Pol β-NTrs was proposed in which the families have rapidly and independently evolved from a common ancestor to fill a particular functional niche (Aravind and Koonin 1999). This would imply that a simple enzyme evolves into a more sophisticated enzyme. In contrast to this, it has recently been shown that when the RNA-binding tail domain of a bacterial PAP was swapped with a tail domain of a CCAtr, the resulting chimera was unexpectedly adding CCA to a pre-tRNA (Betat et al. 2004). Bacterial PAPs and CCAtrs are almost indistinguishable by their protein sequences but have different substrates and products (only the ATP substrate and leaving pyrophosphate are common). These results suggest that PAPs originated from CCAtrs and by acquiring different RNA-binding domains became enzymes that only perform A-addition resembling the last step of the CCA-addition reaction. This supports the proposal that interconversion could have played a role in the evolution of the eubacterial PAPs and CCAtrs (Yue et al. 1996) but is also compatible with branching of eubacterial PAPs from CCAtrs (Fig. 5). Moreover, bacterial PAPs are present mainly in the proteobacterial branch of eubacteria (Martin and Keller 2004), and a more patchy distribution would be expected if gene loss was responsible for the fact that most bacteria do not possess a poly(A) polymerase but rely on polynucleotide phosphorylase (PNPase) for poly(A) addition (see below; Mohanty and Kushner 2000; Rott et al. 2003).
FIGURE 5.
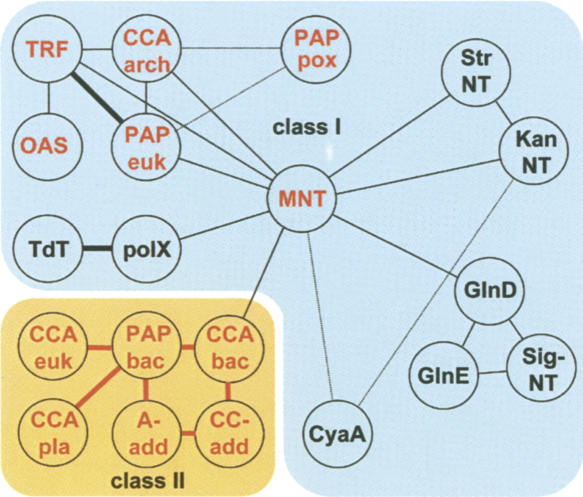
The Pol β-like nucleotidyltransferase superfamily (adapted from Aravind and Koonin 1999). (Red) Protein families relevant to this review. Abbreviations in the circles are (TRF) Trf4-like poly(A) polymerases, including GLD-2 and TUTases; (OAS) 2′-5′-oligo(A) synthetases; (CCA arch) archaeal CCAtrs; (PAP euk) eukaryotic canonical PAPs; (PAP pox) poxviral PAPs; (CCA euk) eukaryotic CCAtrs; (PAP bac) bacterial PAPs; (CCA bac) bacterial CCAtrs; (CCA pla) plant CCAtrs; (CC-add) CC-adding enzymes; (A-add) A-adding enzymes; (MNT) minimal nucleotidyl transferases. Families (in black) not treated in this review are (polX) DNA polymerases of family X, including Pol β; (TdT) terminal deoxynucleotide transferase; (Str NT) streptomycin nucleotidyl transferase and (Kan NT) kanamycin nucleotidyl transferase (antibiotics resistance factors); (GlnD) protein uridylyl-transferases; (GlnE) protein adenylyl-transferases; (Sig-NT) putative signal transducing NTrs in prokaryotes; (CyA) γ-proteobacterial adenylate cyclases (Aravind and Koonin 1999). Thick connection lines indicate high (e-values < 0.01) and broken lines low statistical significance of similarity between two families (black lines are from Aravind and Koonin 1999 and red lines are from G. Martin, unpubl.). The phylogenetic relationships among the groups at the lower left is arbitrary and not statistically proven. (Pale blue background) Class I and (orange background) class II NTrs. Modified from Fig. 1 in Aravind and Koonin (1999) and reprinted with permission from Oxford University Press ©1999.
Nucleotidyl transferases were divided into two classes based on different sequence patterns within the catalytic site and phylogenetic relationships (Yue et al. 1996). Basically, class II includes bacterial and eukaryotic CCAtrs and bacterial PAPs. All the remaining NTrs belong to class I (Fig. 5). For some reason, eukaryotes utilize the bacterial type CCA-adding enzyme (class II), which supports the idea of horizontal gene transfer between bacteria and eukaryotes (Martin and Keller 2004).
One of the structural differences between the two classes lies within the hallmark helix between β-strands 1 and 2 in the protein's catalytic fold where class I helices contain one and class II helices two helical turns (Li et al. 2002; Xiong et al. 2003). Another feature that is different between the two classes is the determinants for nucleotide selection. For example, class II NTrs contain a unique RRD signature that participates in CTP and ATP selection, whereas class I NTrs use a completely different set of ligands for NTP selection. The archaeal CCAtrs are possibly the founders of the canonical and noncanonical rNTrs in eukaryotes, whereas the eubacterial class II CCAtrs are the ancestors of eukaryotic CCAtrs (Fig. 5).
EUKARYOTIC CANONICAL POLY(A) POLYMERASES: ADDITION OF A STABILIZING TAIL
Canonical poly(A) polymerases in eukaryotes are responsible for the addition of poly(A) tails during the processing of the 3′-ends of messenger RNA precursors in the nucleus. The poly(A) tails of the resulting mRNAs serve multiple functions. They are required for the transport of the RNAs to the cytoplasm, and they stimulate the efficiency of protein synthesis by forming RNA–protein complexes that bring the 5′- and the 3′-ends into close proximity. Moreover, the poly(A) tails stabilize the mRNA by preventing premature degradation (Wahle and Rüegsegger 1999; Zhao et al. 1999).
All eukaryotic genomes code for one or several canonical nuclear PAPs. Yeast has one (Lingner et al. 1991), and the genomes of higher eukaryotes have two or three copies. In mammals, PAP α (Raabe et al. 1991; Wahle et al. 1991) and PAP γ (Kyriakopoulou et al. 2001; Perumal et al. 2001; Topalian et al. 2001) have been extensively characterized. Both were found to be up-regulated in tumor tissues. Differences between the two proteins were found in the splicing pattern of their pre-mRNA and in phosphorylation within the C-terminal domain (see below). Both PAP α and γ can participate in pre-mRNA 3′-end formation and were found to be components of the cleavage and polyadenylation complex, where they add poly(A) tails to the upstream cleavage product of pre-mRNAs (Wahle and Rüegsegger 1999). PAP β, the third canonical PAP in mammals, was found to be involved in polyadenylation of testis-specific mRNAs (Kashiwabara et al. 2000). In contrast to PAP α and γ, which both contain two NLS motifs within the C terminus, only one degenerate nuclear localization signal can be identified in PAP β, which predicts it not to be targeted to the nucleus. Nevertheless, PAP β was shown to localize both to the cytoplasm of spermatocytes and spermatids (Kashiwabara et al. 2000) but also to the nucleus (Lee et al. 2000). The gene for PAP β contains no introns, and it is assumed that it was generated by an ancient retrotransposition of a PAP γ cDNA (Kashiwabara et al. 2000; Le et al. 2001).
The structure of the mammalian PAP α in complex with 3′dATP has been determined by X-ray crystallography (Martin et al. 2000, 2004). This structure showed that the enzyme consists of a catalytic domain within the N terminus and an RNA-binding domain overlapping with a bipartite NLS. The catalytic and RNA-binding domains are linked by a central domain (Fig. 2A). These features are conserved in canonical PAPs from yeast to higher eukaryotes and also in PAP β and PAP γ. PAP α was found to exist in numerous isoforms generated by alternative splicing (Zhao and Manley 1996), whereas PAP γ showed no indication of splice variants (Topalian et al. 2001).
Canonical PAPs in vertebrates have an extended C terminus rich in serines and threonines. The C terminus also features several phosphorylation sites for the cdc2/cyclin B (MPF) complex, and PAP α was found to be gradually phosphorylated at the M phase of the cell cycle first at three consensus phosphorylation sites and later at four nonconsensus sites (Colgan et al. 1996). Only when hyperphosphorylation is reached at these sites does PAP α become catalytically inactive (Colgan et al. 1998). PAP α is therefore part of a concerted shutdown pathway of cellular activities directed by the MPF levels before cell division. In contrast, no indication of phosphorylation was found in PAP γ despite the presence of an MPF-binding site, and it was proposed that the two types of PAPs are differentially phosphorylated (Topalian et al. 2001).
In addition, the extreme end of the PAP α C terminus was found to be involved in regulation of the enzyme's catalytic activity and also to stimulate the activity of a nearby splicing complex. First, it was shown that when a dimer of the splicing factor U1A binds to a specific stem–loop structure on its own pre-mRNA and interacts by a domain rich in alternating basic/acidic residues with the PAP C terminus, it is able to suppress PAP catalytic activity (Gunderson et al. 1994, 1997). Moreover, the U1 snRNP protein U1–70K and also SRP75, a member of the SR family of splicing regulators, were found to interact with the PAP α C terminus by a similar mechanism and to suppress polyadenylation of the bound pre-mRNA (Gunderson et al. 1998). Furthermore, an interaction of the 20 C-terminal amino acids of PAP α with the splicing factor U2AF65 was demonstrated to stimulate the splicing reaction by enhancing U2AF65 binding to the intron, confirming that the splicing and polyadenylation reactions are linked (Vagner et al. 2000).
The C-terminal motif of PAP α that interacts with splicing factors is also conserved in PAP γ (Kyriakopoulou et al. 2001). When PAP γ was characterized, binding of U1A could be completed efficiently by a synthetic peptide corresponding to the interaction domain of U1A (Kyriakopoulou et al. 2001). In contrast, PAP β has a shorter C terminus and is lacking the U1A-binding motif.
EUBACTERIAL POLY(A) POLYMERASES: TAGGING FOR DEGRADATION
Bacterial poly(A) polymerase (bact-PAP) was first identified in Escherichia coli as an enzyme that synthesized polyadenylate requiring an RNA primer, ATP, and Mg2+ as cofactor (August et al. 1962). For simplicity, we use the terms “archaeal” and “bacterial” for “archaebacterial” and “eubacterial,” respectively. Later, the E. coli bact-PAP was found to be the product of the pcnB gene and to be involved in RNA decay (Hajnsdorf et al. 1995; O'Hara et al. 1995). There exist excellent reviews on bacterial RNA decay (Kushner 2002; Deutscher 2003; Carpousis 2007); therefore, the subject will only be covered briefly at the end of this section.
Bact-PAPs catalyze the template-independent addition of homopolymeric tails consisting of AMP residues to the 3′-hydroxyl group of RNAs, which by this modification become marked for degradation. The error rate in this poly(A) addition reaction was found to be higher than with canonical PAPs (Yehudai-Resheff and Schuster 2000). Bact-PAPs are very similar in protein sequence to CCAtrs, and only one PAP-specific motif has been identified so far (Martin and Keller 2004). This motif forms a predicted β-loop near the catalytic center that could be involved in binding the 3′-end of the RNA substrate. No structure has been determined for bact-PAPs to date, but strong structural homology with published structures of CCAtrs can be assumed (see below). Bact-PAPs are not found in all eubacteria, and database searches in sequenced genomes detect bact-PAPs only in the β, γ, and δ subdivisions of the proteobacteria and some Chlamydiales and Spirochaetales, whereas Gram-positive bacteria and bacteria that have diverged before the Gram-positives do not contain bact-PAPs (Martin and Keller 2004). Also, no bact-PAP homologs could be detected in archaea and eukaryotes with the exception of plants, where chloroplasts seem to host bact-PAP homologs encoded in the nucleus (Martin and Keller 2004). Bacterial PNPase, a component of a large complex involved in RNA degradation called the “degradosome,” is an enzyme that can add and also degrade heteropolymeric RNAs and can functionally replace bact-PAP. This could explain why bact-PAP is not an essential gene in E. coli (Mohanty and Kushner 1999, 2000).
Polyadenylation of RNAs by bact-PAP plays a significant role in mRNA decay and RNA quality control, although many aspects are still poorly understood (for review, see Kushner 2002; Deutscher 2003). The role of RNA polyadenylation in bacteria has often been underestimated because it was assumed that only few mRNAs are modified post-transcriptionally. By comparing the transcriptomes of wild-type and pcnB deletion strains with macroarray analysis, it was demonstrated that 90% of E. coli RNAs transcribed during exponential growth underwent some degree of polyadenylation by bact-PAP, either as full-length transcripts or as decay intermediates (Mohanty and Kushner 2006). These investigators also found that Rho-independent transcription terminators serve as polyadenylation signals but that RNAs generated by Rho-dependent termination are most likely not substrates for bact-PAP. In turn, the latter can be modified by the addition of heteropolymeric tails by PNPase.
In prokaryotes, RNA turnover is primarily executed by the degradosome complex, a multiprotein assembly that includes RNase E, PNPase, and the RNA helicase RhlB, among many other proteins (for review, see Carpousis 2007). Polyadenylation of RNAs and of their decay intermediates stimulates the degradation activity of the degradosome. Thus, poly(A) tails in prokaryotes seem to function as a platform that can facilitate the attack by the 3′-exonucleolytic activity of the degradosome. In eukaryotes, poly(A) tails made by canonical PAPs stabilize RNAs, whereas the polyadenylation of RNAs destined for degradation is catalyzed by noncanonical PAPs (see below).
CCA-ADDING ENZYMES
CCA:tRNA nucleotidyl transferases (CCAtrs; also called CCA-adding enzymes) catalyze the post-transcriptional addition of a CCA trinucleotide to the 3′-end of pre-tRNAs with CTP and ATP as cosubstrates (for review, see Xiong and Steitz 2006). The CCA end is universally conserved and is essential for the function of tRNAs as acceptors of amino acids. CCA is only encoded in the tRNA genes of some eubacterial species, where the CCAtrs are thought to be involved in the repair of defective tRNAs lacking part or the entire CCA tail. In most other organisms, including eukaryotes, the CCA is not encoded in the genome and is therefore added to tRNAs de novo and is essential for cell viability. CCA addition occurs independently of a nucleic acid template, and information from crystal structures indicates that the catalytic mechanism to generate the correct CCA end depends on a protein template.
CCAtrs were found to belong to the superfamily of Pol β-NTrs (Martin and Keller 1996; Aravind and Koonin 1999). Based on specific sequence patterns in the catalytic sites, CCAtrs and related NTrs were then divided into two classes that assigned archaeal CCAtrs to class I and eubacterial and eukaryotic CCAtrs to class II NTrs (see above; Yue et al. 1996). The two lineages of CCAtrs may have been derived independently from a common ancestor and have developed slightly different mechanisms for CCA addition (Fig. 5).
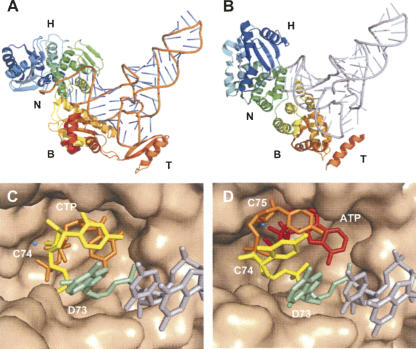
CCAtrs use a very intriguing mechanism to add a CCA sequence to the 3′-end of tRNAs, and this is nearly as fascinating as the far more complex mechanisms of template-dependent RNA polymerases. Although several laboratories contributed a considerable amount of biochemical information to solve the mystery of CCA addition, it was structural biology that found the solution to the enigma. Several crystal structures were initially solved of the apo form or of binary complexes between class I and II CCAtrs and CTP or ATP ribonucleotides (Li et al. 2002; Augustin et al. 2003; Xiong et al. 2003). CCAtrs were seen to consist of a head structure containing the active site with a nucleotide in place followed by a neck, body, and tail domain (Fig. 6A,B). In the class II CCAtr from Bacillus stearothermophilus, specific selection for CTP or ATP was obvious (Li et al. 2002), whereas in the structures of Archaeoglobus fulgidus class I CCAtr CTP and ATP were found to be bound nonspecifically (Xiong et al. 2003). Structures of ternary complexes of the Aquifex aeolicus class II A-adding enzyme (in some ancient bacteria CCA addition is carried out by separate CC- and A-adding enzymes) with a tRNA minisubstrate that show a preinsertion stage were determined subsequently (Fig. 6B; Tomita et al. 2004). At the same time, structures of the A. fulgidus CCAtr were obtained for each step of CCA addition (Fig. 6A; Xiong and Steitz 2004). The transferase was complexed to a minihelix substrate representing the top half of a tRNA (a mimic of the tRNA's acceptor stem, which is the double-stranded region close to the tRNA's 3′- and 5′-ends) and to a nucleotide, and this allowed the prediction of a mechanism for CCA formation. Although the addition of the first C (C74) could not be visualized, recognition of the correct nucleotides for incorporation at positions C75 and A76 was shown to be achieved by the formation of a single nucleotide-binding pocket in which the specificity for CTP and ATP is determined by the side chain of an arginine residue and backbone phosphates of the tRNA. These interactions with the Watson–Crick face of the nucleotide base are not complementary to UTP and GTP and thus exclude these nucleotide triphosphates as substrates. After addition of C75, the binding pocket is enlarged enough to allow binding of the purine base of ATP followed by the covalent addition of AMP to complete the CCA chain. The CCA end is finally stabilized in a stacked conformation, and the 3′-terminal A76 is no longer positioned in the primer-binding site to prevent further nucleotide addition. This way, the CCA end is scrunched into the catalytic pocket by sequential refolding of the newly added nucleotides (Fig. 6C,D). The authors also demonstrate that structural alterations for the reformation of the binding pocket specific for CTP or ATP recognition or for termination are accompanied by a progressive repositioning of the head domain. Furthermore, a conserved β-turn motif located close to the active site was found to play an important role during CCA synthesis. The flexible β-turn participates in all steps of CCA addition to direct the consecutive addition of all three nucleotides (Xiong and Steitz 2004; Cho et al. 2006; Tomita et al. 2006). These structural observations confirm results of UV cross-linking experiments that demonstrated that the tRNA remains firmly bound to the enzyme and does not translocate during CCA synthesis but that the 3′-end is repositioned at every step of the reactions (Shi et al. 1998a; Cho et al. 2006).
FIGURE 6.
Two structure models of class I and class II CCAtrs complexed with tRNAs and the reshaping active site of A. fulgidus CCAtr with bound tRNA 3′-end and CTP. (A) The class I A. fulgidus structure model in complex with tRNA (orange; pdb accession 1SZ1) (Xiong and Steitz 2004); (H) head domain, (N) neck, (B) body, and (T) tail. The polypeptide is colored in rainbow colors from dark blue at the N terminus via green, yellow, orange to red at the C terminus. (B) Class II CCAtr from Aquifex aeolicus with (gray) tRNA modeled instead of the cocrystallized minihelix (pdb accession 1VFG) (Tomita et al. 2004). Coloring of the polypeptide is as in D. (C) Active-site pocket with tRNA-DC74 and CTP. Two gray residues of the tRNA (at the right) followed by (pale green) D73 (the discriminator base), C74, and (orange) adjacent CTP. (Cyan sphere) A metal ion. (D) (pale green) Discriminator base D73 followed by (yellow) C74, (orange) C75, and with (red) bound ATP and (cyan sphere) a metal ion.
A more complete sequential analysis of CCA addition including six structures of the A. fulgidus CCAtr has recently been reported (Tomita et al. 2006). Each step of the CCA addition is shown with and without CTP or ATP in position, except for the structure with the D73 minihelix (D73 is the discriminator base). Unexpectedly, when the A. fulgidus CCAtr was cocrystallized with the D73 minihelix, the RNA was seen with two melted base pairs and extrusion of two other bases in the tRNA acceptor stem. At the same time, the 3′-end of the acceptor stem expanded and the discriminator base was positioned directly into the active site (Tomita et al. 2006). Although such a mechanism would in principle be possible, all these rearrangements in a folded RNA could be energetically too expensive. The base pairs melted in the minihelix, in fact, have no counterparts in a natural tRNA but are located at the junction between the acceptor stem and the rest of the tRNA. Moreover, other reports indicate that no translocation of the tRNA takes place during all steps of CCA synthesis (Cho et al. 2006) and that the TΨC loop is not needed for efficient and faithful C74 addition (Shi et al. 1998a). Further work is needed to clarify the early steps of CCA synthesis.
A critical aspect in the CCA-addition reaction is the contact of the tRNA substrate with regions outside the active site, namely, with the neck, body, and tail domains of the CCAtr (Fig. 6A,B). It was shown earlier that addition of C75 and C76 can be performed efficiently and faithfully even if the tRNA is locked onto the enzyme by UV cross-linking (Shi et al. 1998a). These contacts were also studied in deletion mutants and by swapping domains between class II E. coli PAP and CCAtr (Betat et al. 2004). The authors identified a distinct region in the body domain between residues 219 and 254 of the CCAtr that seemed responsible for faithful CCA addition. When these residues were exchanged with the C terminus of PAP containing the corresponding residues such that the enzyme now consisted of the CCAtr head and neck domain and the PAP body and tail domain, the chimera generated poly(CCA). Moreover, in the reverse swap with the N terminus coming from PAP and the C terminus with the critical residues provided by CCAtr, the chimera still added CCA to the tRNA. The proposal that a distinct helix corresponding to helix M in the B. stearothermophilus structure was responsible for regulation of the nucleotide specificity in the active site (Li et al. 2002) is now rather unlikely in light of several recently published structures of class I and class II CCAtrs complexed with tRNAs, where this particular helix is not seen to contact the tRNA. It rather seems that helices M, N, and O within the body domain are responsible for the tight binding of the tRNA's acceptor stem and helices G and J are contacting the 3′- and 5′-ends of the acceptor stem close to the active site. The firm contact between the body domain of CCAtrs and the center of the acceptor stem appears to be responsible for ensuring the addition of a single CCA. In contrast, the analogous region in a predicted body domain of the bact-PAPs binds RNA more weakly to allow the emerging poly(A) tail to repeatedly translocate on its surface. Furthermore, the tail domain of CCAtr and its interaction with the tRNA's TΨC loop does not seem to be necessary for CCA synthesis (Shi et al. 1998a,b; Betat et al. 2004; Xiong and Steitz 2004).
The structural information also made it possible to reengineer the B. stearothermophilus CCA-adding enzyme by introducing mutations in the catalytic site such that the enzyme incorporated UTP and GTP instead of CTP and ATP, leading to the addition of UUG instead of CCA to the tRNA's 3′-end (Cho et al. 2007). Likewise, the enzyme could be converted into a dCdCdA-adding enzyme by mutating an arginine that interacts with the 2′-hydroxyl of the incoming ribose. In addition, the related A. aeolicus CC- and A-adding enzymes were modified to become UU- and G-adding enzymes, and the E. coli poly(A) polymerase was transformed into a poly(G) polymerase. These experiments confirm the proposed determinants of nucleotide selection from crystal structures of class II CCAtrs.
TERMINAL RNA URIDYLYL TRANSFERASES
Terminal RNA uridylyl transferases catalyze the transfer of UMP residues to the 3′-hydroxyl group of RNAs in eukaryotes (for review, see Aphasizhev 2005; Stuart et al. 2005). Several recently described TUTases were termed poly(U) polymerases (PUPs) because these processive enzymes add up to several hundred U residues to RNAs (Kwak and Wickens 2007; Rissland et al. 2007). Therefore, the distinction between TUTases and PUPs is not clear yet because some protist TUTases also make long poly(U) tails (see below). Most studies on TUTases were done in the parasitic protists Trypanosoma brucei, Leishmania tarentolae, and Leishmania major, where TUTases are involved in the editing of mitochondrial mRNAs (Bakalara et al. 1989; Simpson et al. 2004).
The primary transcripts of numerous protist mitochondrial genes undergo post-transcriptional editing by the insertion or deletion of U residues resulting in the correction of frameshifts, creation of start and stop codons, and sometimes contributing a large part of the mRNA coding sequence. The reaction is initiated by annealing of a guide RNA (gRNA) to a pre-mRNA, followed by endonucleolytic cleavage at a non-base-paired editing site next to the RNA duplex and addition or deletion of Us at the 3′-end of the upstream cleavage fragment, and is completed by ligation of the gap. Editing by U-insertion is probably regulated at the level of differential transcription of the gRNAs for the different genes (Blum and Simpson 1990).
Two proteins with U-adding activity have been studied extensively. These are associated with large multiprotein complexes, one of which is the editosome. RNA-editing TUTase 1 (RET1) is a processive enzyme that can add poly(U) tails of up to several hundred residues to RNAs and in addition adds ∼15 Us to gRNAs, indicating that these U-adding enzymes have two distinct functions in editing. In contrast, RNA-editing TUTase 2 (RET2) is a distributive enzyme, which only adds one U per binding event and is involved in U-insertion into mRNAs cleaved and stabilized by gRNAs at editing sites (McManus et al. 2000). In addition, TUT3, a protein homologous to RET1 and RET2, was identified in T. brucei and L. major (Aphasizhev et al. 2004). Editosomes contain exonucleases, helicases, and RNA ligases in addition to TUTases (Stuart et al. 2005).
Recently reported crystal structures of RET2 (Figs. 2C and 4B) and TUT4, a minimal catalytically active RNA uridylyl transferase, both demonstrate specific UTP selection by a network of protein ligands and water molecules in the active site (Deng et al. 2005; Stagno et al. 2007).
RNA editing involving U-insertion/deletion is unique to kinetoplastid mitochondria and is not found in other organisms. However, TUTase activity resulting in U-addition to the 3′-ends of RNAs is common also in metazoa and humans. For example, a human U6 snRNA-specific TUTase was recently reported (Trippe et al. 2006). This essential enzyme adds or restores four U residues at the 3′-end of U6 snRNA as its sole substrate. These four U residues are not single-stranded but form an intramolecular double strand with a stretch of As within the U6 molecule. So far no data are available on the possible mechanism of U-addition to U6 snRNA except that an RNA templating mechanism can be excluded for (U)4 synthesis (Trippe et al. 2003). It is possible that U6-TUTase uses a mechanism similar to CCA-adding enzymes to scrunch and reposition the 3′-end of the U6 snRNA for the next U-addition and to stop the reaction when the (U)4 tail is complete.
Some trypanosomatid TUTases and the human U6-specific TUTase contain sequences that predict a domain protruding from the catalytic site similar to that found in the trypanosomal RET2 structure (Fig. 2C; Deng et al. 2005). This structure, which was named “middle domain” (MD) in the RET2 protein, is topologically similar to an RNA recognition motif (RRM) of the spliceosomal U1A protein and might be responsible for binding of the U6 snRNA substrate. In addition, the U6-specific TUTase contains sequence homology with a second RRM at its N terminus (Fig. 1). It is possible that one of the RRMs interacts with RNA and the other with a protein.
Recently, the S. pombe noncanonical rNTr Cid1 (Cid1 stands for caffeine-induced death protein 1; see below for further details) was found to be a poly(U) polymerase that adds up to several hundred U residues onto poly(A) tails of mRNAs after cell cycle arrest (Rissland et al. 2007). In addition, rNTrs that add poly(U) tails to RNA were identified in Arabidopsis, C. elegans, and humans (Kwak and Wickens 2007). These surprising results indicate that U-addition probably plays a more general role in eukaryotes. One of the possible functions of poly(U) tails is RNA turnover. This is supported by recent evidence, in which poly(U) tails were proposed to be involved in the decay of micro-RNA-directed cleavage products in species including Arabidopsis, mouse, and EB virus (Shen and Goodman 2004).
THE REGULATORY CYTOPLASMIC POLY(A) POLYMERASE GLD-2
The germline development gene gld-2 was initially identified as a regulator of the mitosis/meiosis decision in the C. elegans germline (Kadyk and Kimble 1998). Mutants of the gene resulted in defects of germline development (Wang et al. 2002). The Gld-2 protein was found to be required for progression through meiotic prophase and is needed to complete both spermatogenesis and oogenesis in the nematode. In the mouse, GLD-2 was proposed to be a positive regulator in the progression of metaphase I to metaphase II during oocyte maturation (Nakanishi et al. 2006). Sequence comparisons suggest that GLD-2 belongs to the Pol β-NTr superfamily as a noncanonical PAP (Figs. 1 and 3; Wang et al. 2002). GLD-2 was found by Wang et al. to require the collaboration of GLD-3, a bicaudal-C family RNA-binding protein. GLD-3 forms a complex with GLD-2 and binds specific mRNAs, which are then polyadenylated by GLD-2, leading to translational activation of dormant mRNAs by extending their short poly(A) tails (Wang et al. 2002).
Supporting evidence for the mRNA-activating function of GLD-2/GLD-3 came from the work of Suh et al. (2006), who found the gld-1 transcript to be one of the target mRNAs of GLD-2. Gld-1 is another gene acting as a germline regulator (Kadyk and Kimble 1998). The investigators propose that after gld-1 mRNA is activated through polyadenylation by GLD-2, the resulting GLD-1 protein contributes to induce the cell to enter meiosis. Moreover, it was demonstrated that when a MS2-luciferase mRNA fusion was tethered to GLD-2 via a MS2 coat protein, this RNA was very efficiently polyadenylated and translated to active protein (Kwak et al. 2004).
In Xenopus oocytes, a GLD-2 homolog was found to be involved in regulated cytoplasmic polyadenylation as a component of the CPEB/CPSF complex (cytoplasmic polyadenylation element-binding protein and cleavage and polyadenylation specificity factor complex) together with the scaffold protein symplekin (Barnard et al. 2004). After symplekin and GLD-2 have assembled with the complex, a poly(A) tail of ∼200 nt is synthesized. This activity can be suppressed by the RNA-binding proteins maskin, Pumilio, and Nanos (for review, see Mendez and Richter 2001). Furthermore, the poly(A) nuclease PARN was shown to reside in the CPEB/CPSF complex together with symplekin and GLD-2 (Kim and Richter 2006). If PARN is present, the poly(A) tails are kept short, and when oocytes mature, PARN is released such that the poly(A) tails can be elongated by GLD-2.
THE CID1 FAMILY OF NONCANONICAL RNTRS OF FISSION YEAST
The Cid1 family of noncanonical rNTrs defines a group of enzymes found in S. pombe in addition to the canonical PAP pla1 (for review, see Stevenson and Norbury 2006). Sequence comparison classifies this family with the noncanonical rNTrs because of the presence of a nucleotide recognition motif of type NMR 2 (Fig. 3; see above).
The cid1 gene was identified in a screen for suppressors of “checkpoint Rad” mutants in fission yeast. These mutants are sensitive to hydroxyurea (HU), an inhibitor of ribonucleotide reductase (Wang et al. 1999b). Deletion of cid1 also causes sensitivity to the combination of HU and caffeine (therefore the name for Cid 1, caffeine-induced death 1) and cid1Δ mutants affect S–M checkpoints if combined with mutants of DNA polymerases δ and ε, two DNA polymerases involved in chromosomal DNA replication (Wang et al. 2000a).
Surprisingly, recombinant Cid1 expressed in bacteria was initially found to have both poly(A) and poly(U) polymerase (PUP) activity in vitro (Read et al. 2002). However, a native Cid1 complex purified from S. pombe showed only poly(U)-adding activity (Rissland et al. 2007).
Cid13 was identified as a noncanonical PAP located both in the nucleus and in the cytoplasm (Read et al. 2002; Saitoh et al. 2002; Matsuyama et al. 2006). Similar to Cid1, Cid13 was found to partially suppress HU sensitivity of rad3 mutants but was not able to suppress the S–M checkpoint defect of rad3Δ. It was also reported that Cid13 elongates the short poly(A) tails of dormant suc22 mRNA encoding the small subunit of ribonucleotide reductase (Saitoh et al. 2002). However, these results could not be confirmed (Read et al. 2002). It is therefore believed that Cid1 and Cid13 target still-unknown mRNAs to establish bypass pathways for the restoration of dNTP levels after HU treatment or to rescue replication blocks (Stevenson and Norbury 2006).
Cid14 was found to be a functional homolog of Trf4 and Trf5 in Baker's yeast (two PAPs involved in RNA turnover; see below) (Win et al. 2006). cid14 + could even complement a trf4ts top1Δ yeast mutant to grow at restrictive temperatures. Deletion of cid14 did not impair sister-chromatid separation, although Trf4 was reported to be required for sister-chromatid cohesion during S phase to ensure faithful chromosome segregation. Nevertheless, the cid14 mutation showed an increased rate of chromosome segregation failure. Also, Cid14-dependent polyadenylation of some rRNAs was found; these RNAs were then directed to degradation by the exosome (Win et al. 2006). Therefore, Cid14, like Trf4 and Trf5, appears to be involved in RNA turnover.
So far no function has been reported for Cid11 and Cid16. Cid16 is predicted to be a mitochondrial PAP because it was localized to speckles in the cytoplasm (Matsuyama et al. 2006). The fact that fission yeast contains at least six different noncanonical rNTrs compared to S. cerevisiae supports the view that S. pombe is a more complex organism than budding yeast. The higher number of Cid1 PAPs provides the fission yeast with more flexibility for the control of gene expression.
THE NONCANONICAL RNTRS TRF4 AND TRF5 OF S. CEREVISIAE
Trf4 was initially isolated in a screen for mutants the growth defects of which are relieved by the overexpression of topoisomerase I (Top1) (Sadoff et al. 1995). The resulting mutant genes were named trf for topoisomerase I-requiring function. One particularly interesting mutant, trf4, was found to be hypersensitive to the microtubule poison thiabendazole but to have little sensitivity to hydroxyurea. trf4 and top1 single mutants have similar phenotypes such as increased recombination of rDNA and failure to shut off RNA polymerase II transcription during stationary phase. Also, Trf5 was shown to be a homolog of Trf4, and the two genes together are essential for growth and are needed for proper mitosis (Castano et al. 1996b). Moreover, top1 trf4-ts double mutants were found to affect mitotic events such as chromosome condensation, spindle elongation, and nuclear segregation, but not DNA replication (Castano et al. 1996a). In addition, these investigators demonstrated that the top1 trf4-ts mutant is deficient in chromosome condensation of rDNA at mitosis and that Trf4 associates physically with both Smcl and Smc2, the yeast homologs of Xenopus proteins that are required for mitotic chromosome condensation in vitro. Trf4 has also been reported to have DNA polymerase activity and, in fact, to be the replicative DNA polymerase κ (Wang et al. 2000b). These results were challenged by more recent work that showed Trf4 to be a poly(A) polymerase (Kadaba et al. 2004; LaCava et al. 2005; Vanacova et al. 2005; Wyers et al. 2005).
Several reports have demonstrated a role for Trf4 in RNA turnover and quality control acting on rRNA, tRNAs, snoRNAs, and cryptic RNA polymerase II transcripts (LaCava et al. 2005; Vanacova et al. 2005; Wyers et al. 2005; for review, see Vanacova and Stefl 2007). Trf4 was found in a complex with the zinc knuckle proteins Air1 and Air2 and the RNA helicase Mtr4 (a component of the nuclear exosome) and was therefore called Trf4/Air1–2/Mtr4 polyadenylation (TRAMP) complex (LaCava et al. 2005). RNAs bound by the TRAMP complex were polyadenylated, delivered to the exosome, and degraded to small fragments or single nucleotides. The zinc finger proteins Air1 and Air2 are thought to act as specific RNA-binding cofactors similar to GLD-3, which is assisting the cytoplasmic PAP GLD-2 in binding specific mRNAs for polyadenylation (Wang et al. 2002; see above).
In a screen for suppressors of mutants of the essential tRNA m1A58 methyltransferase Trm6, two yeast mutants were identified that suppress the trm6-504 mutant phenotype and restore hypomodified tRNAi Met to near normal levels (Kadaba et al. 2004), whereas the absence of the A58 modification causes tRNAi Met instability in wild-type cells. The investigators identified one suppressor gene to encode Dis3/Rrp44, a 3′-5′-exoribonuclease as a member of the multi-subunit exosome complex and another suppressor to be Trf4. Whereas deletion of the trf4 gene was found to stabilize tRNAi Met, overexpression of Trf4 destabilized the hypomodified tRNAi Met in trm6-504 cells. These results suggested that a tRNA surveillance pathway exists in yeast that polyadenylates and degrades hypomodified tRNAi Met and for that purpose requires Trf4 and the exosome.
These in vivo results have been confirmed by Vanacova and colleagues, who tested the proposed quality-control system in vitro by combining TAP-tagged Trf4 and Rrp6 complexes isolated from yeast and incubating them together with unmodified, in vitro transcribed tRNAi Met or with native tRNAi Met isolated from cells (Vanacova et al. 2005). Rrp6 is a component of the nuclear exosome, which is missing in the cytoplasmic form of the exosome (for review, see Vanacova and Stefl 2007). The results showed that the TRAMP complex added a poly(A) tail only to unmodified but not to native tRNAi Met. Such polyadenylated tRNAs were subsequently degraded to mono- or oligonucleotides. The authors also demonstrated that simply adding a poly(A) tail was not sufficient to promote degradation of the tRNA by the exosome but that continuous readenylation was needed because a Trf4 mutation that impairs catalytic activity no longer stimulated the degradation process. How the TRAMP complex can distinguish modified from nonmodified tRNAi Met remains to be elucidated. Air1 and Air2 are thought to be specificity factors for the recognition of incorrectly folded tRNAs. More recently, it has been shown that recombinant Rrp44 and the TRAMP polyadenylation complex each specifically recognizes tRNAi Met lacking a single m1A58 modification (Schneider et al. 2007).
In a mass spectrum analysis of the TRAMP complex, Vanacova et al. (2005) identified the putative hect E3 ubiquitin ligase Hul4, in addition to Air1, Air2, and Mtr4. Hul4 was found to be a nonessential ubiquitin ligase gene in yeast (Wang et al. 1999a). It is not clear whether Hul4 represents a link between Trf4 and its role in mitotic and meiotic cell division because the E3 ubiquitin ligase complex is also involved in chromosome segregation and in the metaphase-to-anaphase (M–A) transition in the cell cycle (for review, see Pines 2006). The M–A transition is controlled by the anaphase-promoting complex (APC), which targets many proteins for ubiquitylation and subsequent degradation by the 26S proteasome. It is possible that the TRAMP complex participates in the degradation of RNAs that were associated with these proteins and have to be disposed of as well.
The TRAMP complex was also found to participate in the processing or the degradation of U14 snoRNA, U5 snRNA, and pre-ribosomal RNA (LaCava et al. 2005). In addition, it was shown that several supposedly silent intergenic regions in the genome of S. cerevisiae are actually transcribed by RNA polymerase II, and that RNAs originating from these regions are rapidly degraded by the combined action of the exosome and Trf4 together with Air1 or Air2 (Wyers et al. 2005). In addition, the investigators could detect degradation of several RNA polymerase I and RNA polymerase III transcripts, and they have proposed that the TRAMP-mediated quality-control mechanism reverses inappropriate expression of genetic information.
These findings were quite surprising because polyadenylation in eukaryotes was considered to stabilize RNAs, and RNA degradation stimulated by poly(A) tails was known only in bacteria. The new findings indicate that eukaryotes have more than one pathway for the polyadenylation of RNA and that the addition of poly(A) tails is not limited to pre-mRNAs. Depending on the type and the cellular location of the polyadenylation machinery involved, the poly(A) tails can have different functions.
CANONICAL AND NONCANONICAL RIBONUCLEOTIDYL TRANSFERASES IN HUMANS
Figure 1 lists known and predicted human rNTrs. We refer to the respective sections above for descriptions of the canonical, some noncanonical rNTrs, and the CCAtrs. Here, we discuss the mitochondrial PAP, predicted noncanonical PAPs, and TUTases that have not been well characterized yet.
Polyadenylation of mitochondrial RNAs in humans is catalyzed by a recently described mitochondrial noncanonical PAP (Fig. 1; Tomecki et al. 2004; Nagaike et al. 2005). It appears that mitochondrial PAP and human polynucleotide phosphorylase (hPNPase) are both involved in mitochondrial polyadenylation. The results suggest that the activity of human mitochondrial PAP leads to increased stability of mitochondrial mRNA, whereas polyadenylation by hPNPase is rather involved in mRNA deadenylation and RNA turnover. It has been suggested that additional factors are involved in the regulation of polyadenylation of pre-mRNAs in mitochondria (Nagaike et al. 2005).
Recently, a 200-kDa protein with homology with two copies of Pol β-NTrs was isolated and named ZCCHC11L (Minoda et al. 2006). The N-terminal copy of PAP in this protein is predicted to be inactive because some metal chelating residues are missing (Fig. 1). ZCCHC11 was found to interact with TIFA (TRAF-interacting protein with a forkhead-associated domain) in a macrophage cell line (Minoda et al. 2006). Overexpression and knockdown by siRNA indicated that ZCCHC11 functions as a negative regulator of Toll-like receptor-mediated NF-κ B activation. However, the N-terminal region including the CCHC-type Zn-finger motif was sufficient for suppression of NF-κ B, and it is unclear what role the predicted poly(A) polymerase homologies of this unusual protein plays in these pathways (Minoda et al. 2006).
The databases contain another predicted protein of 1495 amino acids with strong homology with ZCCHC11, named ZCCHC6 (Fig. 1). This protein also contains three Zinc-finger motifs and homologies to two copies of noncanonical rNTrs of which the N-terminal copy is also likely to be inactive. ZCCHC6 was recently shown to have poly(U) polymerase activity (Rissland et al. 2007).
The two predicted Trf4 and Trf5 homologs are not characterized. Based on their similarity to yeast Trf4 and Trf5, they are assumed to have similar functions and to be required for the repair of camptothecin-mediated damage to DNA (Walowsky et al. 1999).
2′-5′-OLIGO(A) SYNTHETASES
2′-5′-Oligoadenylate synthetases (OASs; also named 2′-nucleotidyl transferases) are interferon (IFN)-induced antiviral enzymes that play an important role in the mechanisms of interferon-mediated antiviral activity (for review, see Justesen et al. 2000). OAS was also found to be involved in other cellular processes such as apoptosis and growth control (Justesen et al. 2000). Induction by IFNs works via membrane receptors followed by a rapid transcriptional activation of a specific set of cellular genes including several OAS genes (Benech et al. 1987). Both IFN-I and IFN-II are thought to induce the antiviral effect of OASs. When activated by double-stranded RNA, OASs oligomerize ATP into a 2′-5′-linked oligoadenylate (2–5A) of two to ∼30 residues. 2–5A is bound by the endoribonuclease RNase L and thereby activates the degradation of viral and cellular RNAs.
The structure of human OAS-3 has recently been reported (Hartmann et al. 2003). The enzyme has similarity with other noncanonical rNTrs (Figs. 1 and 2B) and suggests a similar catalytic mechanism in 2′- and 3′-specific nucleotidyl transferases. Comparison with structures of other rNTrs indicates that the nucleotide is bound by conserved active-site ligands, whereas the RNA substrates are recognized by nonconserved regions (Hartmann et al. 2003).
This enzyme differs strongly from the conventional rNTrs in two respects. First, it synthesizes 2–5A de novo from ATP without an RNA primer. Second, the nucleotide is not transferred to the 3′-end of the recipient substrate but to the 2′-OH. This may be needed to distinguish 2–5A RNA from other RNAs for the recognition by RNase L.
POXVIRAL POLY(A) POLYMERASE
The prototype of the poxvirus family is vaccinia virus. Polyadenylation of viral mRNAs, which is crucial for virion maturation, is carried out by a poly(A) polymerase heterodimer composed of the catalytic component VP55 and by VP39, which acts as a processivity factor. VP39 also functions as a cap-specific nucleoside-2′-O-methyl transferase at the 5′-ends of viral mRNAs (Schnierle et al. 1992).
The recently reported crystal structure of vaccinia PAP complexed with ATP (pdb accession 2GA9) reveals an unusual architecture for VP55 that comprises N-terminal, catalytic, and C-terminal domains that differ from the nomenclature used for eukaryotic poly(A) polymerases (Moure et al. 2006). The N-terminal domain of VP55 does not exist in other PAPs, the catalytic domain corresponds to the catalytic domain in eukaryotic PAPs, and the C-terminal domain corresponds to the central domain of eukaryotic PAPs (Fig. 2D). The different domains in VP55 also do not seem to be flexible as in the case of yeast and mammalian PAPs and thus do not suggest closure of the active site upon substrate binding. In addition, an RNA path was proposed in the VP55/VP39 complex that involves both subunits of the heterodimer. Vaccinia VP55 PAP was classified in the extra group 8 of Aravind and Koonin (Fig. 5) and is thought to have evolved independently of the bacterial and the canonical and noncanonical rNTrs of eukaryotes (Aravind and Koonin 1999). Surprisingly, inspection of the VP55 structure reveals the presence of two helical turns between β-strands 1 and 2, reminiscent of the class II NTrs (Yue et al. 1996), although the protein sequences strongly differ. In addition, the structure of VP55 suggests a structural homology between its central domain and the central domains of other canonical and noncanonical rNTrs (Fig. 2D). However, although the function of the central domain in vaccinia PAP appears to be nucleotide recognition, the protein sequence in this region shows no homology with other PAPs, and its evolutionary origin is not known. The structure indicates that the nucleotide in the active site of VP55 is specifically recognized by several amino acid side chains and water molecules (Moure et al. 2006). This is in contrast to canonical PAPs from eukaryotes, where only semispecific selection was found in crystal structures. VP55 seems to employ a different mode of poly(A) synthesis, which does not include an induced-fit mechanism such as in canonical PAPs. Kinetic studies could reveal the nature of the catalytic mechanism in this enzyme.
MINIMAL NUCLEOTIDYL TRANSFERASES
The “minimal nucleotidyl transferases” (MNTs) were previously defined as a group of Pol β-NTrs in eubacteria and archaea. For example, up to 13 copies of MNTs per genome exist in Archaeoglobus (classified in group 5 by Aravind and Koonin 1999). In Haemophilus influenza, the two open reading frames HI0073 and HI0074 form an operon coding for a two-protein MNT. The recently determined structure of the catalytic domain (pdb:1NO5) displays a four-stranded β-sheet with a helical turn between β-strands 2 and 3 and three metal-chelating Asp residues characteristic of class I Pol β-type polymerases (Lehmann et al. 2005). The structure of HI0074 (pdb:1JOG) predicted to represent the substrate-binding subunit of the MNT dimer is a four-helix bundle and resembles the substrate-binding domain of kanamycin nucleotidyl transferase (KanNT), an antibiotics resistance factor (Pedersen et al. 1995). Nevertheless, no substrates are known for the HI0073/74 MNT, and no function for the protein has been described. Structural homology has been found to the catalytic domains of PAPs, CCAtrs, and to KanNT with the DALI server (Holm and Sander 1993). In addition, an unusually high number of pseudogenes are coding for MNTs present in the genomes of both eubacteria and archaea, suggesting that these genes represent duplications that could be activated any time during evolution. Thus, MNTs are only some of many examples that show how much we still have to learn about the function and evolution of the fascinating, diverse, and ubiquitous family of nucleotidyl transferases.
ACKNOWLEDGMENTS
We thank Sylvie Doublié (University of Vermont at Burlington) and Christiane Rammelt for reading the manuscript and for many useful suggestions. We also thank the two referees for their constructive criticism and very helpful suggestions. This work has been supported by the University of Basel and the Swiss National Science Fund.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.652807.
REFERENCES
- Aphasizhev, R. RNA uridylyltransferases. Cell. Mol. Life Sci. 2005;62:2194–2203. doi: 10.1007/s00018-005-5198-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aphasizhev, R., Aphasizheva, I., Simpson, L. Multiple terminal uridylyltransferases of trypanosomes. FEBS Lett. 2004;572:15–18. doi: 10.1016/j.febslet.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Aravind, L., Koonin, E.V. DNA polymerase β-like nucleotidyltransferase superfamily: Identification of three new families, classification, and evolutionary history. Nucleic Acids Res. 1999;27:1609–1618. doi: 10.1093/nar/27.7.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- August, J.T., Ortiz, P.J., Hurwitz, J. Ribonucleic acid-dependent ribonucleotide incorporation. I. Purification and properties of the enzyme. J. Biol. Chem. 1962;237:3786–3793. [PubMed] [Google Scholar]
- Augustin, M.A., Reichert, A.S., Betat, H., Huber, R., Mörl, M., Steegborn, C. Crystal structure of the human CCA-adding enzyme: Insights into template-independent polymerization. J. Mol. Biol. 2003;328:985–994. doi: 10.1016/s0022-2836(03)00381-4. [DOI] [PubMed] [Google Scholar]
- Bakalara, N., Simpson, A.M., Simpson, L. The Leishmania kinetoplast-mitochondrion contains terminal uridylyltransferase and RNA ligase activities. J. Biol. Chem. 1989;264:18679–18686. [PubMed] [Google Scholar]
- Balbo, P.B., Meinke, G., Bohm, A. Kinetic studies of yeast polyA polymerase indicate an induced fit mechanism for nucleotide specificity. Biochemistry. 2005;44:7777–7786. doi: 10.1021/bi050089r. [DOI] [PubMed] [Google Scholar]
- Balbo, P.B., Toth, J., Bohm, A. X-Ray crystallographic and steady state fluorescence characterization of the protein dynamics of yeast polyadenylate polymerase. J. Mol. Biol. 2007;366:1401–1415. doi: 10.1016/j.jmb.2006.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bard, J., Zhelkovsky, A.M., Helmling, S., Earnest, T.N., Moore, C.L., Bohm, A. Structure of yeast poly(A) polymerase alone and in complex with 3′-dATP. Science. 2000;289:1346–1349. doi: 10.1126/science.289.5483.1346. [DOI] [PubMed] [Google Scholar]
- Barnard, D.C., Ryan, K., Manley, J.L., Richter, J.D. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell. 2004;119:641–651. doi: 10.1016/j.cell.2004.10.029. [DOI] [PubMed] [Google Scholar]
- Benech, P., Vigneron, M., Peretz, D., Revel, M., Chebath, J. Interferon-responsive regulatory elements in the promoter of the human 2′,5′-oligo(A) synthetase gene. Mol. Cell. Biol. 1987;7:4498–4504. doi: 10.1128/mcb.7.12.4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betat, H., Rammelt, C., Martin, G., Mörl, M. Exchange of regions between bacterial poly(A) polymerase and the CCA-adding enzyme generates altered specificities. Mol. Cell. 2004;15:389–398. doi: 10.1016/j.molcel.2004.06.026. [DOI] [PubMed] [Google Scholar]
- Blum, B., Simpson, L. Guide RNAs in kinetoplastid mitochondria have a nonencoded 3′ oligo(U) tail involved in recognition of the preedited region. Cell. 1990;62:391–397. doi: 10.1016/0092-8674(90)90375-o. [DOI] [PubMed] [Google Scholar]
- Carpousis, A.J. The RNA degradosome of Escherichia coli: A multiprotein mRNA-degrading machine assembled on RNase E. Annu. Rev. Microbiol. 2007;6:71–87. doi: 10.1146/annurev.micro.61.080706.093440. [DOI] [PubMed] [Google Scholar]
- Castano, I.B., Brzoska, P.M., Sadoff, B.U., Chen, H., Christman, M.F. Mitotic chromosome condensation in the rDNA requires TRF4 and DNA topoisomerase I in Saccharomyces cerevisiae . Genes & Dev. 1996a;10:2564–2576. doi: 10.1101/gad.10.20.2564. [DOI] [PubMed] [Google Scholar]
- Castano, I.B., Heath-Pagliuso, S., Sadoff, B.U., Fitzhugh, D.J., Christman, M.F. A novel family of TRF (DNA topoisomerase I-related function) genes required for proper nuclear segregation. Nucleic Acids Res. 1996b;24:2404–2410. doi: 10.1093/nar/24.12.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, H.D., Chen, Y., Varani, G., Weiner, A.M. A model for C74 addition by CCA-adding enzymes: C74 addition, like C75 and A76 addition, does not involve tRNA translocation. J. Biol. Chem. 2006;281:9801–9811. doi: 10.1074/jbc.M512603200. [DOI] [PubMed] [Google Scholar]
- Cho, H.D., Verlinde, C.L., Weiner, A.M. Reengineering CCA-adding enzymes to function as (U,G)- or dCdCdA-adding enzymes or poly(C,A) and poly(U,G) polymerases. Proc. Natl. Acad. Sci. 2007;104:54–59. doi: 10.1073/pnas.0606961104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colgan, D.F., Murthy, K.G., Prives, C., Manley, J.L. Cell-cycle related regulation of poly(A) polymerase by phosphorylation. Nature. 1996;384:282–285. doi: 10.1038/384282a0. [DOI] [PubMed] [Google Scholar]
- Colgan, D.F., Murthy, K.G., Zhao, W., Prives, C., Manley, J.L. Inhibition of poly(A) polymerase requires p34cdc2/cyclin B phosphorylation of multiple consensus and nonconsensus sites. EMBO J. 1998;17:1053–1062. doi: 10.1093/emboj/17.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano, W.L. The PyMOL Molecular Graphics System. 2002 http://www.pymol.org
- Delarue, M., Boule, J.B., Lescar, J., Expert-Bezancon, N., Jourdan, N., Sukumar, N., Rougeon, F., Papanicolaou, C. Crystal structures of a template-independent DNA polymerase: Murine terminal deoxynucleotidyltransferase. EMBO J. 2002;21:427–439. doi: 10.1093/emboj/21.3.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, J., Ernst, N.L., Turley, S., Stuart, K.D., Hol, W.G. Structural basis for UTP specificity of RNA editing TUTases from Trypanosoma brucei . EMBO J. 2005;24:4007–4017. doi: 10.1038/sj.emboj.7600861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher, M.P. Degradation of stable RNA in bacteria. J. Biol. Chem. 2003;278:45041–45044. doi: 10.1074/jbc.R300031200. [DOI] [PubMed] [Google Scholar]
- Edmonds, M. A history of poly A sequences: From formation to factors to function. Prog. Nucleic Acid Res. Mol. Biol. 2002;71:285–389. doi: 10.1016/s0079-6603(02)71046-5. [DOI] [PubMed] [Google Scholar]
- Edmonds, M., Abrams, R. Polynucleotide biosynthesis: Formation of a sequence of adenylate units from adenosine triphosphate by an enzyme from thymus nuclei. J. Biol. Chem. 1960;235:1142–1149. [PubMed] [Google Scholar]
- Gunderson, S.I., Beyer, K., Martin, G., Keller, W., Boelens, W.C., Mattaj, L.W. The human U1A snRNP protein regulates polyadenylation via a direct interaction with poly(A) polymerase. Cell. 1994;76:531–541. doi: 10.1016/0092-8674(94)90116-3. [DOI] [PubMed] [Google Scholar]
- Gunderson, S.I., Vagner, S., Polycarpou-Schwarz, M., Mattaj, I.W. Involvement of the carboxyl terminus of vertebrate poly(A) polymerase in U1A autoregulation and in the coupling of splicing and polyadenylation. Genes & Dev. 1997;11:761–773. doi: 10.1101/gad.11.6.761. [DOI] [PubMed] [Google Scholar]
- Gunderson, S.I., Polycarpou-Schwarz, M., Mattaj, I.W. U1 snRNP inhibits pre-mRNA polyadenylation through a direct interaction between U1 70K and poly(A) polymerase. Mol. Cell. 1998;1:255–264. doi: 10.1016/s1097-2765(00)80026-x. [DOI] [PubMed] [Google Scholar]
- Hajnsdorf, E., Braun, F., Haugel-Nielsen, J., Regnier, P. Polyadenylylation destabilizes the rpsO mRNA of Escherichia coli . Proc. Natl. Acad. Sci. 1995;92:3973–3977. doi: 10.1073/pnas.92.9.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann, R., Justesen, J., Sarkar, S.N., Sen, G.C., Yee, V.C. Crystal structure of the 2′-specific and double-stranded RNA-activated interferon-induced antiviral protein 2′-5′-oligoadenylate synthetase. Mol. Cell. 2003;12:1173–1185. doi: 10.1016/s1097-2765(03)00433-7. [DOI] [PubMed] [Google Scholar]
- Holm, L., Sander, C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- Holm, L., Sander, C. DNA polymerase β belongs to an ancient nucleotidyltransferase superfamily. Trends Biochem. Sci. 1995;20:345–347. doi: 10.1016/s0968-0004(00)89071-4. [DOI] [PubMed] [Google Scholar]
- Justesen, J., Hartmann, R., Kjeldgaard, N.O. Gene structure and function of the 2′-5′-oligoadenylate synthetase family. Cell. Mol. Life Sci. 2000;57:1593–1612. doi: 10.1007/PL00000644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba, S., Krueger, A., Trice, T., Krecic, A.M., Hinnebusch, A.G., Anderson, J. Nuclear surveillance and degradation of hypomodified initiator tRNAMet in S. cerevisiae . Genes & Dev. 2004;18:1227–1240. doi: 10.1101/gad.1183804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyk, L.C., Kimble, J. Genetic regulation of entry into meiosis in Caenorhabditis elegans . Development. 1998;125:1803–1813. doi: 10.1242/dev.125.10.1803. [DOI] [PubMed] [Google Scholar]
- Kashiwabara, S., Zhuang, T., Yamagata, K., Noguchi, J., Fukamizu, A., Baba, T. Identification of a novel isoform of poly(A) polymerase, TPAP, specifically present in the cytoplasm of spermatogenic cells. Dev. Biol. 2000;228:106–115. doi: 10.1006/dbio.2000.9894. [DOI] [PubMed] [Google Scholar]
- Kaufmann, I., Martin, G., Friedlein, A., Langen, H., Keller, W. Human Fip1 is a subunit of CPSF that binds to U-rich RNA elements and stimulates poly(A) polymerase. EMBO J. 2004;23:616–626. doi: 10.1038/sj.emboj.7600070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J.H., Richter, J.D. Opposing polymerase-deadenylase activities regulate cytoplasmic polyadenylation. Mol. Cell. 2006;24:173–183. doi: 10.1016/j.molcel.2006.08.016. [DOI] [PubMed] [Google Scholar]
- Kühn, U., Nemeth, A., Meyer, S., Wahle, E. The RNA binding domains of the nuclear poly(A)-binding protein. J. Biol. Chem. 2003;278:16916–16925. doi: 10.1074/jbc.M209886200. [DOI] [PubMed] [Google Scholar]
- Kühn, U., Wahle, E. Structure and function of poly(A) binding proteins. Biochim. Biophys. Acta. 2004;1678:67–84. doi: 10.1016/j.bbaexp.2004.03.008. [DOI] [PubMed] [Google Scholar]
- Kushner, S.R. mRNA decay in Escherichia coli comes of age. J. Bacteriol. 2002;184:4658–4665. doi: 10.1128/JB.184.17.4658-4665.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak, J.E., Wickens, M. A family of poly(U) polymerases. RNA. 2007;13:860–867. doi: 10.1261/rna.514007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak, J.E., Wang, L., Ballantyne, S., Kimble, J., Wickens, M. Mammalian GLD-2 homologs are poly(A) polymerases. Proc. Natl. Acad. Sci. 2004;101:4407–4412. doi: 10.1073/pnas.0400779101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyriakopoulou, C.B., Nordvarg, H., Virtanen, A. A novel nuclear human poly(A) polymerase (PAP), PAP γ. J. Biol. Chem. 2001;276:33504–33511. doi: 10.1074/jbc.M104599200. [DOI] [PubMed] [Google Scholar]
- LaCava, J., Houseley, J., Saveanu, C., Petfalski, E., Thompson, E., Jacquier, A., Tollervey, D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- Le, Y.J., Kim, H., Chung, J.H., Lee, Y. Testis-specific expression of an intronless gene encoding a human poly(A) polymerase. Mol. Cells. 2001;11:379–385. [PubMed] [Google Scholar]
- Lee, Y.J., Lee, Y., Chung, J.H. An intronless gene encoding a poly(A) polymerase is specifically expressed in testis. FEBS Lett. 2000;487:287–292. doi: 10.1016/s0014-5793(00)02367-x. [DOI] [PubMed] [Google Scholar]
- Lehmann, C., Pullalarevu, S., Krajewski, W., Willis, M.A., Galkin, A., Howard, A., Herzberg, O. Structure of HI0073 from Haemophilus influenzae, the nucleotide-binding domain of a two-protein nucleotidyl transferase. Proteins. 2005;60:807–811. doi: 10.1002/prot.20586. [DOI] [PubMed] [Google Scholar]
- Li, F., Xiong, Y., Wang, J., Cho, H.D., Tomita, K., Weiner, A.M., Steitz, T.A. Crystal structures of the Bacillus stearothermophilus CCA-adding enzyme and its complexes with ATP or CTP. Cell. 2002;111:815–824. doi: 10.1016/s0092-8674(02)01115-7. [DOI] [PubMed] [Google Scholar]
- Lingner, J., Kellermann, J., Keller, W. Cloning and expression of the essential gene for poly(A) polymerase from S. cerevisiae . Nature. 1991;354:496–498. doi: 10.1038/354496a0. [DOI] [PubMed] [Google Scholar]
- Martin, G., Keller, W. Mutational analysis of mammalian poly(A) polymerase identifies a region for primer binding and catalytic domain, homologous to the family X polymerases, and to other nucleotidyltransferases. EMBO J. 1996;15:2593–2603. [PMC free article] [PubMed] [Google Scholar]
- Martin, G., Keller, W. Sequence motifs that distinguish ATP(CTP):tRNA nucleotidyl transferases from eubacterial poly(A) polymerases. RNA. 2004;10:899–906. doi: 10.1261/rna.5242304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, G., Keller, W., Doublié, S. Crystal structure of mammalian poly(A) polymerase in complex with an analog of ATP. EMBO J. 2000;19:4193–4203. doi: 10.1093/emboj/19.16.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, G., Möglich, A., Keller, W., Doublié, S. Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) polymerase. J. Mol. Biol. 2004;341:911–925. doi: 10.1016/j.jmb.2004.06.047. [DOI] [PubMed] [Google Scholar]
- Matsuyama, A., Arai, R., Yashiroda, Y., Shirai, A., Kamata, A., Sekido, S., Kobayashi, Y., Hashimoto, A., Hamamoto, M., Hiraoka, Y., et al. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe . Nat. Biotechnol. 2006;24:841–847. doi: 10.1038/nbt1222. [DOI] [PubMed] [Google Scholar]
- McManus, M.T., Adler, B.K., Pollard, V.W., Hajduk, S.L. Trypanosoma brucei guide RNA poly(U) tail formation is stabilized by cognate mRNA. Mol. Cell. Biol. 2000;20:883–891. doi: 10.1128/mcb.20.3.883-891.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez, R., Richter, J.D. Translational control by CPEB: A means to the end. Nat. Rev. Mol. Cell Biol. 2001;2:521–529. doi: 10.1038/35080081. [DOI] [PubMed] [Google Scholar]
- Minoda, Y., Saeki, K., Aki, D., Takaki, H., Sanada, T., Koga, K., Kobayashi, T., Takaesu, G., Yoshimura, A. A novel zinc finger protein, ZCCHC11, interacts with TIFA and modulates TLR signaling. Biochem. Biophys. Res. Commun. 2006;344:1023–1030. doi: 10.1016/j.bbrc.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Mohanty, B.K., Kushner, S.R. Residual polyadenylation in poly(A) polymerase I (pcnB) mutants of Escherichia coli does not result from the activity encoded by the f310 gene. Mol. Microbiol. 1999;34:1109–1119. doi: 10.1046/j.1365-2958.1999.01674.x. [DOI] [PubMed] [Google Scholar]
- Mohanty, B.K., Kushner, S.R. Polynucleotide phosphorylase functions both as a 3′ > 5′ exonuclease and a poly(A) polymerase in Escherichia coli . Proc. Natl. Acad. Sci. 2000;97:11966–11971. doi: 10.1073/pnas.220295997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty, B.K., Kushner, S.R. The majority of Escherichia coli mRNAs undergo post-transcriptional modification in exponentially growing cells. Nucleic Acids Res. 2006;34:5695–5704. doi: 10.1093/nar/gkl684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moure, C.M., Bowman, B.R., Gershon, P.D., Quiocho, F.A. Crystal structures of the Vaccinia virus polyadenylate polymerase heterodimer: Insights into ATP selectivity and processivity. Mol. Cell. 2006;22:339–349. doi: 10.1016/j.molcel.2006.03.015. [DOI] [PubMed] [Google Scholar]
- Nagaike, T., Suzuki, T., Katoh, T., Ueda, T. Human mitochondrial mRNAs are stabilized with polyadenylation regulated by mitochondria-specific poly(A) polymerase and polynucleotide phosphorylase. J. Biol. Chem. 2005;280:19721–19727. doi: 10.1074/jbc.M500804200. [DOI] [PubMed] [Google Scholar]
- Nakanishi, T., Kubota, H., Ishibashi, N., Kumagai, S., Watanabe, H., Yamashita, M., Kashiwabara, S., Miyado, K., Baba, T. Possible role of mouse poly(A) polymerase mGLD-2 during oocyte maturation. Dev. Biol. 2006;289:115–126. doi: 10.1016/j.ydbio.2005.10.017. [DOI] [PubMed] [Google Scholar]
- O'Hara, E.B., Chekanova, J.A., Ingle, C.A., Kushner, Z.R., Peters, E., Kushner, S.R. Polyadenylylation helps regulate mRNA decay in Escherichia coli . Proc. Natl. Acad. Sci. 1995;92:1807–1811. doi: 10.1073/pnas.92.6.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen, L.C., Benning, M.M., Holden, H.M. Structural investigation of the antibiotic and ATP-binding sites in kanamycin nucleotidyltransferase. Biochemistry. 1995;34:13305–13311. doi: 10.1021/bi00041a005. [DOI] [PubMed] [Google Scholar]
- Pelletier, H., Sawaya, M.R., Kumar, A., Wilson, S.H., Kraut, J. Structures of ternary complexes of rat DNA polymerase β, a DNA template-primer, and ddCTP. Science. 1994;264:1891–1903. [PubMed] [Google Scholar]
- Perumal, K., Sinha, K., Henning, D., Reddy, R. Purification, characterization, and cloning of the cDNA of human signal recognition particle RNA 3′-adenylating enzyme. J. Biol. Chem. 2001;276:21791–21796. doi: 10.1074/jbc.M101905200. [DOI] [PubMed] [Google Scholar]
- Pines, J. Mitosis: A matter of getting rid of the right protein at the right time. Trends Cell Biol. 2006;16:55–63. doi: 10.1016/j.tcb.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Raabe, T., Bollum, F.J., Manley, J.L. Primary structure and expression of bovine poly(A) polymerase. Nature. 1991;353:229–234. doi: 10.1038/353229a0. [DOI] [PubMed] [Google Scholar]
- Ramadan, K., Shevelev, I., Hübscher, U. The DNA-polymerase-X family: Controllers of DNA quality? Nat. Rev. Mol. Cell Biol. 2004;5:1038–1043. doi: 10.1038/nrm1530. [DOI] [PubMed] [Google Scholar]
- Read, R.L., Martinho, R.G., Wang, S.W., Carr, A.M., Norbury, C.J. Cytoplasmic poly(A) polymerases mediate cellular responses to S phase arrest. Proc. Natl. Acad. Sci. 2002;99:12079–12084. doi: 10.1073/pnas.192467799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissland, O.S., Mikulasova, A., Norbury, C.J. Efficient RNA polyuridylation by noncanonical poly(A) polymerases. Mol. Cell. Biol. 2007;27:3612–3624. doi: 10.1128/MCB.02209-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogozin, I.B., Aravind, L., Koonin, E.V. Differential action of natural selection on the N and C-terminal domains of 2′-5′ oligoadenylate synthetases and the potential nuclease function of the C-terminal domain. J. Mol. Biol. 2003;326:1449–1461. doi: 10.1016/s0022-2836(03)00055-x. [DOI] [PubMed] [Google Scholar]
- Rott, R., Zipor, G., Portnoy, V., Liveanu, V., Schuster, G. RNA polyadenylation and degradation in cyanobacteria are similar to the chloroplast but different from Escherichia coli . J. Biol. Chem. 2003;278:15771–15777. doi: 10.1074/jbc.M211571200. [DOI] [PubMed] [Google Scholar]
- Sadoff, B.U., Heath-Pagliuso, S., Castano, I.B., Zhu, Y., Kieff, F.S., Christman, M.F. Isolation of mutants of Saccharomyces cerevisiae requiring DNA topoisomerase I. Genetics. 1995;141:465–479. doi: 10.1093/genetics/141.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh, S., Chabes, A., McDonald, W.H., Thelander, L., Yates, J.R., Russell, P. Cid13 is a cytoplasmic poly(A) polymerase that regulates ribonucleotide reductase mRNA. Cell. 2002;109:563–573. doi: 10.1016/s0092-8674(02)00753-5. [DOI] [PubMed] [Google Scholar]
- Sawaya, M.R., Pelletier, H., Kumar, A., Wilson, S.H., Kraut, J. Crystal structure of rat DNA polymerase β: Evidence for a common polymerase mechanism. Science. 1994;264:1930–1935. doi: 10.1126/science.7516581. [DOI] [PubMed] [Google Scholar]
- Sawaya, M.R., Prasad, R., Wilson, S.H., Kraut, J., Pelletier, H. Crystal structures of human DNA polymerase β complexed with gapped and nicked DNA: Evidence for an induced fit mechanism. Biochemistry. 1997;36:11205–11215. doi: 10.1021/bi9703812. [DOI] [PubMed] [Google Scholar]
- Schneider, C., Anderson, J.T., Tollervey, D. The exosome subunit rrp44 plays a direct role in RNA substrate recognition. Mol. Cell. 2007;27:324–331. doi: 10.1016/j.molcel.2007.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnierle, B.S., Gershon, P.D., Moss, B. Cap-specific mRNA (nucleoside-O2′-)-methyltransferase and poly(A) polymerase stimulatory activities of vaccinia virus are mediated by a single protein. Proc. Natl. Acad. Sci. 1992;89:2897–2901. doi: 10.1073/pnas.89.7.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, B., Goodman, H.M. Uridine addition after microRNA-directed cleavage. Science. 2004;306:997. doi: 10.1126/science.1103521. [DOI] [PubMed] [Google Scholar]
- Shi, P.Y., Maizels, N., Weiner, A.M. CCA addition by tRNA nucleotidyltransferase: Polymerization without translocation? EMBO J. 1998a;17:3197–3206. doi: 10.1093/emboj/17.11.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, P.Y., Weiner, A.M., Maizels, N. A top-half tDNA minihelix is a good substrate for the eubacterial CCA-adding enzyme. RNA. 1998b;4:276–284. [PMC free article] [PubMed] [Google Scholar]
- Simpson, L., Aphasizhev, R., Gao, G., Kang, X. Mitochondrial proteins and complexes in Leishmania and Trypanosoma involved in U-insertion/deletion RNA editing. RNA. 2004;10:159–170. doi: 10.1261/rna.5170704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagno, J., Aphasizheva, I., Rosengarth, A., Luecke, H., Aphasizhev, R. UTP-bound and Apo structures of a minimal RNA uridylyltransferase. J. Mol. Biol. 2007;366:882–899. doi: 10.1016/j.jmb.2006.11.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz, T.A. A mechanism for all polymerases. Nature. 1998;391:231–232. doi: 10.1038/34542. [DOI] [PubMed] [Google Scholar]
- Stevenson, A.L., Norbury, C.J. The Cid1 family of non-canonical poly(A) polymerases. Yeast. 2006;23:991–1000. doi: 10.1002/yea.1408. [DOI] [PubMed] [Google Scholar]
- Stuart, K.D., Schnaufer, A., Ernst, N.L., Panigrahi, A.K. Complex management: RNA editing in trypanosomes. Trends Biochem. Sci. 2005;30:97–105. doi: 10.1016/j.tibs.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Suh, N., Jedamzik, B., Eckmann, C.R., Wickens, M., Kimble, J. The GLD-2 poly(A) polymerase activates gld-1 mRNA in the Caenorhabditis elegans germ line. Proc. Natl. Acad. Sci. 2006;103:15108–15112. doi: 10.1073/pnas.0607050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thai, T.H., Kearney, J.F. Isoforms of terminal deoxynucleotidyltransferase: Developmental aspects and function. Adv. Immunol. 2005;86:113–136. doi: 10.1016/S0065-2776(04)86003-6. [DOI] [PubMed] [Google Scholar]
- Tomecki, R., Dmochowska, A., Gewartowski, K., Dziembowski, A., Stepien, P.P. Identification of a novel human nuclear-encoded mitochondrial poly(A) polymerase. Nucleic Acids Res. 2004;32:6001–6014. doi: 10.1093/nar/gkh923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita, K., Fukai, S., Ishitani, R., Ueda, T., Takeuchi, N., Vassylyev, D.G., Nureki, O. Structural basis for template-independent RNA polymerization. Nature. 2004;430:700–704. doi: 10.1038/nature02712. [DOI] [PubMed] [Google Scholar]
- Tomita, K., Ishitani, R., Fukai, S., Nureki, O. Complete crystallographic analysis of the dynamics of CCA sequence addition. Nature. 2006;443:956–960. doi: 10.1038/nature05204. [DOI] [PubMed] [Google Scholar]
- Topalian, S.L., Kaneko, S., Gonzales, M.I., Bond, G.L., Ward, Y., Manley, J.L. Identification and functional characterization of neo-poly(A) polymerase, an RNA processing enzyme overexpressed in human tumors. Mol. Cell. Biol. 2001;21:5614–5623. doi: 10.1128/MCB.21.16.5614-5623.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trippe, R., Richly, H., Benecke, B.J. Biochemical characterization of a U6 small nuclear RNA-specific terminal uridylyltransferase. Eur. J. Biochem. 2003;270:971–980. doi: 10.1046/j.1432-1033.2003.03466.x. [DOI] [PubMed] [Google Scholar]
- Trippe, R., Guschina, E., Hossbach, M., Urlaub, H., Lührmann, R., Benecke, B.J. Identification, cloning, and functional analysis of the human U6 snRNA-specific terminal uridylyl transferase. RNA. 2006;12:1494–1504. doi: 10.1261/rna.87706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagner, S., Vagner, C., Mattaj, I.W. The carboxyl terminus of vertebrate poly(A) polymerase interacts with U2AF 65 to couple 3′-end processing and splicing. Genes & Dev. 2000;14:403–413. [PMC free article] [PubMed] [Google Scholar]
- Vanacova, S., Stefl, R. The exosome and RNA quality control in the nucleus. EMBO Rep. 2007;8:651–657. doi: 10.1038/sj.embor.7401005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacova, S., Wolf, J., Martin, G., Blank, D., Dettwiler, S., Friedlein, A., Langen, H., Keith, G., Keller, W. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahle, E. A novel poly(A)-binding protein acts as a specificity factor in the second phase of messenger RNA polyadenylation. Cell. 1991;66:759–768. doi: 10.1016/0092-8674(91)90119-j. [DOI] [PubMed] [Google Scholar]
- Wahle, E., Rüegsegger, U. 3′-End processing of pre-mRNA in eukaryotes. FEMS Microbiol. Rev. 1999;23:277–295. doi: 10.1111/j.1574-6976.1999.tb00400.x. [DOI] [PubMed] [Google Scholar]
- Wahle, E., Martin, G., Schiltz, E., Keller, W. Isolation and expression of cDNA clones encoding mammalian poly(A) polymerase. EMBO J. 1991;10:4251–4257. doi: 10.1002/j.1460-2075.1991.tb05003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walowsky, C., Fitzhugh, D.J., Castano, I.B., Ju, J.Y., Levin, N.A., Christman, M.F. The topoisomerase-related function gene TRF4 affects cellular sensitivity to the antitumor agent camptothecin. J. Biol. Chem. 1999;274:7302–7308. doi: 10.1074/jbc.274.11.7302. [DOI] [PubMed] [Google Scholar]
- Wang, G., Yang, J., Huibregtse, J.M. Functional domains of the Rsp5 ubiquitin-protein ligase. Mol. Cell. Biol. 1999a;19:342–352. doi: 10.1128/mcb.19.1.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S.W., Norbury, C., Harris, A.L., Toda, T. Caffeine can override the S–M checkpoint in fission yeast. J. Cell Sci. 1999b;112:927–937. doi: 10.1242/jcs.112.6.927. [DOI] [PubMed] [Google Scholar]
- Wang, S.W., Toda, T., MacCallum, R., Harris, A.L., Norbury, C. Cid1, a fission yeast protein required for S–M checkpoint control when DNA polymerase δ or ε is inactivated. Mol. Cell. Biol. 2000a;20:3234–3244. doi: 10.1128/mcb.20.9.3234-3244.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z., Castano, I.B., De Las Penas, A., Adams, C., Christman, M.F. Pol κ: A DNA polymerase required for sister chromatid cohesion. Science. 2000b;289:774–779. doi: 10.1126/science.289.5480.774. [DOI] [PubMed] [Google Scholar]
- Wang, L., Eckmann, C.R., Kadyk, L.C., Wickens, M., Kimble, J. A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans . Nature. 2002;419:312–316. doi: 10.1038/nature01039. [DOI] [PubMed] [Google Scholar]
- Win, T.Z., Draper, S., Read, R.L., Pearce, J., Norbury, C.J., Wang, S.W. Requirement of fission yeast Cid14 in polyadenylation of rRNAs. Mol. Cell. Biol. 2006;26:1710–1721. doi: 10.1128/MCB.26.5.1710-1721.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyers, F., Rougemaille, M., Badis, G., Rousselle, J.C., Dufour, M.E., Boulay, J., Regnault, B., Devaux, F., Namane, A., Seraphin, B., et al. Cryptic Pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- Xiong, Y., Steitz, T.A. Mechanism of transfer RNA maturation by CCA-adding enzyme without using an oligonucleotide template. Nature. 2004;430:640–645. doi: 10.1038/nature02711. [DOI] [PubMed] [Google Scholar]
- Xiong, Y., Steitz, T.A. A story with a good ending: tRNA 3′-end maturation by CCA-adding enzymes. Curr. Opin. Struct. Biol. 2006;16:12–17. doi: 10.1016/j.sbi.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Xiong, Y., Li, F., Wang, J., Weiner, A.M., Steitz, T.A. Crystal structures of an archaeal class I CCA-adding enzyme and its nucleotide complexes. Mol. Cell. 2003;12:1165–1172. doi: 10.1016/s1097-2765(03)00440-4. [DOI] [PubMed] [Google Scholar]
- Yehudai-Resheff, S., Schuster, G. Characterization of the E. coli poly(A) polymerase: nucleotide specificity, RNA-binding affinities and RNA structure dependence. Nucleic Acids Res. 2000;28:1139–1144. doi: 10.1093/nar/28.5.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue, D., Maizels, N., Weiner, A.M. CCA-adding enzymes and poly(A) polymerases are all members of the same nucleotidyltransferase superfamily: Characterization of the CCA-adding enzyme from the archaeal hyperthermophile Sulfolobus shibatae . RNA. 1996;2:895–908. [PMC free article] [PubMed] [Google Scholar]
- Zhao, W., Manley, J.L. Complex alternative RNA processing generates an unexpected diversity of poly(A) polymerase isoforms. Mol. Cell. Biol. 1996;16:2378–2386. doi: 10.1128/mcb.16.5.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, J., Hyman, L., Moore, C. Formation of mRNA 3′ ends in eukaryotes: Mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhelkovsky, A.M., Kessler, M.M., Moore, C.L. Structure–function relationships in the Saccharomyces cerevisiae poly(A) polymerase. Identification of a novel RNA binding site and a domain that interacts with specificity factor(s) J. Biol. Chem. 1995;270:26715–26720. doi: 10.1074/jbc.270.44.26715. [DOI] [PubMed] [Google Scholar]