Abstract
Motor dependent transport of mRNA is a key mechanism in axis specification during development. Apical transport and anchoring of wingless and pair-rule transcripts in the Drosophila syncytial blastoderm embryo is mediated by cytoplasmic Dynein, the major minus end directed microtubule dependent molecular motor. Here, we show that, despite apical transport of mRNA being highly directional, mRNA particles often pause and move backward toward the plus ends of microtubules. We suggest that this retrograde movement helps overcome cellular obstructions. We show that the plus end movement of apical mRNA is independent of the major plus end microtubule motors Kinesin-1 and Kinesin-2. In contrast, Dynactin, a Dynein processivity factor, is required to suppress retrograde mRNA movements, as well as for efficient minus end motility. We propose that Dynein itself, rather than the activity of a plus end motor, is responsible for the plus end movements of the mRNA and that Dynactin is involved in preventing short reverse movements of the Dynein motor, known to occur in vitro.
Keywords: Drosophila, intracellular mRNA localization, Dynein, microtubules, actin, Dynein heavy chain, Dynactin, Glued/p150
INTRODUCTION
RNA is asymmetrically distributed in diverse organisms and cell types in order to target proteins to their site of function (Lopez de Heredia and Jansen 2004), thus playing a key role in a variety of cellular processes, including cell polarity, axis specification, and synaptic activity (St Johnston 2005). In Drosophila, mRNA localization sets up the primary embryonic axes, which provide the basis of later patterning through the action of a cascade of transcription factors. Pair-rule segmentation genes are one class of transcription factors in this cascade, which define the pattern of segmentation. Pair-rule transcripts, which include fushi tarazu (ftz), hairy (h), even skipped (eve), and runt (run), are all localized to the apical cytoplasm in syncytial blastoderm embryos (Davis and Ish-Horowicz 1991). Apical mRNA localization concentrates the protein close to the nuclei from which they are synthesized, thus ensuring the fidelity of segmentation (Bullock et al. 2004). The transcripts of the segment polarity gene, wingless, are also apically localized and are believed to concentrate the Wingless/Wnt signal to the apical membrane (Simmonds et al. 2001). Injected pair-rule and wingless transcripts form particles that are transported by the major cellular minus end directed microtubule (MT) motor, cytoplamic Dynein (Dynein), to the apical cytoplasm above the nuclei (Wilkie and Davis 2001). Dynein is also required for the static anchoring of pair-rule and wingless transcripts in the apical cytoplasm (Delanoue and Davis 2005). In late Drosophila oogenesis, gurken (grk) mRNA utilizes the same motor machinery to localize to the dorso-anterior corner (MacDougall et al. 2003; Bullock and Ish-Horowicz 2001) and establishes the dorso-ventral axis.
Dynein is a well conserved motor protein involved in the transport of a diverse range of cytoplasmic cargoes (Vallee et al. 2004). It is an AAA ATPase that hydrolyzes ATP to generate mechanical force that leads to nanometer steps toward the minus ends of microtubules (MTs). In contrast to the highly processive Kinesin-1 motor that moves hand-over-hand by coordinating its two motor heads (Vale and Milligan 2000), Dynein is not as robust in vitro. A single Dynein motor complex exerts limited force and only travels short distances before dissociating from MTs (Wang et al. 1995; Mallik et al. 2005). Dynein performs better in vivo (Ashkin et al. 1990; Presley et al. 1997; Gross et al. 2002), possibly due to the deployment of multiple Dynein motors within a single cargo/motor complex (Gross et al. 2000; Mallik et al. 2005) and the action of accessory proteins. One such factor that increases Dynein's processivity is Dynactin (King and Schroer 2000), which is required for most Dynein dependent motility (Holleran et al. 1998).
Dynactin is a multimeric complex, consisting of at least seven subunits ranging in size from 22 to 150 kDa. These include p45/Arp-1/centractin, the actin related proteins Arp-11, p150/Glued, and p50/dynamitin, capping protein, and several other less well characterized polypeptides (Holleran et al. 1998). Dynamitin is positioned in the shoulder of Dynactin and is crucial for its structural integrity (Eckley et al. 1999). At least in a few cases, Arp-1 is thought to link the cargo to the Dynein/Dynactin complex (Muresan et al. 2001), through its interaction with the C-terminal part of Glued (Waterman-Storer et al. 1995). Glued binds to both MTs and the Dynein intermediate chain (Karki and Holzbaur 1995). These interactions are thought to stabilize Dynein's attachment to the MTs or to keep Dynein in close proximity with the MT once it dissociates, allowing it to reattach more readily.
Many cargos of molecular motors have been demonstrated to exhibit bidirectional saltatory motion along MTs, characterized by movement toward one direction, punctuated by frequent pauses or runs toward the opposite direction. Cargo displaying such bidirectional movements include mitochondria, lipid droplets, pigment granules, viruses, intermediate filaments, and mRNPs (Gross 2004; Welte 2004). In most of these systems, the minus end directed motor is Dynein, and in a few there is evidence implicating Kinesin family members in bidirectional movements. Kinesin-1 is thought to be involved in mitochondrial and lysosomal motility in mice extra-embryonic cells (Tanaka et al. 1998), as well as retrograde transport of mitochondria in Drosophila motor axons (Pilling et al. 2006). Kinesin-2 is thought to mediate frog melanosomes' motility (Deacon et al. 2003). In most other cases, the proposed plus end directed motor remains unidentified. Nevertheless, a common theme for most of these examples is that, despite the frequent reversals in direction, one type of motion dominates over each direction or phase of movement. It has been proposed that net displacement is achieved through the action of a “coordination complex” or “switch,” which coordinates the activity of the opposing motors (Gross et al. 2002). However, the molecular details of how the opposing motor activities are controlled remains unclear.
Here, we show that apical transport of mRNA in the Drosophila blastoderm embryo is bidirectional, with a strong net bias to minus end transport. In wild-type embryos, mRNA particles pause frequently and often move in a plus end directed manner toward the basal cytoplasm. Our observations are consistent with the retrograde movements allowing the RNA cargo to overcome temporary obstacles in reaching the apical cytoplasm. We show that the retrograde plus end movements are Kinesin-1 and Kinesin-2 independent and are enhanced by the disruption of Dynactin. We suggest that Dynein itself is responsible for the plus end movements we observe and propose a model for Dynactin dependent repression of Dynein's retrograde transport of pair-rule mRNA in the blastoderm embryo.
RESULTS AND DISCUSSION
ftz RNA particles are transported bidirectionally along MTs
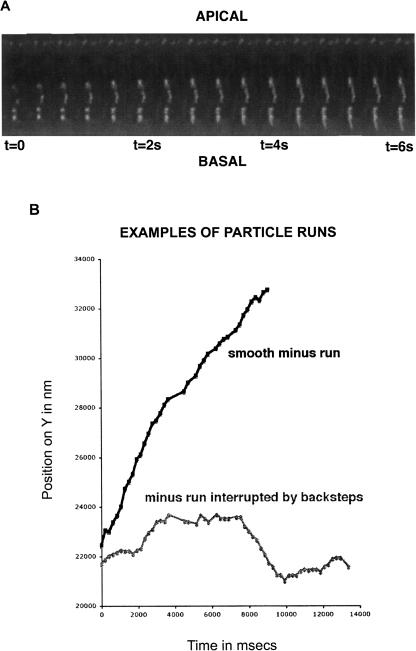
Injected fluorescent pair-rule and wingless RNA form particles that are transported predominantly to the minus ends of MTs (Wilkie and Davis 2001). In order to test whether, like many other Dynein dependent cargos, apical RNA also moves significant distances toward the plus end of MTs, we improved the time resolution of our imaging and the spatial resolution of the analysis. We used a semi-automated tracking method followed by centroid analysis of particle position leading to subpixel accuracy of measurement (see Materials and Methods). Using these methods, we observed that individual particles frequently pause or step backward. Occasionally, the particles display long runs toward the plus ends of MTs before resuming their predominant minus end directed runs (Fig. 1; Supplemental Fig. 1; Supplemental Movies 1,2).
FIGURE 1.
Apical transport of RNA to the minus ends of MTs is punctuated by frequent pauses and plus end runs. Bidirectional motility of RNA in wild-type syncytial blastoderm embryos. (A) Particles of fluorescently labeled, injected ftz RNA travel toward the peripheral apical cytoplasm (minus runs) or away from the periphery, toward the interior (plus runs) of the embryo. (B) Examples of a smooth minus-end run and a run interrupted by pauses and plus end retrograde steps.
Bidirectional transport of cargoes by molecular motors is widespread and has been proposed to allow flexible regulation of cargo destination, a mechanism for rapid widespread distribution of cargoes within the cytoplasm or to overcome obstacles in the cell (Gross 2004). In the case of apical transport of RNA, we observe that the plus end motility of apical RNA particles is relatively short and infrequent. Therefore, the retrograde transport does not perturb significantly the overall progress toward the minus ends of MTs in the apical cytoplasm. Nevertheless, we have observed a number of RNA particles pausing in a given position for some time, moving backward for a short time, reversing direction again, and resuming forward movement on the same path, leading to motility directly past the original stalled site. In such cases, we occasionally observe, a little later, other particles pausing or changing direction at the exact same position (Supplemental Movie 1). We propose that by reversing their course, the RNA particles are able to avoid obstacles in the cytoplasm or “traffic jams” on MTs.
The plus end directed movements we observe could be due to the cargo detaching from the motor or the cargo/motor complex detaching from the MT track and drifting toward the basal cytoplasm. Alternatively, the plus end runs could reflect plus end motor activity or one-dimensional diffusion along MTs. We excluded the first possibility by characterizing in detail the physical properties of the backward movement of the RNA particles. We found that plus end run paths are linear and often cover long distances of up to 2.5 μm (Fig. 3A, below). Furthermore, we often observed that after reversal of direction, RNA particles move backward along the same exact path that they followed during minus end movement (Supplemental Movie 1). Taking these results together with previous published data from Bullock et al. (2006), showing bidirectional movement of hairy transcripts in Drosophila blastoderm embryos, we conclude that the plus end runs of ftz transcripts reflect movement along MT tracks rather than diffusion of the cargo or the motor–cargo complex after detachment from the MTs.
FIGURE 3.
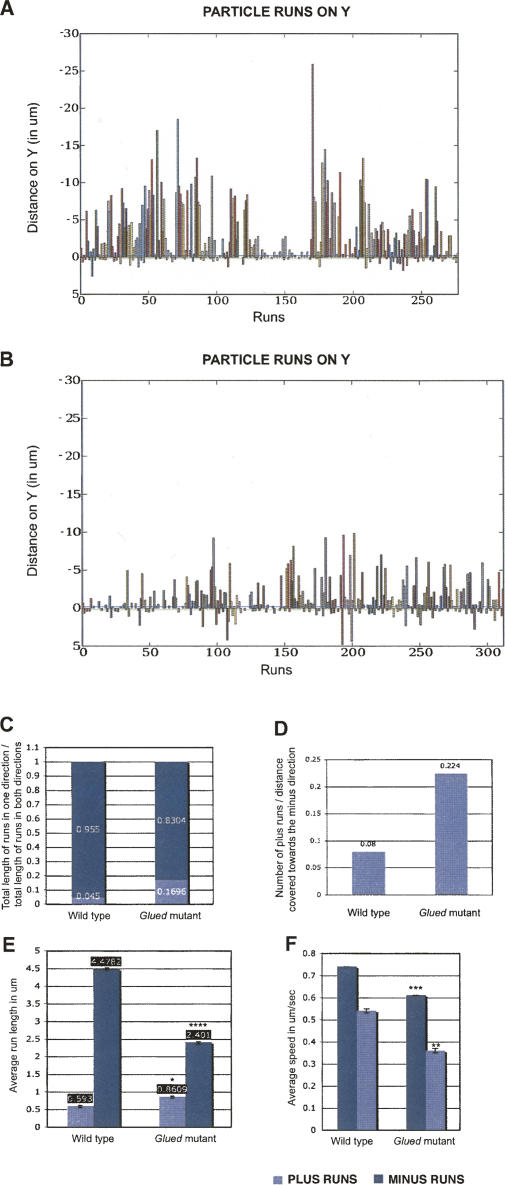
Plus end retrograde RNA motility is enhanced in Glued mutant embryos. In Glued1 mutant embryos, ftz RNA particles display shorter minus runs, longer plus runs, and travel more frequently toward the plus ends compared with wild type. (A) Graph of minus and plus runs in wild type. (B) Graph of minus and plus runs distribution in Glued1. The runs of each RNA particle are grouped and displayed in a distinct color along the X-axis, and pauses are represented as gaps between the runs. The Y values represent microns of distance covered in both directions, along the Y-axis. To achieve this, the images were first rotated so that the Y axis was parallel to the apico-basal axis of the embryos. Runs toward the minus ends of MTs (forward runs) are shown above the X axis and runs toward the plus ends of MTs (backward runs) are shown below the X axis. (C) Proportion of distance covered toward the minus and plus directions in wild type and Glued1 embryos. (D) Frequency of plus runs in wild type and Glued1 embryos. (E) Average length of minus and plus runs in wild type (4.48 ± 0.023 μm and 0.592 ± 0.0034 μm, respectively) and Glued1 embryos (2.401 ± 0.014 μm, P = 0 and 0.86 ± 0.003 μm, P = 0.033) (F) Mean speed of minus and plus runs in wild type (0.74 and 0.54 ± 0.01 μm/sec, respectively) and Glued1 embryos (0.61 μm/sec, P = 0.048 and 0.36 μm/sec, P = 0.067, respectively). Values shown are mean values for all runs analyzed in a given genetic background ± SEM. P shows t-test probability compared to wild type. *P < 0.05, **P < 0.01, ***P < 0.005, ****P < 0.001.
Kinesin-1 and Kinesin-2 are not required for the plus end movement of ftz RNA
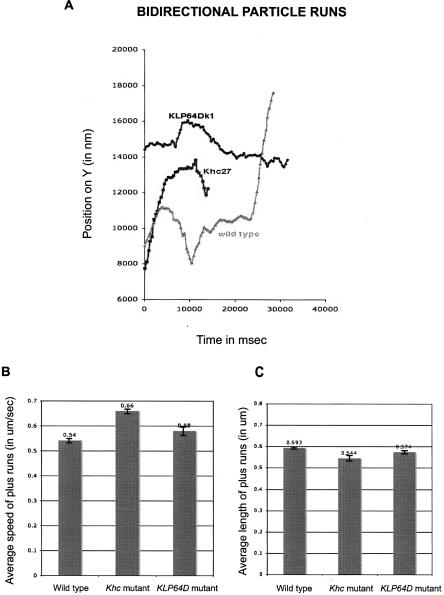
Kinesin-1 (Kin 1) is known to mediate the transport of various cargoes in a variety of systems. To test whether this motor is involved in the plus end transport of RNA in the Drosophila blastoderm embryo, we disrupted the function of Khc, which is the motor subunit of the tetrameric Kinesin-1 motor complex. We used the Dominant Female Sterile technique (Chou et al. 1993) to make Khc 27 germline clones. Khc27 is a null allele with a nonsense (Q65-stop) mutation (Brendza et al. 1999). We found that ftz RNA in Khc27 null embryos moves backward in a similar manner to control wild-type embryos (Fig. 2; Supplemental Movie 3). The average run length of the plus runs is very similar to wild type. The speed of the plus runs is not reduced in the mutant embryos, as we would expect if Kin 1 mediated them. The observed increase in speed is not statistically significant (t-test).
FIGURE 2.
Retrograde RNA motility is not dependent on Kinesin-1 and Kinesin-2. ftz RNA moves bidirectionally in Khc27 and KLP64Dk1 syncytial blastoderm embryos with kinetics similar to wild type. (A) Examples of bidirectional runs in wild-type, Khc27, and KLP64Dk1 embryos. (B) Mean speed of plus runs in Khc27 and KLP64Dk1 embryos is not significantly different from wild type. Mean speed of plus runs in wild-type (0.54 ± 0.01 μm/sec), Khc27 (0.66 ± 0.01 μm/sec, P = 0.2098), and KLP64Dk1 embryos (0.58 ± 0.02 μm/sec, P = 0.763). (C) Average length of plus runs in Khc27 and KLP64Dk1 embryos is not significantly different from wild type. Average plus run length in wild-type (0.592 ± 0.0034 μm), Khc27 (0.544 ± 0.0058 μm, P = 0.5375), and KLP64Dk1 embryos (0.574 ± 0.0105 μm, p = 0.8179). Values shown are mean values for all runs analyzed in a given genetic background ± SEM. P shows t-test probability compared to wild type.
We then tested whether Kinesin-2 could be responsible for the plus end movements we observe. The Kinesin-2 superfamily consists of heterotrimeric complexes with two different motor subunits responsible for force generation and a third polypeptide, known as Kinesin associated protein or KAP. The Drosophila subunits are referred to as KLP64D, KLP68D, and KAP3. We disrupted KLP64D function by generating KLP64Dk1 mutant germline clones. KLP64Dk1 is most likely to be a null allele since it bears an EMS induced point mutation that causes a stop at codon 13 of the gene (Ray et al. 1999). ftz RNA injected into KLP64Dk1 null embryos shows backward motility similar to wild type (Fig. 2; Supplemental Movie 4).
Among all the members of the Kinesin superfamily, Kinesin-1, Kinesin-2, and Kinesin-3 are considered as the major cargo transporters in the cell (Vale 2003). Although we did not specifically inhibit Kinesin-3, we consider its involvement in RNA motility unlikely. Kinesin-3 has a well-documented general role in membrane trafficking in lower eukaryotes, and its metazoan orthologs are less abundant than Kinesin-1 and Kinesin-2 and are considered to be specialized in the transport of synaptic vehicles in neurons, rather than having a more general function in cargo transport (Vale 2003). The Drosophila ortholog, also known as Kinesin-73, has not been implicated in cellular transport so far.
Early Drosophila embryos contain various Kinesin related proteins with a reported motor activity, such as KLP3A, KLP67A, KLP61F, KLP59C, KLP38B, and Pavarroti. However, none of these motors has a documented role in cargo transport, and their role is most likely limited to their already known functions in spindle assembly, chromosome motility, or MT depolymerization. Similar functions have also been proposed for their orthologs in other organisms.
To rule out potential redundancy between Kinesin-1 and -2 and the involvement of other Kinesins, we inhibited Kinesin activity with AMP-PNP, a nonhydrolyzable ATP analog that preferentially inhibits Kinesin superfamily members rather than Dynein (Murray et al. 2000; Ross et al. 2006). Preinjection of a high volume of 100 mM of AMP-PNP in blastoderm embryos does not block the transport of ftz RNA toward the minus ends of MTs and allows normal apical localization of the transcript, showing that Dynein's motor activity is not severely impaired. Moreover, the physical properties (run length and speed) of Dynein motion are indistinguishable from wild type (Supplemental Fig. 2). ftz RNA particles move bidirectionally after injection of 100 mM AMP-PNP (Supplemental Movie 5), whereas control injection of similar volumes of 10 mM concentration of the inhibitor in oocytes is enough to completely abolish fast ooplasmic streaming (not shown). Interestingly, the frequency of plus reversals is very similar between injected (0.1 plus reversals/μm of minus-directed runs) and noninjected embryos (0.08 plus reversals/μm of minus runs). The kinetics (run lengths and velocities) of plus motion in embryos preinjected with the inhibitor are also indistinguishable from wild type (Supplemental Fig. 2).
Dynactin suppresses the frequency of plus end runs
We previously showed that Dynactin was required for efficient Dynein dependent minus end transport of the RNA particles in the embryo (Wilkie and Davis 2001). In order to determine whether the Dynein motor complex itself has a role in the plus end motility we observe, we studied the effect of disrupting Dynactin. We used Glued 1 (Gl1), a spontaneous gain of function mutation (Plough and Ives 1934) caused by the insertion of a transposable element near the 3′ end of the gene (Swaroop et al. 1985). Glued 1 encodes a truncated product (McGrail et al. 1995) that acts as a dominant negative.
We injected ftz RNA into embryos laid by Gl 1 heterozygous mothers and examined different parameters of motion, namely, the frequency, length, and speed of minus and plus end runs compared with controls (Supplemental Movie 6; Fig. 3; Supplemental Fig. 3; Supplemental Table 1). Our results show that in Gl 1 mutant embryos, particles pause or reverse their direction significantly more frequently than in wild type. The average length of the plus end runs is significantly increased in Glued1. The average length of the minus runs is severely reduced, compared with wild type because of the known reduction in processivity of Dynein caused by disrupting the function of Dynactin. We also found that the speed of minus end and plus end runs are significantly reduced compared with wild type (Fig. 3). We conclude that Dynactin has a role in suppressing the plus end motility of the RNA, in addition to the previously known role in promoting minus end motility of the Dynein motor.
The requirement for Dynactin in the prevention of retrograde transport of RNA, can be interpreted in two ways. Dynactin could be involved in coordinating the activity of Dynein with a plus end motor, yet to be discovered. In which case, the increase in the frequency and length of plus end runs that we observe could be caused by improper regulation or a shift of balance of the motors resulting from the disruption in Dynactin. Alternatively, Dynein itself could be responsible for the plus end runs we observe. We favor the second interpretation for the following reasons. First, we have excluded an involvement of any combination of Kinesins. We used Kinesin-1 and Kinesin-2 nulls as well as inhibiting the activity of all Kinesin family members with AMP-PNP. Second, in cases where two opposing motor activities are thought to be coordinated, perturbing the coordinating complex leads to an impairment of motion in both directions, whereas in our case disruption of Dynactin impairs minus end directed motility, while it increases plus end directed motility. Third, in cases where Kinesins have been implicated in the bidirectional motility of cargo, the plus motion does not simply punctuate the minus motion briefly, but instead is the dominant motion at some phase of the cargo's journey, such as retrograde transport of mitochondria in Drosophila axons (Pilling et al. 2006) and dispersal of melanophores in the form of melanosomes (Tuma et al. 1998). In contrast, apical RNA does not undergo a net plus end transport at any stage. Finally, a recent in vitro study has shown that Dynein frequently undergoes unproductive intervals of pauses and reversals in direction (Mallik et al. 2005). A comparison between our data in different genetic backgrounds and the in vitro study of single or multiple Dyneins by Mallik et al. (2005) shows that in all cases, the minus motion is predominant with higher frequency, velocity, and run lengths than the plus motion. Taking all these arguments together we conclude that the plus end motility we observe for apical transcripts does not require Kinesin motors and is most likely a consequence of retrograde motility of the Dynein motor itself, kept in check by the Dynactin complex. It is interesting that a recently identified MT binding domain in chicken Glued (Culver-Hanlon et al. 2006) can “skate” along MTs in the absence of molecular motors. However, this skating is almost an order of magnitude slower and much shorter in length than the plus motility we observe, and even if Drosophila Dynactin can undergo skating events, these could not account for our plus runs.
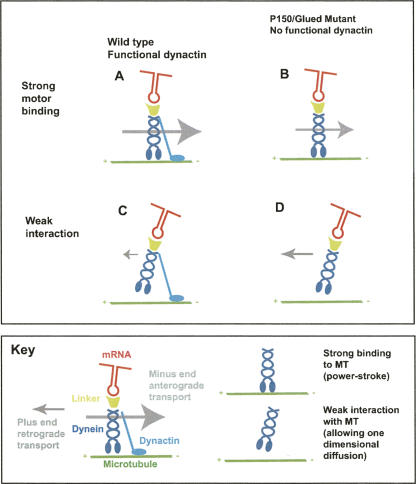
The Dynein motor is thought to drive active motility through its strong binding to MTs that mediates the “power stroke.” When the strong binding fails, Dynein is still able to diffuse one-dimensionally along MTs. During this diffusion, the motor is thought to be held on the MTs by a weak force (Wang and Sheetz 1999). The Dynein processivity factor, Dynactin, provides an additional site of interaction with MTs (Karki and Holzbaur 1995). To explain our observations, we propose that during wild-type Dynein based motility, Dynactin reduces the one-dimensional movement of the motor cargo complex, thus preventing most backward motility (Fig. 4). According to this model, perturbing Dynactin function releases the Dynein motor to slip backward by one-dimensional diffusion, as well as decreasing the processivity and efficiency of minus end transport. It will be interesting to discover whether the suppression of Dynein based backward motility by Dynactin will turn out to be a universal function of the motor cofactor.
FIGURE 4.
Model for dynactin dependent regulation of retrograde Dynein transport. (A) During wild-type Dynein based motility, the motor interacts with the MT strongly, thus mediating the ATP dependent “power stroke.” (B) When the strong force temporarily fails in wild type, the motor's weak interaction with the MT allows one-dimensional diffusion. (C) In a Dynactin mutant, Dynein's processivity is decreased and minus end run lengths are shorter. (D) When the strong attachment of Dynein to MTs fails in a Dynactin mutant, the motor is able to diffuse along the MT much more readily, leading to an enhancement of backward (plus end) motility. The key explains the elements of the model in diagram.
It has been suggested that plus runs of Dynein–Dynactin in vitro result from active steps taken by the motor and not from one-dimensional diffusion on MTs (Ross et al. 2006). In contrast, we do not believe that the plus runs we observe represent active Dynein motility because, in contrast to the in vitro motion of Dynein–Dynactin complexes, the velocities and run lengths we observe are much lower than those of minus runs. Bullock et al. (2006) have suggested that net minus transport of bidirectional motor–RNA complexes is achieved through BicD- and Egl-dependent recruitment of additional Dynein motors by RNA signals. One-dimensional diffusion of Dynein along the MT axis is not considered in their model, which rather favors active motility of Dynein or Kinesin motors toward the plus ends of MTs. However, overexpression of BicD alters transitions between travel states (Bullock et al. 2006) and BicD could in principle regulate backward slippage of Dynein in a manner similar to that we propose for Dynactin. Our results raise the possibility that Dynactin could suppress the retrograde movement of other Dynein cargos in other systems showing bidirectional movements similar to apical RNA in the blastoderm embryo.
MATERIALS AND METHODS
Fly stocks
Stocks were raised on standard cornmeal–agar medium at 25°C. Strains used for injections were Oregon R (wild type), Glued 1, KLP64Dk1 (Bloomington stock center) and Khc27 (W. Saxton). Germline clones were generated using the DFS technique (Chou et al. 1993; Brendza et al. 2000), by inducing mitotic recombination in hsFLP;FRT-Khc27/FRT-ovoD1 and hsFLP;FRT-KLP64D k1/FRT-ovoD1 first instar larvae. Only mutant embryos are laid by mosaic mothers, since OvoD1 egg chambers arrest during early oogenesis.
Injections
Embryos were prepared for injection and capped ftz RNA was transcribed in vitro and injected as previously described (Wilkie and Davis 2001), at concentration of 150–250 ng/μL.
AMP-PNP (Sigma) was injected at 100 mM 2 min before injection of the RNA. ftz RNA localized apically in all embryos that were preinjected with the inhibitor. In the majority of the embryos, the kinetics of minus motion was similar to wild type. However, in rare cases the kinetics of minus-directed transport was affected, which we interpret as an inhibition of Dynein due to the occasional injection of higher volumes of the inhibitor.
Imaging
Embryos were imaged on coverslips in halocarbon oil (series 700), on a widefield DeltaVision microscope (Applied Precision, Olympus 1×70 and Rope Coolsnap HQ), with a 20 × 0.75NA objective, as previously described (Parton and Davis 2006). The images were deconvolved using constrained iterative algorithms (SoftWorx, Applied Precision) based on Sedat/Agard methods (Agard et al. 1989). Processing of images and movies was carried out using SoftWoRx, ImageJ, and QuicktimePro. Movies were compressed with Quicktime Pro (Apple).
Tracking
Individual Alexa-546 (Molecular Probes) labeled RNA particles were followed over time between successive frames in a sequential series of images, with subpixel resolution, using the Metamorph correlation-based, template match method (Metamorph 6.1, Universal Imaging Corporation). Templates containing each individual particle and their search area were defined manually, and then tracking proceeded automatically. We analyzed 236 runs in wild-type embryos (101 particles analyzed from 11 independent embryo injections), 237 runs in Glued1 (73 particles analyzed from 7 independent embryo injections), 110 runs in Khc27 (40 particles analyzed from 4 independent embryo injections), 41 runs in KLP64Dk1 embryos (13 particles analyzed from 3 independent embryo injections), and 59 runs in wild-type embryos preinjected with 100 mM AMP-PNP (25 particles analyzed from 4 independent embryo injections). Images were acquired at a rate of 2–6 frames/sec. (In wild-type embryos, 48 particles were analyzed at 2–3 frames/sec and 53 particles were analyzed at 4–6 frames/sec. In Glued1 embryos, 45 particles were analyzed at 2–3 frames/sec and 27 particles were analyzed at 4–6 frames/sec. In Khc27embryos, 25 particles were analyzed at 2–3 frames/sec and 15 particles were analyzed at 4–6 frames/sec. In KLP64Dk1 embryos, 10 particles were analyzed at 2–3 frames/sec and 3 particles were analyzed at 5–6 frames/sec. In embryos preinjected with AMP-PNP, 25 particles were analyzed at 3–4 frames/sec.)
Analysis
The X and Y subpixel coordinates of the selected tracked particles were recorded into an Excel log file and imported into ParticleStats, a program we wrote to display particle runs and pauses and calculate run lengths and speeds and the statistical significance of differences between selected data sets (t-test). To eliminate errors due to the irregular and changing shapes of the RNA particles, we defined a run as any continuous movement ≥200 nm without a reversal or a pause. A pause is defined as any movement <300 nm that lasts for a duration of 2 sec or longer. ParticleStats is a set of command line scripts written in platform-independent Python programming language and will run on any Linux, Mac OSX, or Windows based computer. Prerequisites for running ParticleStats are Python 2.4 and the following Python modules: Xlrd, RPy, matplotlib, and PIL. In addition, the R statistical computing package (R Development Core Team 2006) must also be installed. ParticleStats is distributed under the terms of the GNU public license (GPL) and can be downloaded from the ParticleStats web page (http://www.ParticleStats.com). The excel data files and details of the parameters used in ParticleStats for this study are provided on the web site. We have currently also been using ParticleStats to analyze the directionality of tracked particles in Drosophila oocytes and nurse cells, and these extended features will be published in a separate technical paper about ParticleStats.
SUPPLEMENTAL DATA
All Supplemental Materials (figures and movies) are available at http://www.ParticleStats.com/Vendra2007RNA.
ACKNOWLEDGMENTS
We are grateful to Daniel Zicha, Aaron Pilling, Steve Gross, William Saxton, David Kelly, and Richard Parton for advice on particle tracking and Alastair Kerr for advice on statistics. We are also grateful to Michael Welte, Simon Bullock, and David Ish-Horowicz for discussions and communication of unpublished data.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.509007.
REFERENCES
- Agard, D.A., Hiraoka, Y., Shaw, P., Sedat, J.W. Fluorescence microscopy in three dimensions. Methods Cell Biol. 1989;30:353–377. doi: 10.1016/s0091-679x(08)60986-3. [DOI] [PubMed] [Google Scholar]
- Ashkin, A., Schutze, K., Dziedzic, J.M., Euteneuer, U., Schliwa, M. Force generation of organelle transport measured in vivo by an infrared laser trap. Nature. 1990;348:346–348. doi: 10.1038/348346a0. [DOI] [PubMed] [Google Scholar]
- Brendza, K.M., Rose, D.J., Gilbert, S.P., Saxton, W.M. Lethal kinesin mutations reveal amino acids important for ATPase activation and structural coupling. J. Biol. Chem. 1999;274:31506–31514. doi: 10.1074/jbc.274.44.31506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendza, R.P., Serbus, L.R., Duffy, J.B., Saxton, W.M. A function for Kinesin I in the posterior transport of oskar mRNA and Staufen protein. Science. 2000;289:2120–2122. doi: 10.1126/science.289.5487.2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock, S.L., Ish-Horowicz, D. Conserved signals and machinery for RNA transport in Drosophila oogenesis and embryogenesis. Nature. 2001;414:611–616. doi: 10.1038/414611a. [DOI] [PubMed] [Google Scholar]
- Bullock, S.L., Stauber, M., Prell, A., Hughes, J.R., Ish-Horowicz, D., Schmidt-Ott, U. Differential cytoplasmic mRNA localisation adjusts pair-rule transcription factor activity to cytoarchitecture in dipteran evolution. Development. 2004;131:4251–4261. doi: 10.1242/dev.01289. [DOI] [PubMed] [Google Scholar]
- Bullock, S.L., Nicol, A., Gross, S.P., Zicha, D. Guidance of bidirectional motor complexes by mRNA cargoes through control of Dynein number and activity. Curr. Biol. 2006;16:1447–1452. doi: 10.1016/j.cub.2006.05.055. [DOI] [PubMed] [Google Scholar]
- Chou, T.B., Noll, E., Perrimon, N. Autosomal P[ovoD1] dominant female-sterile insertions in Drosophila and their use in generating germ-line chimeras. Development. 1993;119:1359–1369. doi: 10.1242/dev.119.4.1359. [DOI] [PubMed] [Google Scholar]
- Culver-Hanlon, T.L., Lex, S.A., Stephens, A.D., Quintyne, N.J., King, S.J. A microtubule-binding domain in dynactin increases Dynein processivity by skating along microtubules. Nat. Cell Biol. 2006;8:264–270. doi: 10.1038/ncb1370. [DOI] [PubMed] [Google Scholar]
- Davis, I., Ish-Horowicz, D. Apical localization of pair-rule transcripts requires 3′ sequences and limits protein diffusion in the Drosophila blastoderm embryo. Cell. 1991;67:927–940. doi: 10.1016/0092-8674(91)90366-7. [DOI] [PubMed] [Google Scholar]
- Deacon, S.W., Serpinskaya, A.S., Vaughan, P.S., Lopez Fanarraga, M., Vernos, I., Vaughan, K.T., Gelfand, V.I. Dynactin is required for bidirectional organelle transport. J. Cell Biol. 2003;160:297–301. doi: 10.1083/jcb.200210066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delanoue, R., Davis, I. Dynein anchors its mRNA cargo after apical transport in the Drosophila blastoderm embryo. Cell. 2005;122:97–106. doi: 10.1016/j.cell.2005.04.033. [DOI] [PubMed] [Google Scholar]
- Eckley, D.M., Gill, S.R., Melkonian, K.A., Bingham, J.B., Goodson, H.V., Heuser, J.E., Schroer, T.A. Analysis of Dynactin subcomplexes reveals a novel actin-related protein associated with the Arp1 minifilament pointed end. J. Cell Biol. 1999;147:307–320. doi: 10.1083/jcb.147.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross, S.P. Hither and yon: A review of bidirectional microtubule-based transport. Phys. Biol. 2004;1:R1–11. doi: 10.1088/1478-3967/1/2/R01. [DOI] [PubMed] [Google Scholar]
- Gross, S.P., Welte, M.A., Block, S.M., Wieschaus, E.F. Dynein-mediated cargo transport in vivo. A switch controls travel distance. J. Cell Biol. 2000;148:945–956. doi: 10.1083/jcb.148.5.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross, S.P., Welte, M.A., Block, S.M., Wieschaus, E.F. Coordination of opposite-polarity microtubule motors. J. Cell Biol. 2002;156:715–724. doi: 10.1083/jcb.200109047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holleran, E.A., Karki, S., Holzbaur, E.L. The role of the Dynactin complex in intracellular motility. Int. Rev. Cytol. 1998;182:69–109. doi: 10.1016/s0074-7696(08)62168-3. [DOI] [PubMed] [Google Scholar]
- Karki, S., Holzbaur, E.L. Affinity chromatography demonstrates a direct binding between cytoplasmic Dynein and the Dynactin complex. J. Biol. Chem. 1995;270:28806–28811. doi: 10.1074/jbc.270.48.28806. [DOI] [PubMed] [Google Scholar]
- King, S.J., Schroer, T.A. Dynactin increases the processivity of the cytoplasmic Dynein motor. Nat. Cell Biol. 2000;2:20–24. doi: 10.1038/71338. [DOI] [PubMed] [Google Scholar]
- Lopez de Heredia, M., Jansen, R.P. mRNA localization and the cytoskeleton. Curr. Opin. Cell Biol. 2004;16:80–85. doi: 10.1016/j.ceb.2003.11.002. [DOI] [PubMed] [Google Scholar]
- MacDougall, N., Clark, A., MacDougall, E., Davis, I. Drosophila gurken (TGFα) mRNA localizes as particles that move within the oocyte in two Dynein-dependent steps. Dev. Cell. 2003;4:307–319. doi: 10.1016/s1534-5807(03)00058-3. [DOI] [PubMed] [Google Scholar]
- Mallik, R., Petrov, D., Lex, S.A., King, S.J., Gross, S.P. Building complexity: An in vitro study of cytoplasmic Dynein with in vivo implications. Curr. Biol. 2005;15:2075–2085. doi: 10.1016/j.cub.2005.10.039. [DOI] [PubMed] [Google Scholar]
- McGrail, M., Gepner, J., Silvanovich, A., Ludmann, S., Serr, M., Hays, T.S. Regulation of cytoplasmic Dynein function in vivo by the Drosophila Glued complex. J. Cell Biol. 1995;131:411–425. doi: 10.1083/jcb.131.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muresan, V., Stankewich, M.C., Steffen, W., Morrow, J.S., Holzbaur, E.L., Schnapp, B.J. Dynactin-dependent, Dynein-driven vesicle transport in the absence of membrane proteins. A role for spectrin and acidic phospholipids. Mol. Cell. 2001;7:173–183. doi: 10.1016/s1097-2765(01)00165-4. [DOI] [PubMed] [Google Scholar]
- Murray, J.W., Bananis, E., Wolkoff, A.W. Reconstitution of ATP-dependent movement of endocytic vesicles along microtubules in vitro: An oscillatory bidirectional process. Mol. Biol. Cell. 2000;11:419–433. doi: 10.1091/mbc.11.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parton, R., Davis, I. Lifting the fog: Image restoration by deconvolution. In: Celis J.E., editor. Cell biology. Academic Press; New York: 2006. pp. 187–200. [Google Scholar]
- Pilling, A.D., Horiuchi, D., Lively, C.M., Saxton, W.M. Kinesin-1 and Dynein are the primary motors for fast transport of mitochondria in Drosophila motor axons. Mol. Biol. Cell. 2006;17:2057–2068. doi: 10.1091/mbc.E05-06-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plough, H.H., Ives, P.T. Heat induced mutations in Drosophila . Proc. Natl. Acad. Sci. 1934;20:268–273. doi: 10.1073/pnas.20.5.268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presley, J.F., Cole, N.B., Schroer, T.A., Hirschberg, K., Zaal, K.J., Lippincott-Schwartz, J. ER-to-Golgi transport visualized in living cells. Nature. 1997;389:81–85. doi: 10.1038/38001. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R Foundation for Statistical Computing; Vienna, Austria: 2006. R: A Language and Environment for Statistical Computing. [Google Scholar]
- Ray, K., Perez, S.E., Yang, Z., Xu, J., Ritchings, B.W., Steller, H., Goldstein, L.S. Kinesin-II is required for axonal transport of choline acetyltransferase in Drosophila . J. Cell Biol. 1999;147:507–518. doi: 10.1083/jcb.147.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross, J.L., Wallace, K., Shuman, H., Goldman, Y.E., Holzbaur, E.L. Processive bidirectional motion of dynein–dynactin complexes in vitro. Nat. Cell Biol. 2006;8:562–570. doi: 10.1038/ncb1421. [DOI] [PubMed] [Google Scholar]
- Simmonds, A.J., dosSantos, G., Livne-Bar, I., Krause, H.M. Apical localization of wingless transcripts is required for Wingless signaling. Cell. 2001;105:197–207. doi: 10.1016/s0092-8674(01)00311-7. [DOI] [PubMed] [Google Scholar]
- St Johnston, D. Moving messages: The intracellular localization of mRNAs. Nat. Rev. Mol. Cell Biol. 2005;6:363–375. doi: 10.1038/nrm1643. [DOI] [PubMed] [Google Scholar]
- Swaroop, A., Paco-Larson, M.L., Garen, A. Molecular genetics of a transposon-induced dominant mutation in the Drosophila locus Glued. Proc. Natl. Acad. Sci. 1985;82:1751–1755. doi: 10.1073/pnas.82.6.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuma, M.C., Zill, A., Le Bot, N., Vernos, I., Gelfand, V. Heterotrimeric kinesin II is the microtubule motor protein responsible for pigment dispersion in Xenopus melanophores. J. Cell Biol. 1998;143:1547–1558. doi: 10.1083/jcb.143.6.1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, Y., Kanai, Y., Okada, Y., Nonaka, S., Takeda, S., Harada, A., Hirokawa, N. Targeted disruption of mouse conventional kinesin heavy chain, kif5B, results in abnormal perinuclear clustering of mitochondria. Cell. 1998;93:1147–1158. doi: 10.1016/s0092-8674(00)81459-2. [DOI] [PubMed] [Google Scholar]
- Vale, R.D. The molecular motor toolbox for intracellular transport. Cell. 2003;112:467–480. doi: 10.1016/s0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]
- Vale, R.D., Milligan, R.A. The way things move: Looking under the hood of molecular motor proteins. Science. 2000;288:88–95. doi: 10.1126/science.288.5463.88. [DOI] [PubMed] [Google Scholar]
- Vallee, R.B., Williams, J.C., Varma, D., Barnhart, L.E. Dynein: An ancient motor protein involved in multiple modes of transport. J. Neurobiol. 2004;58:189–200. doi: 10.1002/neu.10314. [DOI] [PubMed] [Google Scholar]
- Wang, Z., Sheetz, M.P. One-dimensional diffusion on microtubules of particles coated with cytoplasmic Dynein and immunoglobulins. Cell Struct. Funct. 1999;24:373–383. doi: 10.1247/csf.24.373. [DOI] [PubMed] [Google Scholar]
- Wang, Z., Khan, S., Sheetz, M.P. Single cytoplasmic Dynein molecule movements: Characterization and comparison with Kinesin. Biophys. J. 1995;69:2011–2023. doi: 10.1016/S0006-3495(95)80071-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman-Storer, C.M., Karki, S., Holzbaur, E.L. The p150/Glued component of the Dynactin complex binds to both microtubules and the actin-related protein centractin (Arp-1) Proc. Natl. Acad. Sci. 1995;92:1634–1638. doi: 10.1073/pnas.92.5.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welte, M.A. Bidirectional transport along microtubules. Curr. Biol. 2004;14:R525–R537. doi: 10.1016/j.cub.2004.06.045. [DOI] [PubMed] [Google Scholar]
- Wilkie, G.S., Davis, I. Drosophila wingless and pair-rule transcripts localize apically by dynein-mediated transport of RNA particles. Cell. 2001;105:209–219. doi: 10.1016/s0092-8674(01)00312-9. [DOI] [PubMed] [Google Scholar]