Abstract
Each chain of the native trimeric P22 tailspike protein has eight cysteines that are reduced and buried in its hydrophobic core. However, disulfide bonds have been observed in the folding pathway and they are believed to play a critical role in the registration of the three chains. Interestingly, in the presence of sodium dodecyl sulfate (SDS) only monomeric chains, rather than disulfide-linked oligomers, have been observed from a mixture of folding intermediates. Here we show that when the oligomeric folding intermediates were separated from the monomer by native gel electrophoresis, the reduction of intermolecular disulfide bonds did not occur in the subsequent second-dimension SDS–gel electrophoresis. This result suggests that when tailspike monomer is present in free solution with SDS, the partially unfolded tailspike monomer can facilitate the reduction of disulfide bonds in the tailspike oligomers.
Keywords: P22 tailspike protein (TSP), transient disulfide bond, assembly, disulfide shuffling, SDS
P22 tailspike protein is a member of the elongated β-helix family of viral adhesins that participates in polysaccharide binding during viral infection that includes pectate lyase C and pertactin (Mitraki et al. 2002; Simkovsky and King 2006). Tailspike is a homotrimer and each monomer has 666 amino acid residues. Tailspike has four major domains: the N-terminal head-binding domain; the β-helix domain, which is formed by alternating β strands and turns; a β-sheet prism domain, where three chains are interdigitated; and the C-terminal domain (Steinbacher et al. 1994, 1997). Each chain in the native trimer has eight reduced cysteines: six are located in the β-helix domain and two are in the C-terminal domain. Disulfide bonds are formed during tailspike assembly, and they are believed to play critical roles in the registration of the three tailspike chains (Robinson and King 1997; Haase-Pettingell et al. 2001; Danek and Robinson 2003, 2004). Our current model is that intermolecular disulfide bonds form between the C613 on one chain and the C635 on another chain in the C-terminal domain to help align the three subunits. Site-directed mutagenesis of either C613 or C635 significantly decreased the assembly kinetics of tailspike as well as final yields both in vivo and in vitro (Haase-Pettingell et al. 2001; Danek and Robinson 2003). Since the intermolecular disulfide bonds are absent in the native trimer, reduction of those disulfide bonds is a required step in tailspike folding. How does the reduction occur? The nascent chains fold on the ribosome while translation is in progress, and one could imagine many sources of reductants (Brunschier et al. 1993; Gordon et al. 1994; Sather and King 1994; Clark and King 2001). However, in vitro, purified tailspike manages to fold into native trimer in the absence of redox buffers to yields of 80%–90% (Danek and Robinson 2004). A further troubling issue has been the presence of only two detectable bands on SDS-PAGE—the monomer and the trimer—although nondenaturing PAGE consistently showed disulfide-bonded oligomeric species, predominantly as dimers and trimers. The appearance of oligomeric folding intermediates was also observed during size-exclusion chromatography (Lefebvre and Robinson 2003).
In this work, we found that tailspike monomer chains in the presence of 0.1% SDS in solution could reduce transient disulfide bonds in oligomeric intermediates. This provides an explanation for two phenomena: the absence of disulfide-linked intermediates on SDS-PAGE and the reduction of tailspike chains to form the native, nondisulfide-linked trimer. In this study, purified native tailspike trimer (2 mg/mL) was denatured with 8 M urea (pH 3.0) for 1 h at room temperature. Then, this denatured protein solution was diluted 20-fold with ice-cold 50 mM Tris buffer (pH 7.6) such that the final protein concentration was 0.1 mg/mL. After 1 h of refolding on wet ice to populate folding intermediates (Betts and King 1998), aliquots were taken and mixed with 1/2 volume of cold native sample buffer or cold SDS sample buffer in the presence or the absence of DTT prior to electrophoresis.
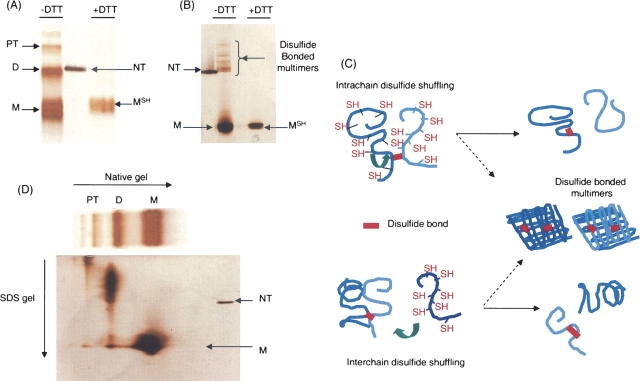
As Figure 1A shows, oligomeric folding intermediates such as dimer and protrimer (the precursor of the native trimer on the folding pathway [Robinson and King 1997]) were observed on nondenaturing gel electrophoresis in addition to the monomer. However, those oligomeric folding intermediates were dissociated into monomer when DTT was present in the native sample buffer, consistent with earlier data showing intermolecular disulfide bonds in oligomeric folding intermediates (Robinson and King 1997; Danek and Robinson 2004). The monomer band formed in the absence of DTT is more diffuse than the one in the reducing conditions and runs with slightly faster mobility. This suggests that intramolecular disulfide bonds may be present in the monomeric species. When these bands are shifted to higher temperatures (20°C), nearly all are competent to form trimers (Betts and King 1998; J. Kim and A.S. Robinson, in prep.). In this experiment, the ±DTT samples were electrophoresed on the same native gel. We have observed that DTT interferes with the silver staining process following native gel electrophoresis. Therefore, the discrepancy in staining is due to this DTT reaction with the silver, although the amounts of samples were identical in concentration and volume.
Figure 1.
(A) Nondenaturing gel electrophoresis of P22 tailspike folding intermediates. Refolding was performed on ice for 1 h and mixed with cold native sample buffer (3 mL glycerol, 18 mg Tris, 86.4 mg glycine, 2.5 mg Bromophenol blue in 10 mL). After native gel electrophoresis (9% acrylamide) at 4°C, silver staining was used to reveal the protein bands. Monomer (M), dimer (D), and protrimer (PT) were predominant folding intermediates in the absence of DTT. In the presence of DTT (10 mM), the reduced monomer (MSH) was the sole species appearing on the gel. NT represents native trimer as a marker. (B) SDS–gel electrophoresis of P22 tailspike-folding intermediates. The identical refolding sample in A was subjected to SDS–gel electrophoresis after mixing with SDS sample buffer (0.23 g Tris at pH 6.8 with HCl, 0.6 g SDS, 3 ml glycerol in 10 mL). Silver staining was used. In the absence of DTT, intense monomer (M) band appeared with several weak multimeric species, which have retarded mobilities compared with native trimer (NT). In the presence of DTT (10 mM), a single monomer band with sharp resolution was the sole species that appeared on the gel. (C) Intrachain and interchain disulfide shuffling models. Intrachain disulfide shuffling model describes the possible interaction between a free thiol group on one chain in dimer and its interchain disulfide. Interchain disulfide shuffling can occur between free monomers and disulfide-linked dimers. The byproducts of these interactions can be monomers with intrachain disulfide bonds and DTT-sensitive multimers, which are observed in B. (D) Nondenaturing–denaturing (SDS) gel electrophoresis of P22 tailspike folding intermediates. The folding intermediates at 0°C of P22 tailspike protein were separated by nondenaturing gel electrophoresis. One gel lane was excised from it and incubated in SDS running buffer (3.03g Tris, 14.1 g glycine, and 1 g SDS in 1 L) for 1.5 h at room temperature. Then, the excised lane was placed on top of the SDS gel and separated again. Monomer in the first-dimension nondenaturing gel appeared as monomer in the second-dimension SDS gel. However, dimer appeared to be assembled into a larger species. The weak monomer band was observed from the dimer lane in the second SDS gel.
In Figure 1A, monomer formed under reducing conditions was labeled as the reduced monomer, MSH (Ausubel et al. 1987; Betts and King 1999). Interestingly, almost all of the oligomeric intermediates were dissociated in SDS–gel electrophoresis, even in samples lacking DTT (Fig. 1B). Several less-intense bands that have a slower migration than native trimer appeared in the SDS gel but were absent when DTT was added to the SDS sample buffer. These bands are apparent only with the highly sensitive silver staining. These bands are below the detection range of Coomassie staining (data not shown). This unexpected reduction of disulfide bond-linked intermediates in the absence of DTT suggests that two potential redox reactions may occur between protein chains in the presence of SDS—intramolecular or intermolecular shuffling between tailspike chains. Figure 1C illustrates those mechanisms. Tailspike dimeric-folding intermediates have one disulfide bond in the C-terminal domain based on the model suggested by Robinson and coworkers (Danek and Robinson 2003, 2004). There are another six cysteines in the β-helix domain, although the redox state of each cysteine during folding is unknown. In the SDS solution, it is likely that the chains are at least partly unfolded, and shuffling from interchain to intrachain disulfide bonds in a dimer is possible and is consistent with the observations of Figure 1B. Alternatively, monomeric chains could act as electron donors and enable interchain shuffling (Fig. 1C, bottom). However, interaction between oligomers cannot be ruled out.
To test the alternatives shown in Figure 1C, we utilized two-dimensional nondenaturing–denaturing gel electrophoresis. If disulfide shuffling occurs between oligomers and monomers, separation of disulfide-linked oligomers from monomers would prevent monomer-driven reduction in the presence of SDS. Refolding of tailspike was performed as described above. Duplicate samples of folding intermediates were separated with 9% nondenaturing gel electrophoresis in the cold. Subsequently, the gel lanes containing the tailspike folding intermediates were excised and subjected to a 1-1/2-h incubation with SDS–gel electrophoresis running buffer. Then, the excised gel lane was placed horizontally on top of an SDS gel and separated by SDS-PAGE in this second dimension.
Interestingly, the oligomers showed little dissociation (Fig. 1D). Instead, the migration of the dimers was reduced, suggesting an increase in the oligomeric state (Fig. 1D). Since SDS can dissociate aggregates into smaller subunits when there are no disulfide bonds, this association may result from adventitious disulfide bonding between oligomeric chains. Interestingly, monomers present in the native gel did not associate during the incubation (Fig. 1D). Shorter incubation times showed similar results. Addition of DTT to the incubation buffer results in reduction of all bands to the mobility of the monomer (data not shown). Although this redox chemistry of tailspike in the SDS solution cannot directly explain how the transient disulfide bonds of tailspike intermediates are reduced under physiological folding conditions, it suggests that cysteine interactions between partially (un)folded chains are a plausible mechanism for disulfide shuffling.
Footnotes
Reprint requests to: Anne Skaja Robinson, 259 Colburn Lab, Department of Chemical Engineering, University of Delaware, Newark, DE 19716, USA; e-mail: Robinson@che.udel.edu; fax: (301) 831-6262.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.062197206.
Abbreviations: SDS, sodium dodecyl sulfate; DTT, dithiothreitol.
References
- Ausubel F.M., Brent R., Kingston R.E., Moore D.D., Seidman J.G., Smith J.A., Struhl K. In Current protocols in molecular biology pp. 10.16.11–10.16.25. 1987. John Wiley & Sons, Inc, New York.
- Betts S.D. and King J. 1998. Cold rescue of the thermolabile tailspike intermediate at the junction between productive folding and off-pathway aggregation. Protein Sci. 7: 1516–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betts S and King J. 1999. There's a right way and a wrong way: In vivo and in vitro folding, misfolding and subunit assembly of the P22 tailspike. Structure 7: R131–R139. [DOI] [PubMed] [Google Scholar]
- Brunschier R., Danner M., Seckler R. 1993. Interactions of phage P22 tailspike protein with GroE molecular chaperones during refolding in vitro. J. Biol. Chem. 268: 2767–2772. [PubMed] [Google Scholar]
- Clark P.L. and King J. 2001. A newly synthesized, ribosome-bound polypeptide chain adopts conformations dissimilar from early in vitro refolding intermediates. J. Biol. Chem. 276: 25411–25420. [DOI] [PubMed] [Google Scholar]
- Danek B.L. and Robinson A.S. 2003. Nonnative interactions between cysteines direct productive assembly of P22 tailspike protein. Biophys. J. 85: 3237–3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danek B.L. and Robinson A.S. 2004. P22 tailspike trimer assembly is governed by interchain redox associations. Biochim. Biophys. Acta 1700: 105–116. [DOI] [PubMed] [Google Scholar]
- Gordon C.L., Sather S.K., Casjens S., King J. 1994. Selective in ;vivo rescue by GroEL/ES of thermolabile folding intermediates to phage P22 structural proteins. J. Biol. Chem. 269: 27941–27951. [PubMed] [Google Scholar]
- Haase-Pettingell C., Betts S., Raso S.W., Stuart L., Robinson A., King J. 2001. Role of cysteine residues in the in vivo folding and assembly of the phage P22 tailspike. Protein Sci. 10: 397–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre B.G. and Robinson A.S. 2003. Pressure treatment of tailspike aggregates rapidly produces on-pathway folding intermediates. Biotechnol. Bioeng. 82: 595–604. [DOI] [PubMed] [Google Scholar]
- Mitraki A., Miller S., van Raaij M.J. 2002. Review: Conformation and folding of novel β-structural elements in viral fiber proteins: The triple β-spiral and triple β-helix. J. Struct. Biol. 137: 236–247. [DOI] [PubMed] [Google Scholar]
- Robinson A.S. and King J. 1997. Disulphide-bonded intermediate on the folding and assembly pathway of a non-disulphide bonded protein. Nat. Struct. Biol. 4: 450–455. [DOI] [PubMed] [Google Scholar]
- Sather S.K. and King J. 1994. Intracellular trapping of a cytoplasmic folding intermediate of the phage P22 tailspike using iodoacetamide. J. Biol. Chem. 269: 25268–25276. [PubMed] [Google Scholar]
- Simkovsky R. and King J. 2006. An elongated spine of buried core residues necessary for in vivo folding of the parallel β-helix of P22 tailspike adhesin. Proc. Natl. Acad. Sci. 103: 3575–3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbacher S., Seckler R., Miller S., Steipe B., Huber R., Reinemer P. 1994. Crystal structure of P22 tailspike: Interdigitated subunits in a thermostable trimer. Science 265: 383–386. [DOI] [PubMed] [Google Scholar]
- Steinbacher S., Miller S., Baxa U., Budisa N., Weintraub A., Seckler R., Huber R. 1997. Phage P22 tailspike protein: Crystal structure of the head-binding domain at 2.3 Å, fully refined structure of the endorhamnosidase at 1.56 Å resolution, and the molecular basis of O-antigen recognition and cleavage. J. Mol. Biol. 267: 865–880. [DOI] [PMC free article] [PubMed] [Google Scholar]