Abstract
The molecular hallmarks of inflammation-mediated lung carcinogenesis have not been fully clarified, mainly due to the scarcity of appropriate animal models. We have used a silica-induced multistep lung carcinogenesis model driven by chronic inflammation to study the evolution of molecular markers and genetic alterations. We analyzed markers of DNA damage response (DDR), proliferative stress, and telomeric stress: γ-H2AX, p16, p53, and TERT. Lung cancer-related epigenetic and genetic alterations, including promoter hypermethylation status of p16(CDKN2A), APC, CDH13, Rassf1, and Nore1A, as well as mutations of Tp53, epidermal growth factor receptor, K-ras, N-ras, and c-H-ras, have been also studied. Our results showed DDR pathway activation in preneoplastic lesions, in association with inducible nitric oxide synthase and p53 induction. p16 was also induced in early tumorigenic progression and was inactivated in bronchiolar dysplasias and tumors. Remarkably, lack of mutations of Ras and epidermal growth factor receptor, and a very low frequency of Tp53 mutations suggest that they are not required for tumorigenesis in this model. In contrast, epigenetic alterations in p16(CDKN2A), CDH13, and APC, but not in Rassf1 and Nore1A, were clearly observed. These data suggest the existence of a specific molecular signature of inflammation-driven lung carcinogenesis that shares some, but not all, of the molecular landmarks of chemically induced lung cancer.
Keywords: lung cancer, inflammation, animal model, preneoplastic lesions, DNA damage response
Introduction
Lung cancer is a complex disease that develops through the progressive accumulation of both genetic and epigenetic alterations. A number of molecular changes potentially leading to lung cancer have already been described [1]. These changes may be induced by several genotoxic carcinogens [2] to which a smoker's airways are continuously exposed. Aside from direct carcinogens, inhaled smoke contains other constituents that can stimulate a chronic inflammatory response in the lung [3]. Strong evidence supports that chronic inflammation promotes tumorigenesis [4]. Inflammatory cellular response can induce cell proliferation and tissue repair, creating a fertile “soil” enriched with cytokines and growth or angiogenic factors. Inflammatory foci are also a continuous source for reactive oxygen species (ROS) and reactive nitrogen species, which may induce DNA damage, including DNA strand breaks and adducts, mismatches, and mutations [5,6]. These events may eventually lead to transformation and tumorigenesis [4].
There are strong epidemiological evidences linking pulmonary chronic inflammation to a higher risk of lung cancer [7]: Chronic obstructive airway disease has been shown in many series of patients to be an independent predictor of lung cancer risk, and various studies have reported increased cancer risk among adults with asthma, tuberculosis, or postinflammatory pulmonary interstitial fibrosis, such as in patients with silicosis and asbestosis [7]. Chronic inflammation may promote some of the molecular changes observed in airway epithelial cells that lead to cancer. Molecules involved in both inflammation and tumorigenesis include proteins of the COX-2 and NF-κB signaling pathways, chemokines and cytokines (TNF-α, IL-1, IL-6, and IL-8), growth and angiogenic factors (PDGF, TGF-β, and EGF), proteolytic enzymes, and cytotoxic agents such as ROS and nitric oxide (NO) [4,8,9]. The role of inflammation in lung carcinogenesis has not yet been clarified mainly due to the scarcity of appropriate chronic inflammation models of lung cancer. Several animal models have been developed in the last decades to better understand lung carcinogenesis, but most of these do not provide unambiguous information on the specific role of inflammation. Malkinson [10] has studied a mouse model in which chronic inflammation, induced by butylated hydroxytoluene, promotes lung carcinogenesis previously initiated by a chemical carcinogen (methylcholanthrene). Very few animal models of cancer in which chronic inflammation drives carcinogenesis with no other carcinogenic insult have been molecularly characterized [11–13]. In the silica-induced rat lung carcinogenesis model, a single intratracheal instillation of crystalline silica dust suspended in saline leads to silicotic chronic inflammation and progressive proliferative epithelial lesions. Silica leads to the formation of silicotic granulomas, which consist of aggregates of activated macrophages that phagocytose dust mineral, induce collagen deposition, and recruit lymphoid cells. The rat silicotic granulomas remain stable or increase in size over time and, thus, are essentially irreversible. They produce several molecular mediators (interleukins, TNF-α, TGF-β, NO, and so on), which promote further inflammatory stress and fibrogenic responses within the lung and induce progressive proliferative epithelial reactivity in neighboring areas [14]. Therefore, this is a very useful model for studying this type of pure chronic inflammation-driven carcinogenesis. The severity of epithelial lesions grows over time from alveolar and bronchiolar epithelial hyperplasias to advanced preneoplastic lesions, and finally to adenocarcinomas (AC) and squamous cell carcinomas (SCC) [14,15]. According to the literature and our own experiments, lung tumors are already induced by the 11th month in around 40% of the rats. In later stages, after month 17, an increase in incidence (90%) is clearly observed [14]. Most of the tumors found in this model are AC (84%), compared to mixed carcinomas (8%) or SCC (8%). At the histopathological level, this model recapitulates a number of features of the multistep carcinogenic process observed in human peripheral lung cancer and provides access to a wealth of preneoplastic lesions amenable for molecular analysis. In the last decade, several groups have studied the silicotic rat model, focusing mainly on the short-term activation of classic inflammation-related molecular hallmarks, including the activation of the NF-κB and PKC signaling pathways, as well as the production of growth factors IL-1, IL-6, TNF-α, ROS, NO, and so on [16–19]. However, a detailed analysis of molecular alterations in the progression of the preneoplastic and neoplastic epithelial lesions in this model is still lacking.
The aim of our study was to improve our understanding of the molecular pathways involved in inflammation-mediated carcinogenesis and to follow molecular changes along the process from normal epithelial cells to cancer in the silica-induced lung cancer model. We focused our study on the evolution of a variety of molecular markers that are already associated with human lung cancer. Recently, several reports have emphasized the existence of tumorigenesis barriers that slow or inhibit the progression of preneoplastic lesions to neoplasia [20,21]. One such barrier involves DNA replication stress, which leads to activation of the DNA damage checkpoint and, thereby, to apoptosis or cell cycle arrest mediated by either p53 or p16 [22,23]. We thus have analyzed preneoplastic and neoplastic lung lesions for a number of well-characterized markers of DNA damage response (DDR), proliferative stress, and telomeric stress such as Ser139-phosphorylated histone 2AX (γ-H2AX), p53, p16, and the catalytic subunit of telomerase reverse transcriptase (TERT). In order to dissect the molecular profile, we also studied lung cancer-related epigenetic and genetic alterations, including promoter hypermethylation status of p16 (CDKN2A), APC, CDH13, Rassf1, and Nore1A, and mutations of Tp53, epidermal growth factor receptor (EGFR), K-ras, N-ras, and c-H-ras. In brief, our study shows the relevance of a set of molecular and epigenetic alterations, rather than of “classic” mutations, in the multistep progression of an inflammation-mediated lung cancer model.
Materials and Methods
Carcinogenesis Protocol
Fisher F344/NCr female rats fromHarlanUKLimited (Oxon, UK) were used. Rats were housed in specific pathogen-free conditions with access to food and water ad libitum. Procedures were carried out in strict compliance European Union and University of Navarra (Institutional Animal Care and Use Committee) relevant guidelines for the use of laboratory animals. The crystalline silica sample was 99% pure α-quartz (Min-U-Sil 5; US Silica Co., Berkeley Springs, WV), with a particle size of < 5 µm [14]. The silica sample was suspended in sterile neutral-buffered saline and briefly sonicated to provide full dispersion. Eight-week-old rats were anesthetized with a mixture of oxygen and isoflurane, and placed on their backs on a metal board slanted at a 60° angle, with the mouth kept open. In this position, at the end of an expiration, rats received a single intratracheal instillation of 16 mg of quartz in 0.3 ml of saline through a cannula connected to a syringe [14]. Epithelial reactions to silica started with foci of hyperplastic epithelial cells adjacent to silicotic granulomas, including hyperplasias of type II pneumocytes from month 1 and hyperplasias of bronchiolar epithelial cells from month 4. Preneoplastic adenomatoid lesions (AL) and dysplastic bronchiolar lesions were observed from month 8. AL in large clusters of completely lined alveolar spaces that had progressed to become more conspicuous and include areas of adenomatoid proliferation separated by thick fibrous septa were formed by alveolar epithelial hyperplastic reaction. The shape of hyperplastic adenomatoid cells was usually less cuboidal and more flattened. AC and SCC were observed from month 11. AC were fibrotic or nonfibrotic, with an acinar, papillary, or alveolar pattern showing varying degrees of differentiation.
Samples
Seventy rats were instilled with 16 mg of quartz, and the lungs were obtained on months 3 to 4 (n = 10), month 6 (n = 5), month 8 (n = 5), month 10 (n = 5), month 12 (n = 5), months 16 to 17 (n = 20), and month 21 (n = 20) after instillation. Several lesions were studied throughout these time periods: hyperplasias of type II pneumocytes, hyperplasias of bronchiolar epithelial cells, preneoplastic AL and dysplastic bronchiolar epithelium, and AC (fibrotic and non-fibrotic) and SCC. Hyperplastic and advanced preneoplastic lesions were also observed in late stages. Tissues from control rats (n = 8), instilled with 0.3 ml of saline without quartz, were collected on month 6 (n = 4) and month 12 (n = 4). Rats were killed by exsanguination under anesthesia with Ketolar (Parke Davis, Madrid, Spain) and Rompun (Bayer AG, Leverkusen, Germany). The trachea was exposed by dissection and ligated during maximal inspiration. The larynx, trachea, bronchi, lungs, lymph nodes, and heart were removed en bloc and fixed by immersion in 4% formaldehyde in phosphate buffer for 24 hours. Lung lobes were sectioned along their main bronchial axis, embedded in paraffin, and sectioned at 4 µm thickness. Paraffin-embedded lungs from treated and control rats were used.
A subset of the tumors was isolated from the fresh unfixed lungs and split into two halves. One was rapidly snap frozen in liquid nitrogen and kept at -80°C for DNA extraction. The other was processed for histologic analysis. Normal lungs from control rats were processed in the same manner.
Methylation Analysis
Quantification of global DNA methylation
Genomic DNA was extracted by conventional methods from frozen control lungs (n = 6); frozen treated lungs obtained on months 3 to 4 (n = 5), months 6 to 8 (n = 5), and months 10 to 12 (n = 5); andfrozen isolated tumorsobtained onmonths16 to 21 (n=9; five AC and four SCC). 5-Methylcytosine (mC) DNA content in all the samples was determined by high-performance capillary electrophoresis, as previously described [24]. Briefly, genomic DNA (3–5 µg) was obtained from the different tissues, and DNA hydrolysis was carried out with 1.25 µl (200 U/ml) of nuclease P1 for 16 hours at 37°C. Subsequently, alkaline phosphatase was added, and mixtures were incubated for 2 hours at 37°C. Hydrolyzed samples were injected under pressure (0.3 psi) for 3 seconds into an uncoated fused-silica capillary in a CE system (P/AC MDQ; Beckman Coulter, Palo Alto, CA). Quantification of the relative methylation of each DNA sample was determined as: %mC = (mC peak area × 100)/(C peak area + mC peak area). All samples were analyzed in duplicate, and three analytical measurements were made per replicate.
Bisulfite genomic analysis of the p16(CDKN2A), APC, CDH13, Rassf1, and Nore1A CpG islands
DNA samples from control lungs (n = 6); treated lungs obtained on months 3 to 4 (n = 5), months 6 to 8 (n = 5), and months 10 to 12 (n = 5); and tumors obtained on months 16 to 21 (n = 9; five AC and four SCC) were treated with sodium bisulfite, as previously described [25]. Primers (Table W1) spanning the CpG island of the rat p16(CDKN2A), APC, H-cadherin (CDH13), Rassf1, and Nore1A promoters were used for bisulfite genomic sequencing. At least six individual clones were sequenced for each sample. A single CpG was considered to be methylated when more than half of the clones retained an unmodified cytosine at that position.
Mutational Analysis
DNA extraction after laser capture microdissection
To obtain near-pure cancer cell populations for mutation analyses, we performed laser capture microdissection (CTRMIC/ASLMD, Leica, Germany). Two consecutive 5-µm sections were cut from formalin-fixed paraffin-embedded tissue blocks. One section was stained with hematoxylin and eosin (H&E), which was used for histologic evaluation and control; another section was stained with methyl green for microdissection (Figure W1). Tumor tissue and adjacent normal lung tissue for control were obtained by microdissection. The microdissected cells were collected in a 0.5-ml microcentrifuge tube containing 40 µl of DNA lysis buffer (10 mM Tris-HCl pH 8.0, 1 mM EDTA, 1% Tween-20, and 20 mg/ml proteinase K). Tubes were then incubated for 14 hours at 55°C. Proteinase was inactivated by incubation at 95°C for 10 minutes.
Polymerase chain reaction exon amplification
Eight microliters of genomic DNA was used for very-high-fidelity polymerase chain reaction (PCR) amplification with AccuPrime Pfx DNA Polymerase System (Invitrogen, Carlsbad, CA). PCR was carried out in a total volume of 30 µl containing 0.2 µM of each primer (Table W2), 1 U of AccuPrime Pfx DNA Polymerase, and 1% AccuPrime Pfx Reaction Mix (50 mM Tris-HCl pH 8.0, 50 mM KCl, 1 mM DTT, 0.1 mM EDTA, 1 mM MgSO4, and 3 mM dNTPs). PCRs were performed on a PTC-100 thermocycler (MJResearch, Watertown, MA) using the following protocol: 95°C for 3 minutes; 37 cycles of 95°C for 30 seconds, primers' specific annealing temperature for 35 seconds, and 68°C for 35 seconds; and 7 minutes at 68°C for extension. PCR products, which were confirmed to have a single target band in a 2% agarose gel, were purified using the Qiagen MinElute PCR Purification Kit (Qiagen, Valencia, CA).
Sequencing and analysis
The purified products were then subjected to direct sequencing by an ABI377 sequencer (Perkin-Elmer Applied Biosystems, Foster City, CA). Exons were sequenced upstream and downstream, and compared to a control sequence. They were carefully analyzed first with ChromasPro 1.33 (Tewantin, Australia) and, later, manually. If a mutation was detected, additional microdissection and sequencing of the tumor and the surrounding normal tissue were carried out to verify the mutation.
Immunostaining
Immunostaining was performed with antibodies to p16 (F-12, 1:50; Santa Cruz Biotechnology, Santa Cruz, CA), p53 (FL-393, 1:100; Santa Cruz Biotechnology; or Pab 240, 1:100; Abcam, Cambridge, UK), EGFR (2232, 1:50; Cell Signaling, Danvers, MA), TERT (Ab-2, 1:500; Calbiochem, San Diego, CA), γ-H2AX (2577, 1:200; Cell Signaling), and inducible nitric oxide synthase (iNOS) (Sc-650, 1:100; Santa Cruz Biotechnology). An indirect avidin-biotin-peroxidase method (Dako, Barcelona, Spain) was used for EGFR, TERT, γ-H2AX, and iNOS analysis. In the case of p53 and p16 immunostaining, we used the EnVision (K4001; Dako) signal enhancement system. Immunohistochemical technique was carried out as follows. Slides were deparaffinized and incubated for 10 minutes with 3% H2O2 in water to quench endogenous peroxidase activity. Heat-mediated antigen retrieval was used for antibodies to EGFR and γ-H2AX (15 minutes at 375 W in 1 mM EDTA, pH 8.0) and p53 (15 minutes at 750 W and 15 minutes at 375 W in 10 mM sodium citrate, pH 6.0). Tissues were incubated with 5% normal rabbit serum in Tris-buffered saline (TBS) (500 mM NaCl and 50 mM Tris-HCl pH 7.4) for 30 minutes at room temperature. After blotting excess serum, sections were incubated at 4°C overnight, with the primary antibody diluted in TBS for p53, p16, telomerase, and iNOS, and diluted in mixed sera for EGFR and γ-H2AX.
Tissues were washed in TBS and incubated with the appropriate secondary antibody. For the indirect avidin-biotin-peroxidase method, biotinylated rabbit-anti-mouse Ig antiserum was added at a 1:200 dilution for 30 minutes at room temperature; after the slides had been washed, they were incubated for 30 minutes at room temperature with the avidin-biotin complex at a 1:100 dilution. For the EnVision signal enhancement system, the secondary monoclonal complex was applied for 30 minutes at room temperature. After the slides had been washed in TBS, development of peroxidase with diaminobenzidine and H2O2 was performed. The slides were counterstained with hematoxylin, dehydrated, and mounted. As negative controls, the primary antibody was replaced with mouse IgG1 or IgG2a specific for Aspergillus niger glucose oxidase (Dako).
Staining criteria
The immunostaining level of the following histologic structures was analyzed in each section: normal bronchioles (NB), normal type II alveolar cells (NA), hyperplastic bronchioles (HB), hyperplastic type II alveolar cells (HA), dysplastic bronchioles (DB), AL, and tumors AC and SCC. The total numbers of normal and pathological structures analyzed by each antibody are indicated in the Results section.
p53, p16, or TERT-immunostained area in a given tissue was expressed as the percentage of positive nuclei and calculated as: (positive nuclei/total nuclei) × 100. This quantitative method has been previously published [26]. For p53 and p16 quantification, although some cytoplasmic staining was found at some stages, only nuclear immunostaining was considered. For p53, we considered overexpression when > 50% of the nuclei were immunostained. The result was scored as negative immunostaining if < 10% of the nuclei were positive. In the case of p16, loss of expression was recorded when < 25% of positive nuclei were found. For telomerase, we considered overexpression when > 75% of the nuclei were positive. The mean percentage of nuclear protein expression and its 95% confidence interval were calculated for p53, p16, and TERT in all the histologic structures studied. In the evaluation of EGFR and iNOS immunostaining, the intensity was classified into four categories: (−) negative, (+) weak, (++) moderate, and (+++) strong. Immunostaining was estimated independently by two experienced researchers.
Statistical analysis
Statistical analysis was performed with the SPSS 13.0 software (SPSS, Inc., Chicago, IL). Chi-square analysis was applied to study the differences of p53, p16, or TERT expression between the normal, preneoplastic, and tumoral tissues. Fisher's exact test was applied to study differences in DNA global hypomethylation analysis. Differences were considered significant when P < .05.
Results
DDR and Oxidative Stress from Early Hyperplastic Tissues to Tumors
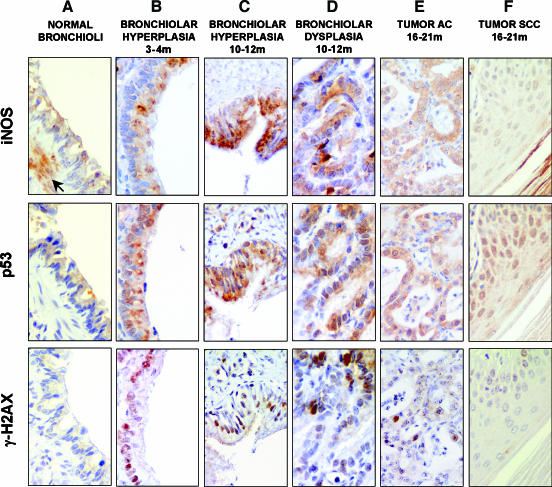
In serial consecutive tissue sections of the silica-induced lung cancer model, we analyzed the expression of p53, iNOS, and γ-H2AX, a marker of activated DDR. We studied the expression of these three markers through five stages (n = 8 for each stage), including normal, preneoplastic, and neoplastic tissues. In the normal bronchiolar epithelial cells of treated animals, p53 and iNOS expressions were found at low levels, whereas there was no signal for γ-H2AX (Figure 1A). iNOS expression was also observed in alveolar and parenchymal macrophages present in the early phases of the silicotic response, as well as in epithelioid macrophages at the core of granulomas. iNOS expression in granulomas was maintained up to the latest stages studied (21 months) (Figure W2). p53, iNOS, and γ-H2AX were clearly induced in the same tissue elements of early preneoplastic lesions such as hyperplastic bronchioloalveolar cells and persisted in advanced preneoplastic stages such as dysplastic bronchiolar cells (Figure 1, B–D). In tumors, colocalization was still present in some areas, although γ-H2AX levels were reduced compared to hyperplastic and advanced preneoplastic tissues (Figure 1, E and F). These data suggest that (nitro)oxidative stress coexists with p53 accumulation in tumorigenic progression, and both are associated with DNA damage measured by the DDR marker γ-H2AX.
Figure 1.
Oxidative stress and DDR in silica-induced lesions measured by immunohistochemistry in serial consecutive sections. The figures in this plate show the activation of the iNOS/p53/γ-H2AX pathway in the multistep progression from normal tissue to tumors. Low levels of iNOS and p53 proteins were found in morphologically normal bronchial epithelial cells, whereas γ-H2AX was completely absent (A). In normal bronchioli, iNOS expression was also found in smooth muscle cells (arrow; A). A clear increase in the coexpression of iNOS/p53/γ-H2AX was observed from hyperplastic (B and C) to dysplastic (D) bronchiolar cells. In tumors, colocalization was still present in some areas, although γ-H2AX levels were reduced compared to hyperplastic and advanced preneoplastic tissues (E and F). Counterstaining by Harris hematoxylin. Original magnifications, × 420 (C, E, and F); × 630 (A, B, and D).
p53 Stabilization and Loss of p16 Expression in Multistage Progression to Silica-Induced Lung Tumors
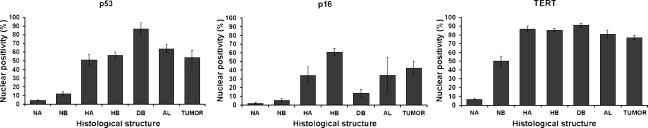
We analyzed the evolution of the p53 and p16 protein expressions along the multistep progression from the normal epithelium to non-small cell lung cancer (NSCLC) tumors (50 AC and 9 SCC). p53 nuclear immunostaining was negative in normal alveoli (mean percentage of positive nuclei: 5 ± 0.1%; n = 35) and bronchiolar epithelial tissues (12 ± 3%; n = 32) (Figures 2A and 3). A significant increase for p53 nuclear expression was observed in hyperplastic bronchiolar (56 ± 4%; n = 40) and alveolar (51 ± 6%; n = 29) epithelial cells (Figures 2, B and C, and 3; Table 1; P < .001), and this overexpression was maintained in epithelial hyperplasias over time, at least up to month 12. In dysplastic bronchiolar lesions (87 ± 7%; n = 12), p53 was significantly higher when compared to hyperplastic bronchiolar epithelium on months 10 to 12 (Figures 2D and 3; Table 1; P < .001). The percentage of cells with nuclear p53 was significantly lower in tumors (54 ± 8%; n = 59) compared to advanced preneoplastic lesions such as bronchiolar dysplasia (P < .001) and adenomatoid lesions (P = .008) (Figure 3; Table 1). In tumors, p53 overexpression was observed in 26 of 50 (52%) AC and in 6 of 9 (67%) SCC (Figure 2, E and F). No statistical differences were observed between the two histologic tumor types.
Figure 2.
p53, p16, and TERT nuclear expression by immunohistochemistry in the multistep progress to lung cancer. (A) Normal lung epithelial cells showed low levels of nuclear protein expression for p53, p16, and TERT. (B and C) Hyperplastic bronchiolar (B) and alveolar (C) cells showed a significant increase for p53, p16, and TERT nuclear immunostaining. (D) DB at 10 to 12 months showed a higher percentage of positive nuclear cells for p53 and TERT. However, p16 was significantly decreased in these advanced preneoplastic lesions. (E and F) p53 and TERT overexpression was commonly observed in AC (E) and SCC (F), whereas p16 overexpression was observed in SCC (F) and in a subset of more fibrotic AC. Loss of p16 protein expression (< 25% positive nuclei) was detected in 44% of AC (p16; E). Counterstaining by Harris hematoxylin. Original magnifications, × 420 (p53-TERT; B) (p16; D, E, and F); × 560 (p16; B); × 700 (A and C) (p53-TERT; D).
Figure 3.
Differential expression for p53, p16, and TERT between different stages of cancer progression. Graphs show the mean percentage of nuclear protein expression and its 95% confidence interval for p53, p16, and TERT immunostaining in all the histologic structures studied. TUMOR, AC and SCC.
Table 1.
Statistical Analysis of the Differences in p53, p16, and TERT Expression between Different Stages of Cancer Progression.
| Comparisons | p53 | p16 | TERT | |
| Normal versus hyperplasia | NA versus HA | χ2 = 74.00, P < .001 | χ2 = 53.79, P < .001 | χ2 = 77.00, P < .001 |
| NB versus HB | χ2 = 89.44, P < .001 | χ2 = 71.38, P < .001 | χ2 = 91.38, P < .001 | |
| Hyperplasia versus advanced preneoplasia | HA versus AL | χ2 = 4.63, P = 2.01 | χ2 = 2.10, P = .718 | χ2 = 4.56, P = .041 |
| HB versus DB | χ2 = 39.74, P < .001 | χ2 = 46.93, P < .001 | χ2 = 1.89, P = .169 | |
| Nonneoplastic versus neoplastic lesions | NA versus TUMOR | χ2 = 78.00, P < .001 | χ2 = 57.48, P < .001 | χ2 = 95.00, P < .001 |
| HB versus TUMOR | χ2 = 54.40, P < .001 | χ2 = 40.12, P < .001 | χ2 = 48.64, P < .001 | |
| HA versus TUMOR | χ2 = 11.39, P = .010 | χ2 = 6.32, P = .177 | χ2 = 12.68, P < .001 | |
| HB versus TUMOR | χ2 = 11.34, P = .010 | χ2 = 28.39, P < .001 | χ2 = 18.22, P < .001 | |
| AL versus TUMOR | χ2 = 11.94, P = .008 | χ2 = 2.43, P = .657 | χ2 = 0.67, P = .412 | |
| DB versus TUMOR | χ2 = 17.31, P < .001 | χ2 = 18.16, P < .001 | χ2 = 19.48, P < .001 |
TUMOR, AC and SCC.
p16 protein expression was negative in normal alveolar (2 ± 0.6%; n = 36) and bronchiolar (5.6 ± 1.8%; n = 35) epithelial tissues (Figures 2A and 3). We observed a significant increase in p16 nuclear overexpression in hyperplastic bronchiolar (61 ± 4%; n = 40) and alveolar (34 ± 10%; n = 29) epithelial cells compared to normal tissues (Figures 2, B and C, and 3; Table 1; P < .001). This overexpression was maintained in bronchiolar and type II hyperplasias from months 6 to 12. Interestingly, p16 was significantly decreased in dysplastic bronchiolar epithelium (13 ± 4%; n = 12) compared to neighboring bronchiolar hyperplasias (Figures 2D and 3; Table 1; P < .001). In tumors, the percentage of cells with nuclear p16 was significantly lower (42 ± 8.4%; n = 59) than in bronchiolar hyperplasias (Figure 3; Table 1; P < .001). Loss of p16 protein expression (< 25% of positive nuclei) was detected in 22 of 50 (44%) AC and in 2 of 9 (22%) SCC (Figure 2E). Nonfibrotic AC showed a significantly lower score for p16 staining (30 ± 8% of positive nuclei) compared to fibrotic AC (58 ± 9%; P = .016) and SCC (76 ± 24%; P = .027).
All these data confirm that p53 nuclear levels increase in epithelial cells during the multistep progression from normal to hyperplastic and advanced preneoplastic tissues, and remain high in most tumors. Conversely, p16 is overexpressed in hyperplastic tissues, but its expression is decreased or lost in advanced bronchiolar dysplasias and in 40% of the tumors.
TERT Protein Overexpression in Multistep Progress to Lung Tumors
Nuclear TERT protein overexpression was observed from months 3 to 12 in hyperplastic bronchiolar (86 ± 2%; n = 19) and alveolar (87 ± 3%; n = 20) tissues compared to normal bronchiolar (50 ± 6%; n = 48) and alveolar (7 ± 1%; n = 38) epithelial cells (Figures 2, A–C, and 3; Table 1; P < .001). A high percentage of immunostained nuclei was also observed from month 10 in advanced preneoplastic adenomatoid lesions (81 ± 5%; n = 9) and dysplastic bronchiolar cells (91 ± 2%; n = 12) compared to normal tissues (Figures 2D and 3). At the late stage, lung tumors also showed a significant overexpression of TERT (77 ± 2%; n = 59) compared to control tissues (Figures 2, E and F, and 3; Table 1; P < .001). The percentage of nuclei stained for TERT at 16 to 21 months was significantly lower in tumors than in hyperplastic bronchioloalveolar and dysplastic bronchiolar lesions (Table 1; P < .001), showing a decrease in TERT nuclear detection from preneoplastic tissues to cancer. No statistical differences were observed between AC and SCC. We conclude that TERT is highly expressed in reactive epithelial tissues during multistep carcinogenesis, with the highest percentage of expression in hyperplastic and dysplastic tissues, and a slight decrease of its expression in tumors.
Mutational Profile of Lung Tumors
Tp53
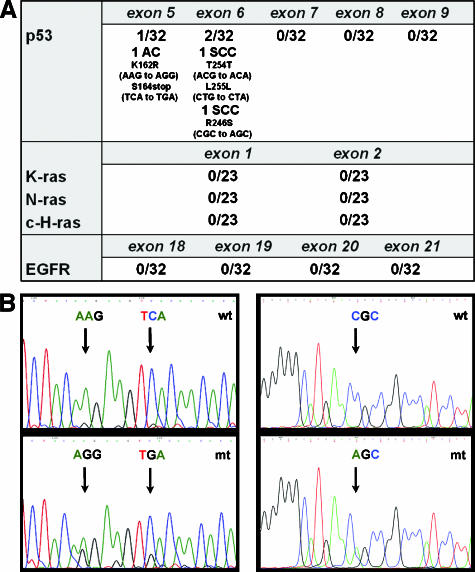
We analyzed the mutational status of the Tp53 gene, the most commonly mutated gene in human cancer. We sequenced exons 5 to 9–those more frequently mutated in human lung cancer and also found mutated in rodent tumors [27,28]. To avoid contamination with nontumor DNA, we microdissected tumors from paraffin sections. We selected a subset of 32 tumors showing either high or low p53 protein nuclear accumulation, as determined by immunohistochemistry. Five mutations were identified in 3 of 32 (9%) tumors (one AC and two SCC). Two mutations were detected in exon 5 of one AC sample: a missense mutation (K162R) and a nonsense mutation in codon 164. In two SCC, three mutations were located in exon 6, involving codons 254 and 255 (both sense mutations), and codon 246 (a missense mutation leading to an arginine-to-serine substitution) (Figure 4, A and B). Therefore, Tp53 mutations are rare events in tumors induced by silica-mediated chronic inflammation.
Figure 4.
Mutational analysis for Ras (n = 23), Tp53, and EGFR (n = 32) in tumor DNA extracts. (A) No mutations were found in K-ras, N-ras, c-H-ras, or EGFR in any of the silica-induced tumors analyzed after microdissection. For Tp53, five mutations were found in three tumors of 32 cases analyzed (9%). (B) Electropherogram of the DNA sequence of the tumor (mt) versus the corresponding adjacent normal tissue (wt) showing the mutations found in exon 5 in AC and exon 6 in SCC.
Ras proto-oncogene family
We then aimed to determine how relevant is the status of the Ras family gene in the development of NSCLC in the rat silica model. First, we analyzed by sequencing the mutational status of exons 1 and 2 of K-ras (codons 6–37 and 44–97), N-ras (codons 1–23 and 48–74), and c-H-ras (codons 1–82) in 23 laser-microdissected tumors (15 AC and 8 SCC). We did not identify any mutation in the tumors (Figure 4A). These data strongly suggest the lack of genetic alterations in these Ras family genes in this chronic inflammation-driven lung cancer model.
EGFR
EGFR mutations and gene amplification, along with protein overexpression, are relatively frequent events in human lung cancer and have been the basis for new molecular targeted therapies [29]. We analyzed the EGFR mutational status and protein pattern expression in 32 NSCLC tumors (25 AC and 7 SCC). First, immunohistochemical analyses using a validated antibody specific to EGFR revealed that 92% of AC and 71% of SCC were positive for this receptor, with both membrane and cytoplasmic localizations (Figure W3). Strong or moderate staining for EGFR was found in 14 of 25 (55%) AC (Figure W3, A and B) and in 1 of 7 (14%) SCC, whereas only 2 of 25 (8%) AC (Figure W3C) and 2 of 7 (29%) SCC were completely negative. Next, we analyzed the DNA mutational status of EGFR through exons 18 to 21 in 32 microdissected tumors previously analyzed for protein expression. We did not find any gene mutation in any of the codons analyzed (Figure 4A). These data suggest that EGFR overexpression, but not mutation, may be a relevant biologic event in the progression of silica-induced tumors.
Epigenetic Alterations in Multistep Progression to Cancer Global DNA methylation status
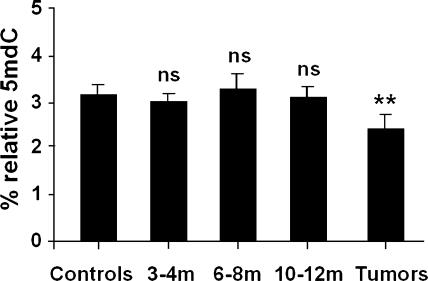
The epigenetic regulation of gene expression has been shown to be very frequent in lung carcinogenesis. As we have previously reported [15], silica-induced tumors show clear global genomic hypomethylation, with an average loss of 25% in mC DNA content. In this study, we extend this analysis to explore whether global hypomethylation was found in previous stages of the lung carcinogenesis process, when only preneoplastic lesions, but not tumors, are present. The global DNA methylation status in the samples obtained on months 3 to 4 (n = 5), months 6 to 8 (n = 5), and months 10 to 12 (n = 5), and in tumors obtained on months 16 to 21 (n = 9) showed that hypomethylation only occurs in tumors, but not in lung extracts containing preneoplastic tissues (Figure 5).
Figure 5.
DNA global hypomethylation analysis in controls (n = 6); preneoplasia-containing lungs on months 3 to 4 (n = 5), months 6 to 8 (n = 5), and months and 10 to 12 (n = 5) after treatment; and in tumors (n = 9). A clear global genomic hypomethylation with an average loss of 25% in mC DNA content only occurs in tumors. Results are expressed as mean ± S.D. Fisher's exact test was applied, and differences were considered significant when P < .05.
CpG island hypermethylation in p16(CDKN2A), APC, and CDH13 gene promoters
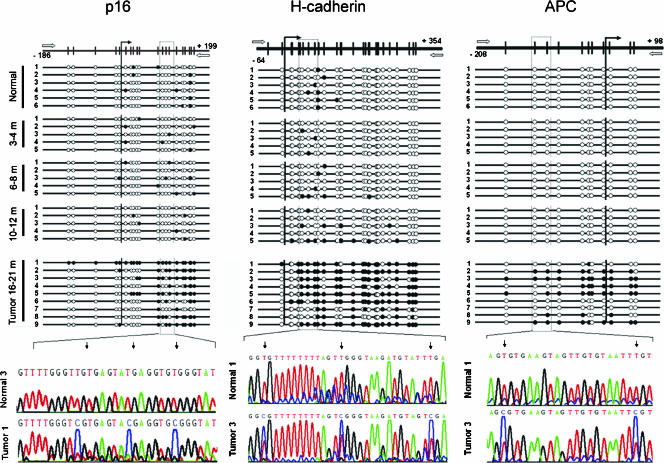
We determined the role of promoter hypermethylation in the transcriptional repression of p16(CDKN2A), APC, and H-cadherin (CDH13) genes, whose promoters are frequently hypermethylated in human lung cancer [30,31]. We analyzed preneoplasia-containing tissues obtained on months 3 to 4 (n = 5), months 6 to 8 (n = 5), and months 10 to 12 (n = 5), and tumors obtained on months 16 to 21 (five AC and four SCC).
All samples analyzed in the time course from months 3 to 12 were unmethylated. However, we observed a strong CpG island hypermethylation in seven of nine tumors in the promoter region of CDH13 (78%); in six of nine tumors in p16(CDKN2A) (67%); and in five of nine tumors in APC (56%) (Figure 6). We found concurrent promoter hypermethylation of all three genes analyzed in four of nine tumors. Furthermore, p16 promoter hypermethylation showed a strong inverse association, with the p16 protein expression measured by immunohistochemistry. Promoter hypermethylation was associated with low or negative p16 protein expression, whereas unmethylated cases showed p16 overexpression (Figures 6 and W4). These data suggest that epigenetic alterations are relevant biologic events in tumors generated in the context of the inflammatory insult.
Figure 6.
Bisulfite genomic sequencing of the p16(CDKN2A), H-cadherin (CDH13), and APC gene promoters in normal tissues (n = 6); preneoplasia-containing tissues on months 3 to 4 (n = 5), months 6 to 8 (n = 5), and months 10 to 12 (n = 5); and nine representative lung tumors. Vertical bars show the distribution of CpG islands at p16, H-cadherin, and APC. The vertical arrow indicates the transcriptional start point. Black dots indicate methylated CpG islands; white dots indicate unmethylated CpG islands. The position of the bisulfite sequencing primers is represented with white horizontal arrows. We observed H-cadherin promoter hypermethylation in seven of nine silica-induced tumors (T1, T2, T3, T4, T5, T6, and T9), and APC promoter hypermethylation in five of nine tumors (T2, T3, T4, T5, and T9). Hypermethylation in the p16 promoter was also an important event in six of nine tumors studied (T1, T2, T3, T5, T8, and T9).
CpG island hypermethylation in Rassf1 and Nore1A gene promoters. We next tested whether the Ras pathway could be activated by promoter hypermethylation of Ras effectors, as has been demonstrated for Rassf1 [32] or Nore1A [33] in human lung cancer. For this approach, we studied the methylation status of the promoters of the rat homologues for Ras effectors Rassf1 and Nore1A in the silica-induced tumors. We did not observe epigenetic alterations in the promoters of these two Ras effectors (Figure W5). These data, together with the mutational Ras status, strongly suggest the lack of genetic and epigenetic alterations in the Ras family members in this inflammation-mediated lung cancer model.
Discussion
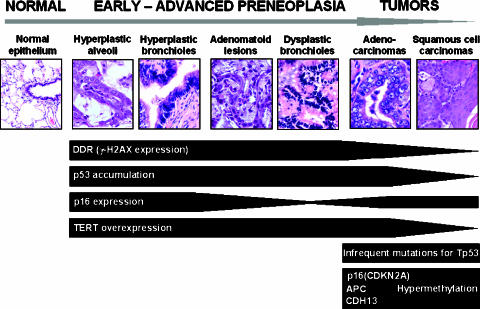
In this report, we have characterized some of the key molecular alterations leading to lung cancer in the context of chronic inflammation (Figure 7). Using a rat silica model of inflammation-induced lung cancer [15], our results showed an activation of the DDR pathway in preneoplastic lesions, in association with proliferative and telomeric stress. Surprisingly, we found a very low frequency of mutations in the Tp53 gene and a total absence of mutations in the main Ras signaling family genes and EGFR. In contrast, epigenetic events in tumor suppressors, including p16, APC, and CDH13, were found with high frequencies in this lung cancer model.
Figure 7.
Summary of the molecular events found in the silica-induced lung carcinogenesis model. DDR, p16, TERT, and p53 overexpressions are observed from early preneoplastic lesions. p16 inactivation occurs in dysplastic bronchiolar lesions and tumors. No mutations were observed in tumors, with the only exception of Tp53 mutations with a very low frequency. Epigenetic alterations were found in tumors in the tumor-suppressor genes p16(CDKN2A), APC, and CDH13.
Our data show that p16 loss is significantly associated with inflammation-induced lung carcinogenesis. In the silica-induced lung cancer model, downregulation of p16 in late preneoplastic lesions could be the landmark for the escape of some epithelial cells from the tight growth-regulatory mechanisms found in preneoplasia to loosely regulated tumorigenic pathways. In fact, in vitro studies of lung cells have already suggested that the loss of p16 expression, rather than the alteration of the ARF/p53 pathway, is the key event in bypassing growth arrest and in acquiring a transformed phenotype with genetic instability and tumoral potential [34]. Accordingly, a significant decrease in p16 protein was also observed in 40% of silica-induced tumors, and p16 protein downregulation correlated with gene promoter hypermethylation. Some, but not all, lung cancer experimental models show p16 promoter hypermethylation [35,36]. Belinsky et al. [11] have shown that tumors produced by instillation of carbon black particles in rats arise, in part, through the epigenetic inactivation of p16, suggesting that exposure to some particular carcinogens may be associated with specific gene inactivation through methylation [37]. More importantly, loss of p16 expression was also found in human lung carcinogenesis, starting at the moderate dysplasia stage [38]. Moreover, p16 loss in preneoplastic lesions occurred exclusively in patients who also showed loss of p16 expression in their related invasive carcinoma [38]. The inactivation of p16 has been reported in approximately 50% of human NSCLC, and this inactivation is frequently associated with p16 promoter hypermethylation [39].
Other classic oncogenic events present in human NSCLC were found at low frequency or were absent in this model. The incidence of Tp53 mutations observed in silica-induced tumors was significantly lower (9%) than the incidence reported for NSCLC in human smokers (50%) and non-smokers (28%) [40]. Studies conducted by several groups have consistently reported a very low frequency or a complete lack of mutations at the Tp53 gene locus in rodent lung tumors [41,42]. The low prevalence of Tp53 mutations in murine versus human lung tumors is striking because the gene is highly conserved and the tumors' morphologic features are very similar. In contrast to Tp53, Ras mutations have been frequently found in chemically induced murine lung tumors and have been strongly correlated with promutagenic adducts generated from the metabolism of chemical carcinogens present in tobacco [43]. In the silica model, mutations in K-ras, N-ras, or H-Ras, including codons 12, 13, and 61 (which are known to transform ras into an oncogenic protein in both human and rodent cells), were completely absent. Ras mutations are also infrequent in other rat lung tumor models induced by particulate carcinogens (diesel exhaust and carbon black) [41]. The lack of mutations in the silica model supports the current view that the existence of Ras mutations is highly dependent on the type of carcinogen. This view is further supported by the lack of mutations in codon 12 of K-ras in lung cancer found in workers with silicosis [44]. Moreover, promoter hypermethylation of Ras effectors Nore1A and Rassf1 present in humans [32,33] was not found in the silica-induced tumors. These data strongly support the view that inflammation-driven carcinogenesis is not mediated by genetic alterations in the Ras pathway. Silica-mediated chronic inflammation may also activate the EGFR pathway. EGFR is expressed in a range from 40% to 70% of human NSCLC [45]. We have observed EGFR expression in 87% of silica-induced tumors, which could reflect the autocrine or paracrine stimulation of tumor growth by several EGFR ligands. NSCLC patients who show clinical responses to EGFR tyrosine kinase inhibitors frequently harbor somatic mutations in the EGFR gene. These EGFR mutations are more commonly found in females, nonsmokers, and AC with bronchioloalveolar features [29]. Moreover, EGFR and Ras mutations seem to be mutually exclusive in NSCLC [46]. We thus looked for EGFR mutations in DNA from microdissected inflammation-induced tumors. Although a great proportion of the tumors arising in the rat silica model are AC with bronchoalveolar-like features, we did not find EGFR mutations in any of the cases studied, suggesting that mutations in Ras or EGFR pathways are not involved in this inflammation-driven lung carcinogenesis model.
Tobacco and nontobacco-related human lung cancers harbor different combinations of mutations and gene promoter methylation events [47]. One of the most interesting findings of this study was the wealth of epigenetic events. Previously, chronic inflammation had been associated with high levels of methylation in ulcerative colitis and chronic gastritis [48,49]. However, a direct causal relationship of promoter hypermethylation has not yet been unequivocally shown in lung tumors. In our study, aberrant methylation events have been detected in the p16(CDKN2A), APC, and CDH13 promoters in the silica-induced tumors but were not present in lung tissues containing preneoplastic lesions. This is the first description of APC gene promoter hypermethylation in a lung cancer animal model. Epigenetic alteration of CDH13 has recently been described in mouse lung tumor models [50]. In human NSCLC, aberrant promoter hypermethylation for these tumor-suppressor genes has been reported by several independent studies [30,31]. An association between CDH13 methylation and tumor progression in human NSCLC has also been suggested [51]. Previously, we have shown a strong epigenetic alteration of E-cadherin and β-catenin accumulation in the context of the epithelial-mesenchymal transition in this model [15]. One could speculate that the loss of function of APC and CDH13 through gene promoter hypermethylation may disrupt cells' ability to regulate adhesion; in turn, APC loss may also activate the Wnt signaling pathway through stabilization of the β-catenin protein [52]. Deregulation of β-catenin causes cells to remain in a less differentiated and more proliferative state [53]. These findings suggest that simultaneous epigenetic inactivation of several putative tumor-suppressor genes (p16, APC, E-cadherin, and CDH13) may have a strong tumorigenic effect. Nevertheless, mutations of other potential oncogenes not analyzed in this study or loss of other putative tumor-suppressor genes cannot be ruled out.
The current view about the acquisition of a tumorigenic phenotype and progression through carcinogenesis describes a strong association between DNA damage events, genomic instability, and selective genomic pressure. Recent studies in several human tumor types have shown that the DNA damage checkpoint was activated in epithelial early preneoplastic lesions either by DNA replication stress or by oncogenic stress [20,21,54]. According to these studies, DDR acts as a tumorigenesis barrier inducing cell cycle arrest, senescence, or apoptosis, thus protecting from progression to malignancy. In the present study, we found an association between the presence of local inflammatory mediators and the activation of the DDR pathway. It is well-known that silica exposure rapidly induces inflammatory mediators in the lung, which results in oxidative stress and DNA alterations, including single-strand breaks, mutagenic base modification by 8-oxoguanine, and various other DNA oxidation products [55,56]. Oxidative stress elicited by crystalline silica was associated with a remarkable increase in the expression of the inflammatory mediator iNOS in the rat airways, in agreement with previous studies [17,57,58]. The reactions to silica particles in the lungs stimulate the activation of a number of molecular mediators, including those that are primarily involved in inflammatory and fibrogenic responses and their immunological aspects, such as interleukins and TNF-α. Silica particles, either by direct surface effects or during phagocytosis, generate ROS, which seem to trigger critical signaling events for both NF-κB and AP-1 activation [16,59]. The pathogenic model proposed for asbestos-induced mesothelioma is similar to this proposal for silica-induced tumors. In both cases, mineral dust/fiber causes accumulation of cytokine-releasing (especially TNF-α) and NO-releasing active macrophages [16,60,61]. A recent report has also suggested that NF-κB-dependent inflammation, combined with asbestos-induced genetic instability, may be a potential cause of mesothelial carcinogenesis [61]. The current hypothesis is that, in iNOS-expressing epithelial cells, chronic exposure to nitrogen and oxygen radicals may induce (nitro)oxidative stress, leading to DNA damage and activation of a p53 response [22,62]. In the lung epithelial lesions found in the rat silica model, iNOS was commonly coexpressed with γ-H2AX and was associated with p53 accumulation. Moreover, this coexpression was more frequently observed in preneoplastic tissues than in tumors, suggesting the relevance of DDR in epithelial cells during chronic inflammation. DNA damage and other cellular stresses can induce the expression of p53 and p16, enforcing cell cycle arrest [22,63].We observed a clear induction of p53 and p16 expression occurring in parallel with chronic inflammatory stress and the replicative activation of hyperplastic lesions. Activated p53 inhibits cell proliferation and protects against genomic instability by controlling cell cycle checkpoints, DNA repair, apoptosis, and senescence [64]. Overexpression of p53 and p16 in hyperplastic lesions may depend on the activation of the DDR and may mutually reinforce the barrier toward tumorigenesis. In contrast, in most advanced preneoplastic lesions, p53 expression remains high, whereas a very significant decrease of p16 occurs. This further emphasizes the potential role of p16 loss as a key molecular event.
Besides oxidative stress, the activation of DDR and cell cycle arrest could also be mediated by telomere shortening due to replicative stress [23]. TERTactivity contributes to the elongation of shortening telomeres in preneoplastic and tumor cells escaping from telomere crisis mediated by critically short telomeres [65]. Previous studies in breast and lung tumors have also shown an increase in telomerase expression in preneoplastic lesions [65,66]. Our data are in accordance with previous studies on rodents where telomerase is detectable in normal tissues and significantly increased in preneoplastic and neoplastic cells [67,68]. We observed a clear induction of TERT expression in the silica-induced multistep progression that was more evident in preneoplastic tissues than in tumoral tissues. The overexpression of telomerase, together with inactivation of the p16-Rb pathway, could bypass DDR-mediated cell cycle arrest and extend the lifespan of tissues. Furthermore, telomerase overexpression may affect the proliferation of epithelial cells not only by stabilizing telomeres but also by affecting the expression of growth-promoting genes. Smith et al. [69] reported that activation of telomerase in p16-inactivated tumoral cells altered the expression of a set of genes that have a profound effect on cell proliferation, including EGFR, which is indeed overexpressed in this model.
In summary, we have explored the specific molecular profile of chronic inflammation-mediated neoplastic transformation of the lung. Our data suggest that induction of iNOS expression in the inflammatory context is associated with activation of the DDR and p53 accumulation in preneoplastic epithelial cells. p16 is also induced in the early steps of tumorigenic progression, followed by a clear loss of expression in late dysplastic bronchiolar lesions. Among the recently reported barriers to preneoplasia-to-neoplasia transition, p16 seems to be the one bypassed in this inflammation-driven model, very likely as a consequence of a strong epigenetic regulation of p16 gene expression. TERT and EGFR are also overexpressed in this model. Lack of mutations of Ras signaling components and EGFR, and a very low frequency for Tp53 suggest that they are not major players in inflammation-driven carcinogenesis, whereas epigenetic alterations in tumor-suppressor genes such as p16(CDKN2A), CDH13, and APC are more relevant. Our results suggest that chronic inflammation-induced tumors have a specific molecular signature that shares some, but not all, of the alterations found in chemically induced lung cancer.
Supplementary Material
Acknowledgements
The authors want to express their special gratitude to Umberto Saffiotti (National Cancer Institute, National Institutes of Health) for his invaluable help in establishing the model at the start of this project, and for his constant advice, encouragement, and suggestions. We thank Paz Zamora, Elena Remirez, and Gabriel de Biurrun for technical assistance.
Abbreviations
- AC
adenocarcinoma
- DDR
DNA damage response
- EGFR
epidermal growth factor receptor
- γ-H2AX
Ser139-phosphorylated histone 2AX
- iNOS
inducible nitric oxide synthase
- mC
5-methylcytosine
- NSCLC
non-small cell lung cancer
- ROS
reactive oxygen species
- SCC
squamous cell carcinoma
- TERT
catalytic subunit of telomerase reverse transcriptase
Footnotes
This article refers to supplementary material, which is designated by “W” (i.e., Table W1, Figure W1) and is available online at www.bcdecker.com.
This work was funded through the “UTE project CIMA” and the Spanish Ministry of Health (ISCIII: RTICC, FIS-PI 04/2128). S.V. is a postdoctoral fellow of the “Jose y Ana Royo” Foundation. C.O. was supported by the Spanish Ministry of Science and Education (MCYT TEC2005-04732), the EU Marie Curie Program (MIRG-CT-2005-028342), and a Ramón y Cajal Fellowship. F.L. was supported by “Fundación Mutua Madrileña,” ISCIII-RTICC RD06/0020, PI042282, and the Spanish Ministry of Industry, Tourism, and Commerce (FIT-090100-2005-46).
References
- 1.Sato M, Shames DS, Gazdar AF, Minna JD. A translational view of the molecular pathogenesis of lung cancer. J Thorac Oncol. 2007;2:327–343. doi: 10.1097/01.JTO.0000263718.69320.4c. [DOI] [PubMed] [Google Scholar]
- 2.Hecht SS. Tobacco carcinogens, their biomarkers and tobacco-induced cancer. Nat Rev Cancer. 2003;3:733–744. doi: 10.1038/nrc1190. [DOI] [PubMed] [Google Scholar]
- 3.Hammond SK, Sorensen G, Youngstrom R, Ockene JK. Occupational exposure to environmental tobacco smoke. JAMA. 1995;274:956–960. [PubMed] [Google Scholar]
- 4.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dalgleish A, Haefner B, editors. Wounds That Do Not Heal. New York, NY.: Springer; 2006. The Link between Inflammation and Cancer. [Google Scholar]
- 6.Jackson AL, Loeb LA. The contribution of endogenous sources of DNA damage to the multiple mutations in cancer. Mutat Res. 2001;477:7–21. doi: 10.1016/s0027-5107(01)00091-4. [DOI] [PubMed] [Google Scholar]
- 7.Schottenfeld D, Beebe-Dimmer J. Chronic inflammation: a common and important factor in the pathogenesis of neoplasia. CA Cancer J Clin. 2006;56:69–83. doi: 10.3322/canjclin.56.2.69. [DOI] [PubMed] [Google Scholar]
- 8.Balkwill F, Mantovani A. Inflammation and cancer: back to Virchow? Lancet. 2001;357:539–545. doi: 10.1016/S0140-6736(00)04046-0. [DOI] [PubMed] [Google Scholar]
- 9.Benhar M, Engelberg D, Levitzki A. ROS, stress-activated kinases and stress signaling in cancer. EMBO Rep. 2002;3:420–425. doi: 10.1093/embo-reports/kvf094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malkinson AM. Role of inflammation in mouse lung tumorigenesis: a review. Exp Lung Res. 2005;31:57–82. doi: 10.1080/01902140490495020. [DOI] [PubMed] [Google Scholar]
- 11.Belinsky SA, Snow SS, Nikula KJ, Finch GL, Tellez CS, Palmisano WA. Aberrant CpG island methylation of the p16(INK4a) and estrogen receptor genes in rat lung tumors induced by particulate carcinogens. Carcinogenesis. 2002;23:335–339. doi: 10.1093/carcin/23.2.335. [DOI] [PubMed] [Google Scholar]
- 12.Bauer AK, Dixon D, DeGraff LM, Cho HY, Walker CR, Malkinson AM, Kleeberger SR. Toll-like receptor 4 in butylated hydroxytoluene-induced mouse pulmonary inflammation and tumorigenesis. J Natl Cancer Inst. 2005;97:1778–1781. doi: 10.1093/jnci/dji403. [DOI] [PubMed] [Google Scholar]
- 13.Altomare DA, Vaslet CA, Skele KL, De Rienzo A, Devarajan K, Jhanwar SC, McClatchey AI, Kane AB, Testa JR. A mouse model recapitulating molecular features of human mesothelioma. Cancer Res. 2005;65:8090–8095. doi: 10.1158/0008-5472.CAN-05-2312. [DOI] [PubMed] [Google Scholar]
- 14.Saffiotti U, Williams A, Daniel L, Kaighn M, Mao Y, Shi X. In: Carcinogenesis by Crystalline Silica: Animal, Cellular and Molecular Studies. Castranova V, Vallyathan V, Wallace WE, editors. Boca Raton, FL: CRC Press; 1996. pp. 345–381. [Google Scholar]
- 15.Blanco D, Vicent S, Elizegi E, Pino I, Fraga MF, Esteller M, Saffiotti U, Lecanda F, Montuenga LM. Altered expression of adhesion molecules and epithelial-mesenchymal transition in silica-induced rat lung carcinogenesis. Lab Invest. 2004;84:999–1012. doi: 10.1038/labinvest.3700129. [DOI] [PubMed] [Google Scholar]
- 16.Castranova V, Porter D, Millecchia L, Ma JY, Hubbs AF, Teass A. Effect of inhaled crystalline silica in a rat model: time course of pulmonary reactions. Mol Cell Biochem. 2002;234–235:177–184. [PubMed] [Google Scholar]
- 17.Porter DW, Millecchia LL, Willard P, Robinson VA, Ramsey D, McLaurin J, Khan A, Brumbaugh K, Beighley CM, Teass A, et al. Nitric oxide and reactive oxygen species production causes progressive damage in rats after cessation of silica inhalation. Toxicol Sci. 2006;90:188–197. doi: 10.1093/toxsci/kfj075. [DOI] [PubMed] [Google Scholar]
- 18.Ding M, Huang C, Lu Y, Bowman L, Castranova V, Vallyathan V. Involvement of protein kinase C in crystalline silica-induced activation of the MAP kinase and AP-1 pathway. Am J Physiol Lung Cell Mol Physiol. 2006;290:L291–L297. doi: 10.1152/ajplung.00053.2005. [DOI] [PubMed] [Google Scholar]
- 19.Williams AO, Flanders KC, Saffiotti U. Immunohistochemical localization of transforming growth factor-beta 1 in rats with experimental silicosis, alveolar type II hyperplasia, and lung cancer. Am J Pathol. 1993;142:1831–1840. [PMC free article] [PubMed] [Google Scholar]
- 20.Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, Zoumpourlis VC, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- 21.Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio RA, Jr, Kastrinakis NG, Levy B, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 22.Helton ES, Chen X. p53 modulation of the DNA damage response. J Cell Biochem. 2007;100:883–896. doi: 10.1002/jcb.21091. [DOI] [PubMed] [Google Scholar]
- 23.Jacobs JJ, de Lange T. p16INK4a as a second effector of the telomere damage pathway. Cell Cycle. 2005;4:1364–1368. doi: 10.4161/cc.4.10.2104. [DOI] [PubMed] [Google Scholar]
- 24.Fraga MF, Uriol E, Borja Diego L, Berdasco M, Esteller M, Canal MJ, Rodriguez R. High-performance capillary electrophoretic method for the quantification of 5-methyl 2′-deoxycytidine in genomic DNA: application to plant, animal and human cancer tissues. Electrophoresis. 2002;23:1677–1681. doi: 10.1002/1522-2683(200206)23:11<1677::AID-ELPS1677>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 25.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang YC, Lin RK, Tan YH, Chen JT, Chen CY. Wild-type p53 overexpression and its correlation with MDM2 and p14ARF alterations: an alternative pathway to non-small-cell lung cancer. J Clin Oncol. 2005;23:154–164. doi: 10.1200/JCO.2005.03.139. [DOI] [PubMed] [Google Scholar]
- 27.Ohno J, Horio Y, Sekido Y, Hasegawa Y, Takahashi M, Nishizawa J, Saito H, Ishikawa F, Shimokata K. Telomerase activation and p53 mutations in urethane-induced A/J mouse lung tumor development. Carcinogenesis. 2001;22:751–756. doi: 10.1093/carcin/22.5.751. [DOI] [PubMed] [Google Scholar]
- 28.Hegi ME, Soderkvist P, Foley JF, Schoonhoven R, Swenberg JA, Kari F, Maronpot R, Anderson MW, Wiseman RW. Characterization of p53 mutations in methylene chloride-induced lung tumors from B6C3F1 mice. Carcinogenesis. 1993;14:803–810. doi: 10.1093/carcin/14.5.803. [DOI] [PubMed] [Google Scholar]
- 29.Mitsudomi T, Kosaka T, Yatabe Y. Biological and clinical implications of EGFR mutations in lung cancer. Int J Clin Oncol. 2006;11:190–198. doi: 10.1007/s10147-006-0583-4. [DOI] [PubMed] [Google Scholar]
- 30.Shivapurkar N, Stastny V, Suzuki M, Wistuba II, Li L, Zheng Y, Feng Z, Hol B, Prinsen C, Thunnissen FB, et al. Application of a methylation gene panel by quantitative PCR for lung cancers. Cancer Lett. 2007;247:56–71. doi: 10.1016/j.canlet.2006.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsou JA, Hagen JA, Carpenter CL, Laird-Offringa IA. DNA methylation analysis: a powerful new tool for lung cancer diagnosis. Oncogene. 2002;21:5450–5461. doi: 10.1038/sj.onc.1205605. [DOI] [PubMed] [Google Scholar]
- 32.Kim DH, Kim JS, Park JH, Lee SK, Ji YI, Kwon YM, Shim YM, Han J, Park J. Relationship of Ras association domain family 1 methylation and K-ras mutation in primary non-small cell lung cancer. Cancer Res. 2003;63:6206–6211. [PubMed] [Google Scholar]
- 33.Irimia M, Fraga MF, Sanchez-Cespedes M, Esteller M. CpG island promoter hypermethylation of the Ras-effector gene NORE1A occurs in the context of a wild-type K-ras in lung cancer. Oncogene. 2004;23:8695–8699. doi: 10.1038/sj.onc.1207914. [DOI] [PubMed] [Google Scholar]
- 34.Petitot F, Lebeau J, Dano L, Lectard B, Altmeyer S, Levalois C, Chevillard S. In vitro aging of rat lung cells. Downregulation of telomerase activity and continuous decrease of telomere length are not incompatible with malignant transformation. Exp Cell Res. 2003;286:30–39. doi: 10.1016/s0014-4827(03)00103-4. [DOI] [PubMed] [Google Scholar]
- 35.Belinsky SA, Nikula KJ, Palmisano WA, Michels R, Saccomanno G, Gabrielson E, Baylin SB, Herman JG. Aberrant methylation of p16(INK4a) is an early event in lung cancer and a potential biomarker for early diagnosis. Proc Natl Acad Sci USA. 1998;95:11891–11896. doi: 10.1073/pnas.95.20.11891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patel AC, Anna CH, Foley JF, Stockton PS, Tyson FL, Barrett JC, Devereux TR. Hypermethylation of the p16(Ink4a) promoter in B6C3F1 mouse primary lung adenocarcinomas and mouse lung cell lines. Carcinogenesis. 2000;21:1691–1700. doi: 10.1093/carcin/21.9.1691. [DOI] [PubMed] [Google Scholar]
- 37.Belinsky SA. Silencing of genes by promoter hypermethylation: key event in rodent and human lung cancer. Carcinogenesis. 2005;26:1481–1487. doi: 10.1093/carcin/bgi020. [DOI] [PubMed] [Google Scholar]
- 38.Brambilla E, Gazzeri S, Moro D, Lantuejoul S, Veyrenc S, Brambilla C. Alterations of Rb pathway (Rb-p16INK4-cyclin D1) in pre-invasive bronchial lesions. Clin Cancer Res. 1999;5:243–250. [PubMed] [Google Scholar]
- 39.Zhu CQ, Shih W, Ling CH, Tsao MS. Immunohistochemical markers of prognosis in non-small cell lung cancer: a review and proposal for a multiphase approach to marker evaluation. J Clin Pathol. 2006;59:790–800. doi: 10.1136/jcp.2005.031351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Husgafvel-Pursiainen K, Kannio A. Cigarette smoking and p53 mutations in lung cancer and bladder cancer. Environ Health Perspect. 1996;104(Suppl 3):553–556. doi: 10.1289/ehp.96104s3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swafford DS, Nikula KJ, Mitchell CE, Belinsky SA. Low frequency of alterations in p53, K-ras, and mdm2 in rat lung neoplasms induced by diesel exhaust or carbon black. Carcinogenesis. 1995;16:1215–1221. doi: 10.1093/carcin/16.5.1215. [DOI] [PubMed] [Google Scholar]
- 42.Kitada H, Tsutsumi M, Tsujiuchi T, Takahama M, Fukuda T, Narita N, Konsihi Y. Frequent mutations of Ki-ras but no mutations of Ha-ras and p53 in lung lesions induced by N-nitrosobis(2-hydroxypropyl)-amine in rats. Mol Carcinog. 1996;15:276–283. doi: 10.1002/(SICI)1098-2744(199604)15:4<276::AID-MC5>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 43.Ross JA, Nesnow S. Polycyclic aromatic hydrocarbons: correlations between DNA adducts and ras oncogene mutations. Mutat Res. 1999;424:155–166. doi: 10.1016/s0027-5107(99)00016-0. [DOI] [PubMed] [Google Scholar]
- 44.Liu B, Guan R, Zhou P, Miao Q, Wang H, Fu D, You B. A distinct mutational spectrum of p53 and K-ras genes in lung cancer of workers with silicosis. J Environ Pathol Toxicol Oncol. 2000;19:1–7. [PubMed] [Google Scholar]
- 45.Nakamura H, Kawasaki N, Taguchi M, Kabasawa K. Survival impact of epidermal growth factor receptor overexpression in patients with non-small cell lung cancer: a meta-analysis. Thorax. 2006;61:140–145. doi: 10.1136/thx.2005.042275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tam IY, Chung LP, Suen WS, Wang E, Wong MC, Ho KK, Lam WK, Chiu SW, Girard L, Minna JD, et al. Distinct epidermal growth factor receptor and KRAS mutation patterns in non-small cell lung cancer patients with different tobacco exposure and clinicopathologic features. Clin Cancer Res. 2006;12:1647–1653. doi: 10.1158/1078-0432.CCR-05-1981. [DOI] [PubMed] [Google Scholar]
- 47.Toyooka S, Tokumo M, Shigematsu H, Matsuo K, Asano H, Tomii K, Ichihara S, Suzuki M, Aoe M, Date H, et al. Mutational and epigenetic evidence for independent pathways for lung adenocarcinomas arising in smokers and never smokers. Cancer Res. 2006;66:1371–1375. doi: 10.1158/0008-5472.CAN-05-2625. [DOI] [PubMed] [Google Scholar]
- 48.Issa JP, Ahuja N, Toyota M, Bronner MP, Brentnall TA. Accelerated age-related CpG island methylation in ulcerative colitis. Cancer Res. 2001;61:3573–3577. [PubMed] [Google Scholar]
- 49.Kang GH, Lee HJ, Hwang KS, Lee S, Kim JH, Kim JS. Aberrant CpG island hypermethylation of chronic gastritis, in relation to aging, gender, intestinal metaplasia, and chronic inflammation. Am J Pathol. 2003;163:1551–1556. doi: 10.1016/S0002-9440(10)63511-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vuillemenot BR, Hutt JA, Belinsky SA. Gene promoter hypermethylation in mouse lung tumors. Mol Cancer Res. 2006;4:267–273. doi: 10.1158/1541-7786.MCR-05-0218. [DOI] [PubMed] [Google Scholar]
- 51.Kim JS, Han J, Shim YM, Park J, Kim DH. Aberrant methylation of H-cadherin (CDH13) promoter is associated with tumor progression in primary nonsmall cell lung carcinoma. Cancer. 2005;104:1825–1833. doi: 10.1002/cncr.21409. [DOI] [PubMed] [Google Scholar]
- 52.Willert K, Jones KA. Wnt signaling: is the party in the nucleus? Genes Dev. 2006;20:1394–1404. doi: 10.1101/gad.1424006. [DOI] [PubMed] [Google Scholar]
- 53.Nathke I. Cytoskeleton out of the cupboard: colon cancer and cytoskeletal changes induced by loss of APC. Nat Rev Cancer. 2006;6:967–974. doi: 10.1038/nrc2010. [DOI] [PubMed] [Google Scholar]
- 54.Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 55.Nehls P, Seiler F, Rehn B, Greferath R, Bruch J. Formation and persistence of 8-oxoguanine in rat lung cells as an important determinant for tumor formation following particle exposure. Environ Health Perspect. 1997;105(Suppl 5):1291–1296. doi: 10.1289/ehp.97105s51291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Seiler F, Rehn B, Rehn S, Bruchs J. Significant differences in the cellular and molecular reactions of rat and hamster lung after quartz exposure. Toxicol Lett. 2001;119:11–19. doi: 10.1016/s0378-4274(00)00289-7. [DOI] [PubMed] [Google Scholar]
- 57.Porter DW, Millecchia L, Robinson VA, Hubbs A, Willard P, Pack D, Ramsey D, McLaurin J, Khan A, Landsittel D, et al. Enhanced nitric oxide and reactive oxygen species production and damage after inhalation of silica. Am J Physiol Lung Cell Mol Physiol. 2002;283:L485–L493. doi: 10.1152/ajplung.00427.2001. [DOI] [PubMed] [Google Scholar]
- 58.Blackford JA, Jr, Antonini JM, Castranova V, Dey RD. Intratracheal instillation of silica up-regulates inducible nitric oxide synthase gene expression and increases nitric oxide production in alveolar macrophages and neutrophils. Am J Respir Cell Mol Biol. 1994;11:426–431. doi: 10.1165/ajrcmb.11.4.7522485. [DOI] [PubMed] [Google Scholar]
- 59.Castranova V. Signaling pathways controlling the production of inflammatory mediators in response to crystalline silica exposure: role of reactive oxygen/nitrogen species. Free Radic Biol Med. 2004;37:916–925. doi: 10.1016/j.freeradbiomed.2004.05.032. [DOI] [PubMed] [Google Scholar]
- 60.Dorger M, Allmeling AM, Kiefmann R, Munzing S, Messmer K, Krombach F. Early inflammatory response to asbestos exposure in rat and hamster lungs: role of inducible nitric oxide synthase. Toxicol Appl Pharmacol. 2002;181:93–105. doi: 10.1006/taap.2002.9388. [DOI] [PubMed] [Google Scholar]
- 61.Yang H, Bocchetta M, Kroczynska B, Elmishad AG, Chen Y, Liu Z, Bubici C, Mossman BT, Pass HI, Testa JR, et al. TNF-alpha inhibits asbestos-induced cytotoxicity via a NF-kappaB-dependent pathway, a possible mechanism for asbestos-induced oncogenesis. Proc Natl Acad Sci USA. 2006;103:10397–10402. doi: 10.1073/pnas.0604008103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Achanta G, Huang P. Role of p53 in sensing oxidative DNA damage in response to reactive oxygen species-generating agents. Cancer Res. 2004;64:6233–6239. doi: 10.1158/0008-5472.CAN-04-0494. [DOI] [PubMed] [Google Scholar]
- 63.Shapiro GI, Edwards CD, Ewen ME, Rollins BJ. p16INK4a participates in a G1 arrest checkpoint in response to DNA damage. Mol Cell Biol. 1998;18:378–387. doi: 10.1128/mcb.18.1.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 65.Blasco MA, Hahn WC. Evolving views of telomerase and cancer. Trends Cell Biol. 2003;13:289–294. doi: 10.1016/s0962-8924(03)00085-0. [DOI] [PubMed] [Google Scholar]
- 66.Lantuejoul S, Soria JC, Moro-Sibilot D, Morat L, Veyrenc S, Lorimier P, Brichon PY, Sabatier L, Brambilla C, Brambilla E. Differential expression of telomerase reverse transcriptase (hTERT) in lung tumours. Br J Cancer. 2004;90:1222–1229. doi: 10.1038/sj.bjc.6601643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Blasco MA. Mice with bad ends: mouse models for the study of telomeres and telomerase in cancer and aging. EMBO J. 2005;24:1095–1103. doi: 10.1038/sj.emboj.7600598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chadeneau C, Siegel P, Harley CB, Muller WJ, Bacchetti S. Telomerase activity in normal and malignant murine tissues. Oncogene. 1995;11:893–898. [PubMed] [Google Scholar]
- 69.Smith LL, Coller HA, Roberts JM. Telomerase modulates expression of growth-controlling genes and enhances cell proliferation. Nat Cell Biol. 2003;5:474–479. doi: 10.1038/ncb985. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.