Abstract
Background
A defect in hypothalamic-pituitary-adrenal (HPA) axis function has been suggested to contribute to susceptibility to rheumatoid arthritis (RA).
Objective
To investigate polymorphisms of the glucocorticoid receptor (GR) gene and determine any associations with RA.
Methods
Three GR polymorphisms that tag 95% of all haplotypes across the GR gene were genotyped. These are an intron B Bcl1 polymorphism, a ttg insertion/deletion within intron F (rs2307674) and the single nucleotide polymorphism (SNP) lying in the 3′ untranslated region of exon 9b (rs6198). The dye terminator-based SNaPshot method or size resolution by capillary electrophoresis was performed. The study population comprised 198 UK Caucasian RA cases and 393 ethnically matched controls.
Results
No significant single point or haplotypic associations were found for GR polymorphisms with RA susceptibility. Furthermore, no evidence for GR polymorphisms with aspects of RA severity was seen.
Conclusion
In this study of the most comprehensive coverage of GR polymorphisms with RA, no significant contributing role for GR polymorphisms with RA was found.
Introduction
Studies from both animals and humans suggest that a defect in the neuroendocrine system may be important in the disease rheumatoid arthritis (RA). Inappropriately normal plasma cortisol and a blunted response of plasma cortisol to surgical stress have been shown for RA patients.1,2 Glucocorticoids (Gcs) are required for the development of streptococcal cell wall-induced arthritis in Lewis rats.3 Furthermore, while many RA patients respond well to exogenous Gc treatment, a proportion fail to do so.4 These observations suggest that hypothalamic-pituitary-adrenal (HPA) axis dysregulation or relative Gc deficiency contributes to the onset of RA.
Gcs exert their effect by binding to the intracellular receptor, the glucocorticoid receptor alpha (GRα). The human GR gene (GCCR, GCR, GRL, NR3C1) (locus 5q31) is 10 exons in length and exons 1-9α are transcribed into GRα mRNA, which is translated into a functional receptor (GRα). Alternative splicing of the primary transcript results in an mRNA containing exons 1-9β, which gives rise to the GRβ. Unlike GRα, GRβ does not bind hormone and is transcriptionally inactive.5,6 Although a dominant negative effect of GRβ on GRα activity has been known for some time, the mechanism by which this occurs is not known and the precise functional significance of GRβ remains unclear. Several associations with GRβ overexpression and autoimmune/inflammatory diseases have been described. It is not known, however, whether this is a cause or consequence of the inflammatory state and whether the increase in the GRβ is sufficient to exert a dominant negative effect on GRα. It has been suggested that GRβ could in part mediate Gc resistance seen in RA.7
Derijk et al. described an A to G polymorphism at position 3736 in exon 9b (now rs6198).8 This polymorphism lies in an AUUUA motif of the 3′ UTR of the mRNA of the GRβ isoform and has been shown to increase mRNA stability and also receptor protein expression.9
Derijk et al. originally detailed the genotype and allele frequencies of the rs6198 polymorphism in a very small sample size of 30 RA cases compared with 24 controls and found carriage of the mutant allele to be associated with significant increased risk of RA.8 Here we report our attempt to replicate and extend these findings. We have previously demonstrated strong linkage disequilibrium across the GR gene locus and suggested haplotype tagging single nucleotide polymorphisms (SNPs).10 In this current study we looked at three polymorphisms that in combination capture 95% of all haplotypes occurring at a frequency of greater than 1% in Caucasians (Fig. 1). These are an intron B Bcl1 polymorphism, which has been associated with increased Gc sensitivity and a lower body mass index,11 a ttg insertion/deletion within intron F (rs2307674) and the SNP lying in the 3′ untranslated region of exon 9b (rs6198). We studied these in a well-characterized panel of RA patients and healthy controls to determine whether polymorphisms of GR contribute to RA susceptibility. Utilizing this combination of SNPs allows for the most comprehensive investigation of GR polymorphisms in RA that has been carried out to date.
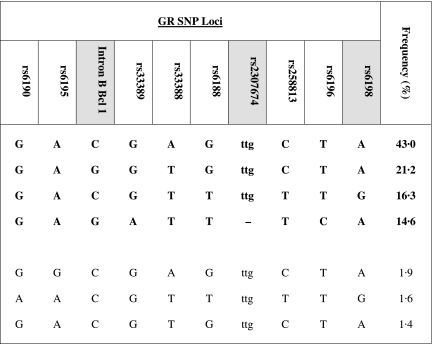
Fig. 1.
Haplotype tagging SNPs. Haplotypes (n = 425) occurring with a frequency of > 1% are shown. Boldface indicates the haplotypes captured by the three haplotype tagging polymorphisms (shaded): intron B Bcl1, C → G, the presence or absence of the ttg insertion (rs2307674) and the rs6198 A → G. (Adapted from Stevens et al.10)
Methods
Patients and controls
Blood samples were obtained with informed written consent. Ethical approval was obtained from Multicentre Research Ethics Committee (MREC) (99/8/84) and the University of Manchester committee on the ethics of research on human beings (8/92/(i)).
All patients and controls were of UK Caucasoid origin.
One hundred and ninety-eight RA cases from the Arthritis Research Campaign National Repository for families with RA were studied. Only one affected case per family was selected, at random, for investigation. All RA cases had disease that met the American College of Rheumatology 1987 criteria, as modified for genetic studies.
DNA was available for 393 control subjects. These were individuals recruited from blood donors and from the records of general practitioners.
Genotyping
SNP genotyping was by the dye terminator-based SNaPshot method (Applied Biosystems, Warrington, UK). In brief, polymerase chain reactions (10 µl final volume) contained 10 ng of genomic DNA, 0·4 µl of each primer (25 pmol/µl), 1 µl of 10 × NH4 buffer (Bioline), 1 µl of dNTPs (Bioline, 2 mM), 0·3 µl MgCl2 (Bioline, 1·5 mM) and 0·2 µl of Taq polymerase (Bioline, 5 U/µl). The primers and probes used were (5′ to 3′): for the intron B Bcl1 SNP: forward primer CTA TTC TTC AAA CTG AAT CTT CTG, reverse primer CAA CAC GTA TAT CTA CAT TTA GAA C, and probe GAC ACC AAT TCC TCT CTT AAA GAG ATT; for the rs6198 SNP: forward primer AGT GTC TTT TTA CCT ACG C, reverse primer ATG TTT CTC CAT ATT TGG C, and probe TGT GGT TTG GTA ATA CCA GAA CAG CAA ATT TAA A.
The presence or absence of a ttg insert (rs2307674) was determined using capillary electrophoresis for size resolution, relative to a ROX 450 internal size standard, on an ABI Prism 310 DNA Genetic Analyser (Applied Biosystems), using a fluorescent dye (HEX)-labelled forward primer. The results were analysed using Genescan analysis and Genotyper 3·6 software (Applied Biosystems). The rs2307674 forward primer was ACC TCA AGT GAT CCA CCC, and the reverse primer GTA CAT GGT TAT ACT CAT ATA TAA C.
The RA cases and a proportion of the controls had been previously genotyped for HLA-DRB1 alleles and shared epitope (SE) status assigned.
Statistical analysis
Associations between the GR polymorphisms and RA were analysed using the χ2-test (Stata, College Station, TX) and correction for multiple testing was performed by Bonferroni's method. Haplotype analysis was carried out using the expectation-maximization algorithm implemented in HelixTree (Golden Helix, Bozeman, MT). Stratification analysis was performed to investigate the effects of gender, disease severity, as assessed by the presence of erosive disease, age at disease onset (the median age at onset was used as the cut-off value), and carriage of SE alleles (HLA-DRB1*0401, *0404, *0101, *0102, *1001 and *1402 were considered positive for the shared epitope).
The study had > 85% power to detect an odds ratio (OR) of 1·8 or greater with RA susceptibility (P = 0·05).
Results
No deviation from Hardy-Weinberg equilibrium was seen for any polymorphism in the cases or the controls. Strong linkage disequilibrium (LD) (r2 = 1) was seen for the polymorphisms studied in both the case and control groups.
No association with any GR polymorphism was found with the RA cases when considered as a whole group (Table 1). Similarly, no significant haplotype associations were seen with RA susceptibility.
Table 1.
Frequency of intron B Bcl1, rs2307674 and rs6198 GR SNP genotypes in control and RA cases
| GR polymorphism | Controls n (%) | RA cases n (%) | P-value |
|---|---|---|---|
| Intron B BCl1 | (n = 392) | (n = 195) | |
| CC | 147 (37·5) | 76 (39·0) | |
| CG | 195 (49·7) | 94 (48·2) | |
| GG | 50 (12·8) | 25 (12·8) | 0·81 |
| (n = 384) | (n = 191) | ||
| rs2307674 | |||
| ttg/ttg | 255 (66·4) | 136 (71·2) | |
| ttg/- | 119 (31·0) | 47 (24·6) | |
| -/- | 10 (2·6) | 8 (4·2) | 0·50 |
| (n = 390) | (n = 178) | ||
| rs6198 | |||
| AA | 256 (65·6) | 129 (72·5) | |
| AG | 121 (31·1) | 44 (24·7) | |
| GG | 13 (3·3) | 5 (2·8) | 0·13 |
On stratified analysis, a weak positive association with an excess of rs6198 AA (wild-type) homozygotes was seen when the SE negative cases were compared with SE negative controls (Table 2). Furthermore, use of Bonferroni's correction for multiple testing (n = 3) rendered this observation nonsignificant (Pcorr = 0·15).
Table 2.
Frequency of intron B BCl1, rs2307674 and rs6198 GR SNP genotypes in controls and in the RA cohort stratified by erosive disease, age at disease onset, and carriage of SE alleles
| SE status | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Erosive disease | Age at disease onset | SE negative | SE positive | ||||||
| Genotype | Controls | Present | Absent | < 41 years | ≥ 42 years | Controls | RA cases | Controls | RA cases |
| Intron B BCl1 | (n = 392) | (n = 139) | (n = 25) | (n = 75) | (n = 72) | (n = 71) | (n = 41) | (n = 86) | (n = 154) |
| CC | 147 (37·5) | 58 (41·8) | 7 (28·0) | 32 (42·7) | 31 (43·1) | 29 (40·8) | 16 (39·1) | 29 (33·7) | 60 (39·0) |
| CG | 195 (49·7) | 63 (45·3) | 14 (56·0) | 30 (40·0) | 28 (38·9) | 35 (49·3) | 19 (46·3) | 47 (54·7) | 75 (48·7) |
| GG | 50 (12·8) | 18 (12·9) | 4 (16·0) | 13 (17·3) | 13 (18·0) | 7 (9·9) | 6 (14·6) | 10 (11·6) | 19 (12·3) |
| P-value | – | 0·55 | 0·37 | 0·95 | 0·98 | – | 0·62 | – | 0·62 |
| Rs2307674 | (n = 384) | (n = 135) | (n = 25) | (n = 73) | (n = 72) | (n = 71) | (n = 41) | (n-86) | (n = 150) |
| ttg/ttg | 255 (66·4) | 101 (74·8) | 15 (60·0) | 57 (78·1) | 31 (43·1) | 51 (71·8) | 27 (65·9) | 56 (65·1) | 109 (72·7) |
| ttg/– | 119 (31·0) | 31 (23·0) | 7 (28·0) | 10 (13·7) | 28 (38·9) | 20 (28·2) | 11 (26·8) | 26 (30·2) | 36 (24·0) |
| –/– | 10 (2·6) | 3 (2·2) | 3 (12·0) | 6 (8·2) | 13 (18·0) | 0 (0·0) | 3 (7·3) | 4 (4·7) | 5 (3·3) |
| P-value | – | 0·10 | 0·16 | 0·38 | 0·38 | – | 0·20 | – | 0·22 |
| rs6198 | (n = 390) | (n = 123) | (n = 23) | (n = 66) | (n = 63) | (n = 70) | (n = 37) | (n = 87) | (n = 141) |
| AA | 256 (65·6) | 87 (70·7) | 19 (82·6) | 45 (68·2) | 41 (65·0) | 44 (62·9) | 30 (81·1) | 56 (64·4) | 99 (70·2) |
| AG | 121 (31·1) | 31 (25·2) | 4 (17·4) | 18 (27·3) | 19 (30·2) | 22 (31·4) | 6 (16·2) | 27 (31·0) | 38 (27·0) |
| GG | 13 (3·3) | 5 (4·1) | 0 (0·0) | 3 (4·5) | 3 (4·8) | 4 (5·7) | 1 (2·7) | 4 (4·6) | 4 (2·8) |
| P-value | – | 0·44 | 0·08 | 0·86 | 0·79 | – | 0·05 | – | 0·30 |
No other single or haplotypic association of GR polymorphisms and no other parameters of RA severity were found.
Discussion
Inappropriately low endogenous cortisol production may be a contributing factor to the onset and progression of RA. How this arises has not been ascertained.12 Alternatively, a normal HPA axis response to inflammation may be compromised in RA patients by a reduction or alteration in GR function. The number of GRα in lymphocytes from RA cases was found to be lower than in controls.13 However, the serum cortisol levels were normal so such apparent downregulation of receptor expression is difficult to explain. Polymorphism of the GR gene may influence GC sensitivity. Certain low-frequency polymorphisms of GR (ER22/23EK) have been found to be associated with parameters of Gc sensitivity in normal individuals.14 These nucleotide changes are in strong LD with the intron B Bcl1 polymorphism in particular, and as such we find no evidence of association of them with RA susceptibility. The possibility of a GR polymorphism conferring risk for RA was raised by Derijk et al.8 This was of particular interest because the nucleotide change occurred in the 3′ UTR of the GRβ molecule. If RA cases had an excessive amount of this variation, which was subsequently found to have functional significance and affect GRβ mRNA stability, then a possible mechanism for relative GR resistance would have been found. Our study aimed to look for variation in GR polymorphism with RA susceptibility. We specifically genotyped for the rs6198 SNP within the 3′ UTR of GRβ, in combination with two other changes, which gave excellent genetic coverage across the GR gene locus. We have accounted for 95% of all haplotypes across the GR gene by using this approach.
No evidence for genetic association with GR polymorphisms and RA susceptibility was observed. This is in agreement with the findings of a Korean study of RA, in which the 149 RA cases and controls were compared and no association with the intron B Bcl1 polymorphism was found with the RA group as a whole, or on stratification for disease severity.15 Although less well powered to look at, we have also stratified our patient cohort by parameters of disease severity. An excess of rs6198 AA (wild-type) homozygotes was seen when the SE negative cases were compared with the SE negative controls (Table 2). However, given the relatively small numbers involved, and the borderline level of significance, which is not maintained once correction for multiple testing is applied, this finding needs to be interpreted with caution.
In summary, we have looked at three changes across the GR gene that capture 95% of all commonly occurring haplotypes, but find no evidence for GR polymorphisms contributing a substantial genetic effect, either as single point or as haplotypic associations, to risk of RA. We conclude that GR polymorphisms do not confer significant risk of RA.
References
- 1.Harbuz MS, Jessop DS. Is there a defect in cortisol production in rheumatoid arthritis? Rheumatology. 1999;38:298–302. doi: 10.1093/rheumatology/38.4.298. [DOI] [PubMed] [Google Scholar]
- 2.Chikanza IC, Petrou P, Kingsley G, Chrousos G, Panayi GS. Defective hypothalamic response to immune and inflammatory stimuli in patients with rheumatoid arthritis. Arthritis and Rheumatism. 1992;35:1281–1288. doi: 10.1002/art.1780351107. [DOI] [PubMed] [Google Scholar]
- 3.Sternberg EM, Hill JM, Chrousos GP, Kamilaris T, Listwak SJ, Gold PW, Wilder RL. Inflammatory mediator-induced hypothalamic-pituitary-adrenal axis activation is defective in streptococcal cell wall arthritis-susceptible Lewis rats. Proceedings of the National Academy of Sciences of the United States of America. 1989;86:2374–2378. doi: 10.1073/pnas.86.7.2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chikanza IC. Mechanisms of corticosteroid resistance in rheumatoid arthritis: a putative role for the corticosteroid receptor beta isoform. Annals of the New York Academy of Sciences. 2002;966:39–48. doi: 10.1111/j.1749-6632.2002.tb04200.x. [DOI] [PubMed] [Google Scholar]
- 5.Hollenberg SM, Weinberger C, Ong ES, Cerelli G, Oro A, Lebo R, Thompson EB, Rosenfeld MG, Evans RM. Primary structure and expression of a functional human glucocorticoid receptor cDNA. Nature. 1985;318:635–641. doi: 10.1038/318635a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Derijk RH, Schaaf M, de Kloet ER. Glucocorticoid receptor variants: clinical implications. Journal of Steroid Biochemistry and Molecular Biology. 2002;81:103–122. doi: 10.1016/s0960-0760(02)00062-6. [DOI] [PubMed] [Google Scholar]
- 7.Goecke A, Guerrero J. Glucocorticoid receptor beta in acute and chronic inflammatory conditions: clinical implications. Immunobiology. 2006;211:85–96. doi: 10.1016/j.imbio.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 8.Derijk RH, Schaaf MJ, Turner G, Datson NA, Vreugdenhil E, Cidlowski J, de Kloet ER, Emery P, Sternberg EM, Detera-Wadleigh SD. A human glucocorticoid receptor gene variant that increases the stability of the glucocorticoid receptor beta-isoform mRNA is associated with rheumatoid arthritis. Journal of Rheumatology. 2001;28:2383–2388. [PubMed] [Google Scholar]
- 9.Schaaf MJ, Cidlowski JA. Molecular mechanisms of glucocorticoid action and resistance. Journal of Steroid Biochemistry and Molecular Biology. 2002;83:37–48. doi: 10.1016/s0960-0760(02)00263-7. [DOI] [PubMed] [Google Scholar]
- 10.Stevens A, Ray DW, Zeggini E, John S, Richards HL, Griffiths CE, Donn R. Glucocorticoid sensitivity is determined by a specific glucocorticoid receptor haplotype. Journal of Clinical Endocrinology and Metabolism. 2004;89:892–897. doi: 10.1210/jc.2003-031235. [DOI] [PubMed] [Google Scholar]
- 11.van Rossum EF, Koper JW, van den Beld AW, Uitterlinden AG, Arp P, Ester W, Janssen JA, Brinkmann AO, de Jong FH, Grobbee DE, Pols HA, Lamberts SW. Identification of the BclI polymorphism in the glucocorticoid receptor gene: association with sensitivity to glucocorticoids in vivo and body mass index. Clinical Endocrinology. 2003;59:585–592. doi: 10.1046/j.1365-2265.2003.01888.x. [DOI] [PubMed] [Google Scholar]
- 12.Jessop DS, Harbuz MS. A defect in cortisol production in rheumatoid arthritis: why are we still looking? Rheumatology. 2005;44:1097–1100. doi: 10.1093/rheumatology/keh644. [DOI] [PubMed] [Google Scholar]
- 13.Schlaghecke R, Kornely E, Wollenhaupt J, Specker C. Glucocorticoid receptors in rheumatoid arthritis. Arthritis and Rheumatism. 1992;35:740–744. doi: 10.1002/art.1780350704. [DOI] [PubMed] [Google Scholar]
- 14.van Rossum EF, Russcher H, Lamberts SW. Genetic polymorphisms and multifactorial diseases: facts and fallacies revealed by the glucocorticoid receptor gene. Trends in Endocrinology and Metabolism. 2005;16:445–450. doi: 10.1016/j.tem.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 15.Lee EB, Kim JY, Lee YJ, Song YW. Glucocorticoid receptor polymorphisms in Korean patients with rheumatoid arthritis. Annals of the Rheumatic Diseases. 2005;64:503–504. doi: 10.1136/ard.2004.023432. [DOI] [PMC free article] [PubMed] [Google Scholar]