Abstract
Cardiac hypertrophy is a major cause of human morbidity and mortality. Although much is known about the pathways that promote hypertrophic responses, mechanisms that antagonize these pathways have not been as clearly defined. Atrogin-1, also known as muscle atrophy F-box, is an F-box protein that inhibits pathologic cardiac hypertrophy by participating in a ubiquitin ligase complex that triggers degradation of calcineurin, a factor involved in promotion of pathologic hypertrophy. Here we demonstrated that atrogin-1 also disrupted Akt-dependent pathways responsible for physiologic cardiac hypertrophy. Our results indicate that atrogin-1 does not affect the activity of Akt itself, but serves as a coactivator for members of the Forkhead family of transcription factors that function downstream of Akt. This coactivator function of atrogin-1 was dependent on its ubiquitin ligase activity and the deposition of polyubiquitin chains on lysine 63 of Foxo1 and Foxo3a. Transgenic mice expressing atrogin-1 in the heart displayed increased Foxo1 ubiquitylation and upregulation of known Forkhead target genes concomitant with suppression of cardiac hypertrophy, while mice lacking atrogin-1 displayed the opposite physiologic phenotype. These experiments define a role for lysine 63–linked ubiquitin chains in transcriptional coactivation and demonstrate that atrogin-1 uses this mechanism to disrupt physiologic cardiac hypertrophic signaling through its effects on Forkhead transcription factors.
Introduction
Factors that increase LV afterload — such as hypertension, aortic stenosis, and age-related arterial stiffness — elicit cardiac hypertrophy as an adaptive mechanism to normalize wall stress. The short-term hemodynamic benefits of hypertrophy occur at a cost: cardiac hypertrophy leads to diastolic dysfunction and heart failure and is a powerful predictor of cardiovascular mortality even in the absence of symptoms (1, 2). At the cellular level, cardiac hypertrophy is a consequence of increased cardiomyocyte cell volume (1, 2), a process that requires coordination of cellular signaling cascades, activation of fetal cardiac gene expression programs, increased protein synthesis, sarcomere assembly, and modulation of cellular energy sources. At the present time, no specific pharmacologic strategies to reverse cardiac hypertrophy have been approved for clinical use, so the delineation of hypertrophic mechanisms (especially those that prevent or reverse hypertrophy) remains a priority.
Although complexity and redundancy exist in the signaling pathways that activate cardiac hypertrophy, 2 independent circuits that elicit distinct manifestations of hypertrophy are now recognized. Hypertrophy in response to stimuli such as pressure overload and adrenergic stimulation activates the calcineurin/nuclear factor of activated T cell–dependent signaling pathway, resulting in so-called “pathological” hypertrophy that is associated with maladaptive features such as fibrosis, chamber dilatation, and hemodynamic decompensation (3). A second, autonomous pathway activated by exercise or insulin/IGF-1 signaling mediates hypertrophy of the heart under more physiologic conditions that occur during development or conditioning. This physiologic form of hypertrophy depends on regulated activation of the kinase Akt, which in turn coordinates protein synthesis and cardiac gene expression to increase cardiomyocyte size and heart mass. Hypertrophy under these circumstances occurs without the propensity to chamber enlargement or heart failure. Recent studies have identified several downstream targets of Akt activation — including mammalian target of rapamycin (mTOR) and glycogen synthase kinase–3β (GSK-3β) (4–7) — that participate in Akt-dependent cardiac hypertrophy in the setting of exercise or IGF-1 stimulation.
We recently found that atrogin-1 (also known as muscle atrophy F-box) blocks pathologic hypertrophy in vivo and in vitro by binding to calcineurin A and inducing its ubiquitylation and degradation (8). Atrogin-1 is a cardiac- and skeletal muscle–specific F-box protein that binds to Skp1, Cul1, and Roc1, the common components of SCF ubiquitin ligase complexes. Atrogin-1 was first identified as a crucial participant in skeletal muscle atrophy programs (9, 10). Interestingly, atrogin-1 expression is carefully regulated at the transcriptional level in skeletal myocytes and in cardiomyocytes by members of the Forkhead family of transcription factors (11, 12). In the setting of cardiomyocyte hypertrophy, atrogin-1 decorates calcineurin with lysine 48–linked ubiquitin chains that target calcineurin for proteasome-dependent degradation. This in turn leads to potent suppression of agonist-induced cardiomyocyte hypertrophy in vitro and increased heart muscle mass after aortic banding in vivo (8), indicating that atrogin-1 plays a critical role in regulating calcineurin-dependent cardiac hypertrophy under pathologic circumstances. To extend these previous studies, we have examined the role atrogin-1 plays in modulating hypertrophic responses that do not depend on calcineurin signaling.
Results
Suppression of IGF-1– and insulin-dependent cardiomyocyte hypertrophy by atrogin-1.
We used an adenovirus-dependent gene delivery system (8) to examine the effects of atrogin-1 on rat neonatal cardiomyocyte hypertrophy elicited by both calcineurin-dependent and calcineurin-independent stimuli. Consistent with our previous observations, phenylephrine stimulation of cardiomyocytes induced a 2-fold increase in cardiomyocyte area, and this hypertrophy was completely blocked by enforced atrogin-1 expression (Figure 1A). Both insulin and IGF-1 similarly increased cardiomyocyte size, and these effects were also efficiently blocked in cardiomyocytes infected with the atrogin-1–expressing adenovirus (Ad–atrogin-1).
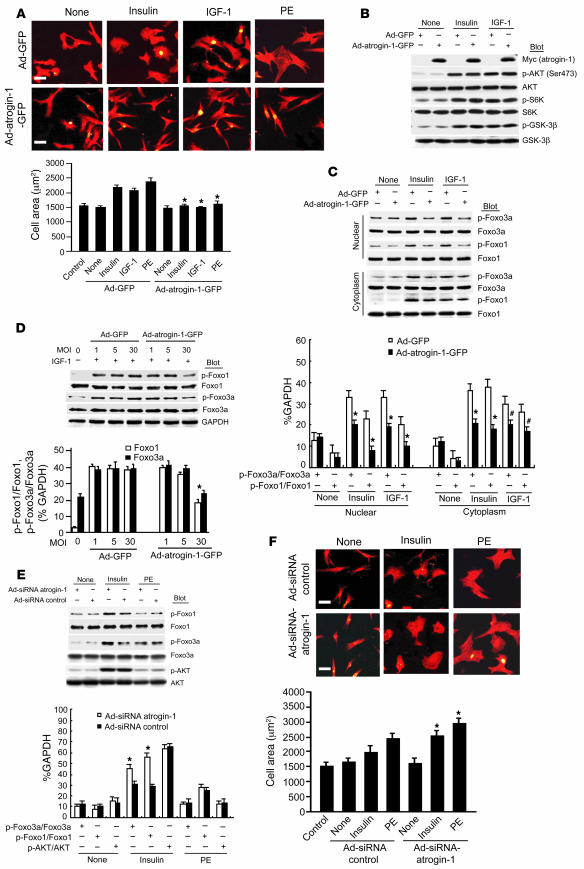
Figure 1. Overexpression of atrogin-1 represses insulin- and IGF-1–induced cardiomyocyte hypertrophy and Foxo1 and Foxo3a dephosphorylation.
(A) Cardiomyocytes were infected with Ad-GFP or Ad–atrogin-1–GFP (MOI 10), and cells were stimulated with insulin (20 μg/ml–1), IGF-1 (20 ng/ml–1), or PE (100 μM) for 24 h and stained with α-actinin antibody (red). Quantitation of cell surface area was performed on 100 cells per group. *P < 0.001 vs. serum-free control and Ad-GFP. (B) Protein levels of endogenous Akt pathway members were determined in cardiomyocytes expressing atrogin-1 ectopically by IB with the indicated antibodies. (C) Foxo1 and Foxo3a expression and phosphorylation in cytoplasmic and nuclear fractions from cardiomyocytes were assessed by IB using anti-total or –phospho-Foxo1 and -Foxo3a antibodies after the indicated treatments. *P < 0.001 vs. Ad-GFP + insulin or IGF-1; #P < 0.01 vs. Ad-GFP + IGF-1. (D) Cardiomyocytes were infected with increasing MOIs of Ad–atrogin-1–GFP and Ad-GFP with IGF-1 (20 μg/ml–1) stimulation for 24 h. Endogenous Foxo1 and Foxo3a levels were determined by IB with the indicated antibodies. Maximal suppression of Foxo1 and Foxo3a occurred at MOI 30. *P < 0.001 vs. Ad-GFP + IGF-1 at MOI 30. (E) Cardiomyocytes were infected with Ad-siRNA–control or Ad-siRNA–atrogin-1 and treated with insulin (20 μg/ml–1) or PE (100 μM) for 24 h. (F) Cardiomyocytes treated as indicated were stained with α-actinin antibody. Quantitation of cell surface area was performed on 100 cells per group. Original magnification, ×200. *P < 0.01 vs. Ad-siRNA control + insulin or PE. Scale bars: 50 μm.
To explore the specific signaling events downstream of insulin and IGF-1 stimulation that are restrained by atrogin-1 in cardiomyocytes, we examined the activation status of Akt and its downstream targets S6 kinase and GSK-3β. Each of these known modulators of cardiomyocyte size was efficiently phosphorylated after insulin and IGF-1 stimulation, but atrogin-1 did not suppress their activation (Figure 1B). In contrast, when we examined the phosphorylation status of the Forkhead transcription factors Foxo1 and Foxo3a, which are downstream targets of Akt that also mediate hypertrophic responses in cardiomyocytes (11–13), we found that atrogin-1 efficiently suppressed insulin- and IGF-1–dependent phosphorylation of both proteins by 55%–60% (Figure 1, C and D). To test the contributions of endogenous atrogin-1 to Forkhead phosphorylation and cardiomyocyte hypertrophy, we knocked down endogenous atrogin-1 expression using an adenoviral siRNA approach that suppresses atrogin-1 mRNA by about 80% (8). Atrogin-1 knockdown augmented Foxo3a phosphorylation in unstimulated cardiomyocytes and increased both Foxo1 and Foxo3a phosphorylation when Akt was activated by insulin (Figure 1E), but had no effect on phosphorylation of Akt itself or on the Forkhead transcription factors when cardiomyocytes were stimulated with phenylephrine. As we have shown previously (8), suppression of atrogin-1 mRNA led to an enhanced hypertrophic response of cardiomyocytes after phenylephrine treatment, and similar consequences were also observed in cells stimulated with insulin to activate Akt (Figure 1F), indicating that endogenous atrogin-1 regulates both Forkhead activation and insulin-dependent hypertrophy in cardiomyocytes.
Interaction of atrogin-1 with Foxo1 and Foxo3a.
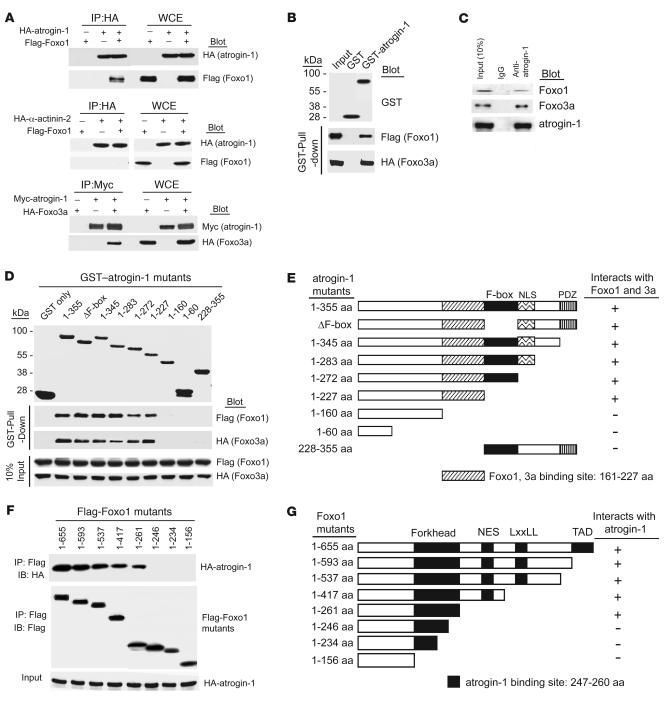
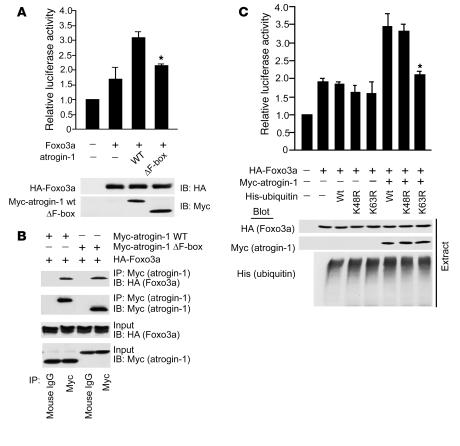
The Forkhead proteins are activated in their dephosphorylated state and trans-activate a transcriptional program that suppresses cardiomyocyte hypertrophy (11–13). To explore whether atrogin-1 mediates its effects on Forkhead phosphorylation via direct interactions, we tested whether mutual interactions existed between these proteins. Using tagged forms of atrogin-1 and Foxo1 or Foxo3a transfected into 293 cells, both Foxo1 and Foxo3a were efficiently coimmunoprecipitated with atrogin-1, but not by similarly tagged α-actinin as a control (Figure 2A). These observations likely reflect direct interactions, because recombinant glutathione-S-transferase–tagged (GST-tagged) atrogin-1 efficiently coprecipitated both Foxo1 and Foxo3a from cell lysates (Figure 2B). Using a specific anti–atrogin-1 antibody, we also found that endogenous atrogin-1 participated in complexes with Foxo1 and Foxo3a in cultured cardiomyocytes (Figure 2C). We mapped the sites of Foxo1 and Foxo3a interactions with atrogin-1 using GST-tagged atrogin-1 fragments and found that both proteins required a 60–amino acid sequence immediately adjacent to the F-box of atrogin-1 for binding (Figure 2, D and E), although our experiments do not exclude the possibility that additional sequences of atrogin-1 also participate in interactions with Foxo proteins. Interestingly, this site is distinct from the calcineurin-binding site of atrogin-1 (8). Conversely, we found that an intact Forkhead domain within HA-tagged Foxo1 fragments was required for atrogin-1 interactions (Figure 2, F and G).
Figure 2. Atrogin-1 interacts with Foxo1 and Foxo3a in vivo and in vitro.
(A) We transfected 293 cells with the indicated plasmids. Equal amounts of protein lysates were immunoprecipitated with anti-HA or anti-Myc antibody and analyzed by IB with the indicated antibodies. WCE, whole cell extract. (B) The ability of Foxo1 and Foxo3a expressed in 293 cells to be retained by GST or a GST–atrogin-1 fusion protein was analyzed by IB after GST pulldown. (C) Endogenous protein interactions were examined in cardiomyocyte cell lysates immunoprecipitated with rabbit IgG or anti–atrogin-1 antibody and analyzed by IB with antibodies to detect Foxo1, Foxo3a, and atrogin-1. (D) Domains of atrogin-1 involved in binding to Foxo1 and Foxo3a expressed in 293 cells were mapped with GST pulldown assays. GST–atrogin-1 proteins were affinity purified and analyzed by SDS-PAGE and Coomassie blue staining. The ability of the truncated atrogin-1 fusion proteins to bind to Foxo1 and Foxo3a from whole-cell lysates was determined by blotting with antibodies against Flag and HA, respectively. (E) Deletion constructs of atrogin-1 in D that bind to Foxo1 and Foxo3a. NLS, nuclear localization sequence; PDZ, PDZ-binding domain. (F) Interaction domains of Foxo1 required for binding to atrogin-1 were mapped by co-IP. We transfected 293 cells with the indicated Flag-Foxo1 and HA–atrogin-1 plasmids. Equal amounts of lysates were prepared for IP with a Flag antibody and immunoblotted with antibodies for HA or Flag. (G) Deletion constructs of Foxo1 in F that bind to atrogin-1. FK, Forkhead domain; NES, nuclear export sequence; LxxLL, nuclear receptor–interacting domain or LxxLL motif; TAD, transactivation domain.
Atrogin-1 is required for optimal transcriptional activity of Forkhead transcription factors in cardiomyocytes.
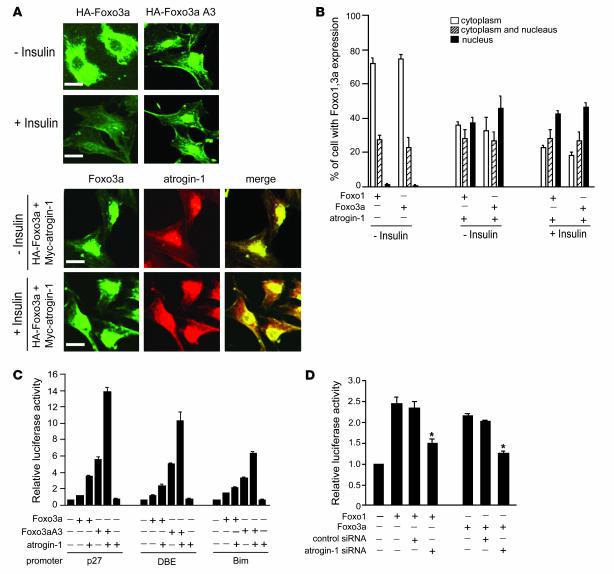
Because atrogin-1 exists in both a sarcomere-associated and a nuclear pool in cardiomyocytes (8), we tested whether the effects of atrogin-1 on Foxo1 phosphorylation coincide with changes in Forkhead localization. Foxo1 and Foxo3a reside primarily in the cytoplasm of insulin-stimulated cardiomyocytes; however, when atrogin-1 was coexpressed, both Foxo1 and Foxo3a relocalized into the nucleus (Figure 3, A and B, and Supplemental Figure 1). It is possible that this enforced nuclear localization prevents Akt-dependent Foxo phosphorylation, accounting for our observations in Figure 1. Coexpression of Foxo1 and Foxo3a with promoter/reporter constructs responsive to Foxo activity (p27kip1, DBE, and Bim; ref. 14) resulted in increased luciferase activity, and in each case coexpression of atrogin-1 with Foxo1 and Foxo3a markedly increased promoter activity (Figure 3C and Supplemental Figure 2). The transcriptional effects of atrogin-1 on reporter activity were even more marked when atrogin-1 was coexpressed with Foxo3aA3 or Foxo1A3, which are constitutively active, nuclear-localized mutants (Figure 3C and Supplemental Figure 2). The ability of atrogin-1 to enhance the constitutively active Forkhead mutants indicates that the transcriptional effects of atrogin-1 do not depend only on increased nuclear accumulation of Forkhead proteins by atrogin-1, which strongly endorses a coactivator role for atrogin-1 on Forkhead activity within the nucleus. To determine whether endogenous atrogin-1 is required for maximal transcriptional activity of Foxo1 and Foxo3a, we tested the activity of these proteins in cardiomyocytes when atrogin-1 expression is knocked down (8). Transcriptional activation of the p27kip1 promoter by Foxo1 and Foxo3a was almost completely suppressed to baseline levels by atrogin-1 knockdown, but not by a scrambled siRNA construct (Figure 3D), indicating that endogenous atrogin-1 is required for optimal Forkhead transactivation of these reporters in cardiomyocytes.
Figure 3. Atrogin-1 induces Foxo1 and Foxo3a nuclear localization and increased transcriptional activity.
(A) Cardiomyocytes were transiently transfected with expression vectors encoding HA-Foxo3a or Myc–atrogin-1. At 24 h after transfection, cells were treated without or with insulin for 12 h and immunostained with antibodies against HA (green) and Myc (red). Staining was assessed by confocal microscopy; representative images are shown. Scale bars: 50 μm. (B) Cardiomyocytes (100 random cells per condition) expressing the indicated proteins were scored for Foxo1 or Foxo3a localization. Data are mean ± SEM of 2 independent experiments. Original magnification, ×200. (C) To assess the role of atrogin-1 on Foxo3a-dependent transcription, cardiomyocytes were transfected with luciferase expression plasmids driven by the indicated promoters and vectors expressing Foxo3a or Foxo3aA3 (a constitutively active form) and atrogin-1. Data were normalized by cotransfection with a plasmid expressing β-gal. Cells were harvested 24 h later for measurement of luciferase activities. Results are expressed relative to the level of expression with the reporter gene alone and representative of 3 independent experiments. Error bars indicate SEM. (D) Cardiomyocytes were transfected with a luciferase reporter driven by the p27kip1 promoter along with Foxo1 or Foxo3a and plasmids expressing siRNA–atrogin-1 or siRNA-control. Luciferase activity was measured 24 h after transfection. *P < 0.01 vs. control siRNA.
Noncanonical ubiquitylation of Forkhead transcription factors by atrogin-1.
Transcriptional activation of Forkhead transcription factors by atrogin-1 raised the question of whether the ubiquitin ligase activity of atrogin-1 participates in the coactivation of Foxo1 and Foxo3a. In pulse-chase experiments using ectopically expressed proteins in 293 cells, both Foxo1 and Foxo3a were stable proteins with t1/2 well in excess of 4 h, and coexpression of atrogin-1 had no effect on the stability of either protein (Figure 4A). Similarly, inhibition of the proteasome with MG-132 had no effect on steady-state levels of Foxo1 or Foxo3a in the presence or absence of atrogin-1 (Figure 4B). Interestingly, steady-state levels of atrogin-1 were consistently higher after MG-132 treatment, suggesting that atrogin-1 expression is itself determined in a proteasome-dependent manner. Collectively, these data indicate that atrogin-1 does not target the Forkhead transcription factors for proteasomal degradation.
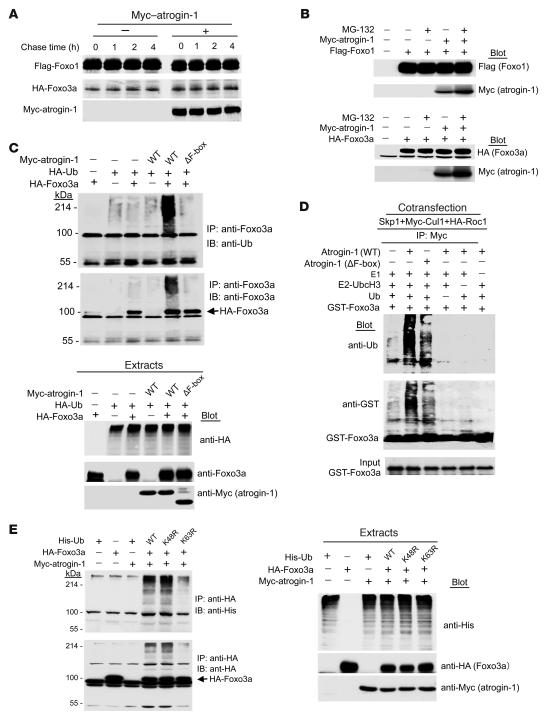
Figure 4. Atrogin-1 induces lysine 63–dependent ubiquitylation of Foxo3a.
(A) Cultured 293 cells were transfected with the indicated plasmids, and pulse-chase analysis was performed. Extracts were subjected to IP using either anti-Flag or anti-HA antibodies. Immunoprecipitates were analyzed by SDS–PAGE followed by autoradiography. (B) The indicated plasmids were cotransfected, and 293 cells were treated for 4 h with DMSO or MG132 (20 μM). The expression levels of the respective proteins were analyzed by IB with anti-Flag, anti-HA, or anti-Myc antibodies. (C) We transfected 293 cells with vectors expressing HA-ubiquitin (HA-Ub), HA-Foxo3a, atrogin-1, or atrogin-1 ΔF-box mutant lacking the E3-ligase activity. Cell extracts were immunoprecipitated with Foxo3a antibody and analyzed by IB with the indicated antibodies. An aliquot of the cell extracts was subjected to direct IB analysis using anti-Foxo3a or anti-Myc antibodies. (D) In vitro ubiquitylation reactions were performed with purified ubiquitin, E1, the E2 UBC13, GST-Foxo3a, and the SCFatrogin-1 complex. Reactions were resolved by SDS-PAGE followed by IB with anti-ubiquitin antibody. (E) We cotransfected 293 cells with the indicated plasmids. At 24 h after transfection, ubiquitin-conjugated proteins were prepared for IP with anti-Foxo3a. Immunoprecipitates were subject to SDS–PAGE followed by IB with anti-ubiquitin, anti-Foxo3a, or anti-Myc antibodies. An aliquot of the cell extracts was subjected to direct IB analysis using anti-Foxo3a or anti-Myc antibodies.
We next asked whether atrogin-1 is capable of adding either mono- or polyubiquitin chains to the Forkhead transcription factors. We performed assays in 293 cells (so we could recover proteins efficiently) after determining that atrogin-1 had similar effects on Forkhead transcriptional activity in 293 cells compared with cardiomyocytes (data not shown). After IP of Foxo3a, we probed immunoprecipitates with an antibody recognizing tagged ubiquitin. Ubiquitylated species of Foxo3a were detected as a high–molecular weight smear only when atrogin-1 was coexpressed with Foxo3a, and the effect of atrogin-1 was abolished by deletion of its F-box (Figure 4C). Similar results were obtained in experiments examining atrogin-1–dependent ubiquitylation of Foxo1 (Supplemental Figure 3A). To determine whether the effects of atrogin-1 on Forkhead multiubiquitylation are direct, we reassembled the components of ubiquitylation in vitro (8) with immunopurified Skp1, Cul1, and Roc1 and recombinant atrogin-1 and Foxo3a or Foxo1. We found that multiubiquitylation of Foxo3a (Figure 4D) and Foxo1 (Supplemental Figure 3B) proceeded efficiently in the presence of atrogin-1, was dependent on the F-box of atrogin-1, and was abolished by deletion of atrogin-1 or any other component of the reaction.
Taken together, these observations indicate that atrogin-1 induces multiubiquitylation of Foxo1 and Foxo3a, yet does not target either protein for proteasome-dependent degradation and instead increases their activity. This suggested that atrogin-1 may not be decorating the Forkhead transcription factors with canonical, lysine 48–linked polyubiquitin chains. To test this, we examined the ubiquitylation of Foxo3a induced by atrogin-1 in the presence of WT ubiquitin or ubiquitin variants that cannot assemble lysine 48 or lysine 63 chains. Mutation of lysine 48 in ubiquitin actually enhanced the polyubiquitylation of Foxo3a by atrogin-1, indicating that canonical ubiquitin chains are not assembled on Foxo3a by atrogin-1 (Figure 4E). In contrast, ubiquitylation of Foxo3a (and also Foxo1; Supplemental Figure 3C) was markedly suppressed by the lysine 63–ubiquitin mutant, indicating that lysine 63–linked chains were the predominant species assembled on Forkhead transcription factors by atrogin-1.
Although lysine 63–linked ubiquitin chains participate in a number of cellular events, such as cell signaling and DNA damage repair (15, 16), they have not yet been clearly associated with transcriptional regulation in mammalian cells, and there are few descriptions of noncanonical ubiquitylation in the context of cardiomyocyte biology. We therefore performed experiments to confirm or refute the importance of lysine 63–linked chains in atrogin-1–dependent coactivation of Foxo1 and Foxo3a. We first asked whether the F-box of atrogin-1 is necessary for coactivation. Indeed, when coexpressed with Foxo3a, deletion of the F-box of atrogin-1 totally abolished its ability to coactivate Foxo3a in transcriptional assays using the p27kip1 promoter (Figure 5A). The F-box requirement in these experiments could indicate that either F-box–dependent ubiquitylation or F-box–dependent binding (independent of ubiquitylation) mediates the transcriptional effects of atrogin-1 on the Forkhead transcription factors. To exclude the latter possibility, we assayed the interactions of Foxo3a and atrogin-1 with and without an intact F-box. The presence of the F-box was not required for Foxo3a interactions (Figure 5B), which is consistent with our in vitro binding assays (Figure 2D) and suggests that the role of the F-box of atrogin-1 in Foxo3a coactivation is to recruit the Skp-Cul1-Roc1 components to atrogin-1 for ubiquitin ligase assembly. If this model is true, then prevention of lysine 63–linked ubiquitin chain assembly on Foxo3a should abolish the coactivation of Foxo3a by atrogin-1. To test this, we examined the transcriptional effects of atrogin-1 on Foxo3a under conditions in which lysine 63–linked ubiquitin linkages are suppressed in reporter gene assays. Under conditions in which atrogin-1 potently coactivates Foxo3a transcription of the p27kip1 promoter, overexpression of lysine 63–ubiquitin completely blocked the transcriptional effects of atrogin-1, whereas lysine 48–ubiquitin had no effect (Figure 5C). Similar results were obtained in studies examining Foxo1 (Supplemental Figure 4). Deletion of the F-box of atrogin-1, or truncation to prevent binding to Foxo proteins, abolished the transcriptional activity of atrogin-1 (Supplemental Figure 5). Collectively, these experiments indicate that atrogin-1 suppresses Akt-dependent cardiomyocyte hypertrophy pathways by favoring nuclear retention of unphosphorylated Forkhead species and that noncanonical ubiquitylation of Foxo1 and Foxo3a via atrogin-1–dependent lysine 63–linked chains is required for transcriptional coactivation.
Figure 5. Lysine 63–linked ubiquitin chains are required for transcriptional coactivation of Foxo3a by atrogin-1.
(A) Cultured cardiomyocytes were transfected with vectors expressing HA-Foxo3a, Myc–atrogin-1, and/or atrogin-1 ΔF-box together with the p27kip1 luciferase reporter and β-gal constructs. At 24 h after transfection, cells were lysed and luciferase activity was measured. Lysates were immunoblotted with anti-HA and Myc. *P < 0.01 vs. atrogin-1 WT. (B) We transfected 293 cells with the indicated plasmids. Equal amounts of protein lysates were immunoprecipitated with anti-Myc or anti-HA antibodies to detect atrogin-1 and Foxo3a, respectively, followed by IB to detect protein-protein interactions. (C) Reporter assays were performed using the p27kip1 promoter in cardiomyocytes cotransfected with plasmids expressing Foxo3a and/or atrogin-1, along with vectors expressing WT ubiquitin or mutant ubiquitins containing mutations of lysine 48 (K48R) or lysine 63 (K63R). At 24 h after transfection, cells were lysed and luciferase activity was measured. Lysates were immunoblotted with anti-HA and Myc. *P < 0.001 vs. WT ubiquitin. Results are expressed relative to the level of expression with the reporter gene alone and representative of 3 independent experiments. Error bars indicate SEM.
Atrogin-1 increases Forkhead protein activity and suppresses IGF-1–dependent cardiac hypertrophy in vivo.
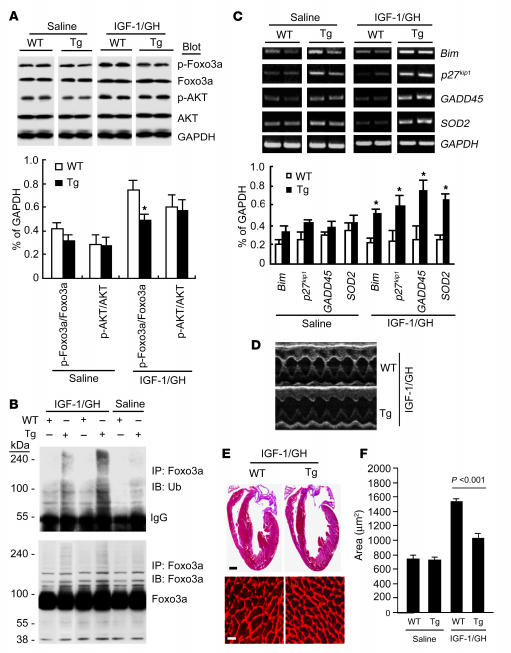
To test the relevance of our observations in vivo, we examined the effects of atrogin-1 on Akt-dependent hypertrophy in Tg mice that overexpress atrogin-1 under control of the α–myosin heavy chain promoter (8). We used injections of IGF-1 and growth hormone (GH) for 14 d to stimulate Akt-dependent cardiac hypertrophy (17–19). Akt phosphorylation was increased by IGF-1/GH treatment in ventricles from both WT and nontransgenic littermates (Figure 6A), as has been previously described (17). However, whereas Foxo1 and Foxo3a phosphorylation were increased after IGF-1/GH treatment in WT mouse hearts, this phosphorylation was suppressed in atrogin-1 Tg mice. To establish whether ubiquitylation of Forkhead proteins is regulated by atrogin-1 in vivo, we immunoprecipitated Foxo3a from heart lysates and probed these precipitates for ubiquitylated species. We found that Foxo3a ubiquitylation was increased in atrogin-1 Tg hearts after IGF-1/GH treatment (Figure 6B). We next measured the mRNA expression of known Forkhead target genes Bim, p27kip1, GADD45, and SOD2 to determine whether the transcriptional activity of Foxo1 and Foxo3a was concomitantly increased in atrogin-1 Tg hearts. Quantitative analysis indicated that the expression of each of these target genes was increased by 2-fold or more in hearts from atrogin-1 Tg mice after IGF-1/GH treatment compared with hearts from WT mice (Figure 6C).
Figure 6. Enhanced Forkhead protein ubiquitylation and activity and suppressed IGF-1/GH-dependent cardiac hypertrophy in cardiac-specific atrogin-1 Tg mice.
(A) Cardiac hypertrophy was induced in 8-wk-old mice with IGF-1/GH injections. Atrogin-1 Tg and WT mice were sacrificed 14 d later; their hearts were freshly isolated; and levels of the indicated total and phospho-proteins were determined by IB. Results were normalized to relative expression levels of phospho-Foxo1, Foxo3a, and Akt (n = 6). *P < 0.001 vs. WT. (B) Equal amounts of lysates from WT and Tg hearts were immunoprecipitated with Foxo3a antibody and analyzed by IB with antibodies against ubiquitin or Foxo3a to detect ubiquitylated forms of Foxo3a in vivo before and after IGF-1/GH treatment. (C) RT-PCR was performed to measure the expression of known Foxo1 and Foxo3a target genes in WT and Tg mouse hearts before and after IGF-1/GH treatment. Total RNA was isolated from mouse hearts, and expression of transcripts for Bim, p27Kip1, GADD45, SOD2, and GAPDH was determined and normalized to GAPDH (n = 6). *P < 0.001 vs. WT. A representative analysis is shown. (D) M-mode echocardiographic analysis of hearts from Tg and WT mice after 2 wk IGF-1/GH injection. (E) Representative macroscopic histologic analysis of H&E-stained hearts from indicated mice after 2 wk IGF-1/GH injection. Histologic sections were also stained with wheat germ agglutinin–TRITC conjugate to determine cell size. Scale bars: 1 mm (top); 50 μm (bottom). (F) Cardiomyocyte size from WT and Tg hearts after IGF-1/GH treatment.
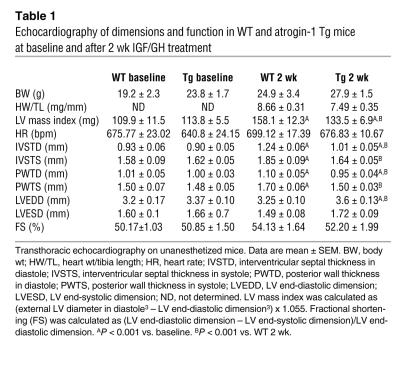
Echocardiography was performed on mice at baseline and after 14 d of IGF-1/GH treatment to determine the physiologic consequences of atrogin-1 on IGF-1/GH–dependent hypertrophy. LV mass increased by 44% in WT hearts and by only 17% in atrogin-1 Tg hearts after IGF-1/GH treatment (Table 1 and Figure 6D). The parameters of hypertrophy, interventricular and posterior wall thickness, also failed to increase in atrogin-1 Tg mouse hearts. Heart weight/tibia length ratios, determined as an additional index of cardiac hypertrophy, were also significantly lower (P < 0.02) in atrogin-1 Tg mice after IGF-1/GH treatment. Cardiac function, as estimated by fractional shortening, was not affected in atrogin-1 Tg mice before or after IGF-1/GH treatment (Table 1), in spite of the lack of a hypertrophic response in these mice. Histologically, cardiomyocyte size increased by 96% in WT mouse hearts but only by 27% in atrogin-1 Tg hearts after IGF-1/GH treatment (Figure 6, E and F).
Table 1 .
Echocardiography of dimensions and function in WT and atrogin-1 Tg mice at baseline and after 2 wk IGF/GH treatment
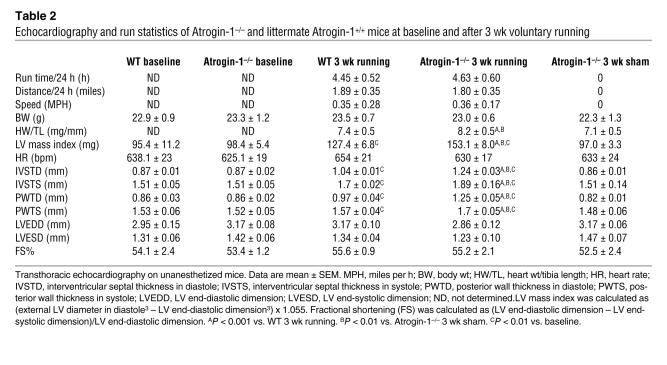
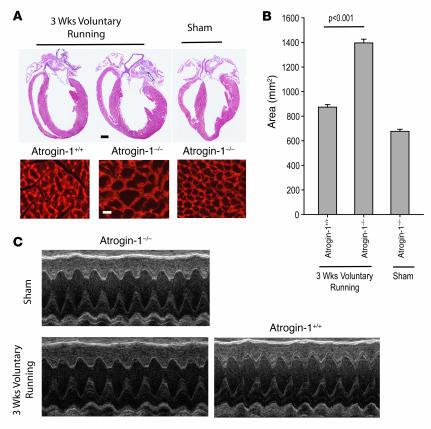
Our analyses using Tg mice allowed us to assess the activity of ectopically expressed atrogin-1 on cardiac responses to IGF-1/GH treatment and to perform biochemical analyses of Forkhead protein activation. However, the role of endogenous atrogin-1 in Akt-dependent hypertrophy, especially under more physiologic conditions, is not addressed by these studies. To answer these questions, we examined the physiologic induction of hypertrophy after 3 wk voluntary wheel exercise in WT and Atrogin-1–/– mice. Run times, total mileage, and average speed were not different between groups (Table 2). Hearts from Atrogin-1–/– mice were grossly larger than those from WT mice after running (Figure 7A and Table 2), and the area of cardiomyocytes from Atrogin-1–/– hearts was 64% greater than control after exercise (Figure 7B). These observations correspond to the increased LV mass index and wall thickness in Atrogin-1–/– mice noted by echocardiography after, but not before, running (Table 2, Figure 7C). Because amounts of ubiquitylated Foxo proteins were at the limits of detection in WT hearts, it was technically not feasible to measure decreases in their modification in Atrogin-1–/– hearts. Nevertheless, our overexpression and underexpression data in vivo collectively correspond closely to our in vitro observations and indicate that atrogin-1 enhances the activation of Forkhead transcription factors Foxo1 and Foxo3a and concomitantly suppresses Akt-dependent cardiomyocyte hypertrophy, indicating what we believe to be a new role for noncanonical ubiquitylation in a pathophysiologically relevant context.
Table 2 .
Echocardiography and run statistics of Atrogin-1–/– and littermate Atrogin-1+/+ mice at baseline and after 3 wk voluntary running
Figure 7. Lack of atrogin-1 expression results in exaggerated cardiac hypertrophy in response to voluntary running exercise.
(A) Representative macroscopic histologic analysis of H&E-stained mouse hearts after 3 wk voluntary wheel exercise. Scale bar: 1 mm. Histologic sections were also stained with wheat germ agglutinin-TRITC to determine cell size. Scale bars: 1 mm (top); 50 μm (bottom). (B) Quantitation of cardiomyocyte size from WT and Atrogin-1–/– heart after 3 wk voluntary wheel exercise and sham controls. (C) M-mode echocardiographic analysis of hearts from WT and Atrogin-1–/– mice after 3 wk voluntary wheel exercise.
Discussion
Because cardiac hypertrophy eventually plateaus even in the setting of continued prohypertrophic stimulation and is reversible when these stimuli abate, it is evident that the regulation of cardiomyocyte size depends on the balance between factors that either promote or antagonize cellular enlargement. Our previous studies identified a role for atrogin-1 in limiting pathological cardiomyocyte hypertrophic responses in vivo and in vitro by targeting calcineurin for proteasome-dependent degradation (8), which provides a regulatory mechanism to counterbalance pro- and antigrowth mechanisms in cardiac hypertrophy (12). The results of our present studies indicate that this counterregulatory role for atrogin-1 exists for calcineurin-independent cardiac growth as well. Akt is a proximal component of the physiologic, calcineurin-independent hypertrophic pathway (1). We found using a combination of knockdown and overexpression approaches that atrogin-1 has no effect on Akt activation in response to IGF-1 or insulin challenge in cardiomyocytes, but nevertheless represses Akt-dependent hypertrophy by activating the Forkhead transcription factors. The transcriptional coactivation of Foxo1 and Foxo3a by atrogin-1 and upregulation of Forkhead-dependent transcriptional targets allows the heart to uncouple the inhibition of Akt-dependent hypertrophic signaling from other essential effects of Akt on cell survival and metabolism.
Activated Forkhead transcription factors participate in repression of both Akt-dependent and calcineurin-dependent hypertrophy (12, 13). Thus, whereas prohypertrophic signaling is segregated exquisitely in physiologic and pathologic circumstances (1), the pathways that antagonize cardiac hypertrophic responses through transcriptional (Forkhead proteins) and protein stability (atrogin-1) mechanisms are common to both types of cardiac hypertrophy. This shared role is consistent with studies implicating both protein synthesis and protein degradation in antigrowth mechanisms in the heart in vivo (20). Importantly, because Forkhead proteins regulate atrogin-1 expression in skeletal and cardiac muscle (11, 12), our results indicate the presence of a feed-forward mechanism in which atrogin-1 is activated by, and in turn coactivates, Foxo3a and Foxo1. The presence of such a mechanism suggests that postnatal cardiac growth is carefully regulated at several levels through inhibitory processes, with atrogin-1 acting as both a transcriptional coactivator and ubiquitin ligase to coordinate multiple steps in this process. These studies also suggest the existence of additional layers of regulation that suppress this feed-forward pathway, and the elucidation of these mechanisms remains a fruitful topic for further inquiry. One potential mechanism to regulate this feedforward loop is through Foxo-dependent activation of Akt itself, which has recently been described (13, 21).
Because canonical ubiquitylation of proteins via lysine 48–linked chains targets proteins for degradation via the proteasome, the observation that atrogin-1 binds and activates the Forkhead transcription factors raised the possibility of non–proteasome-dependent activities for atrogin-1. The Forkhead transcription factors are regulated through several well-defined posttranslational mechanisms. When phosphorylated by Akt, these proteins are retained in the cytoplasm, where they are transcriptionally inactive and susceptible to proteasome-dependent degradation that is triggered by a Skp2-containing ubiquitin ligase (22). In contrast, the dephosphorylated forms of Forkhead proteins are nuclear localized and transcriptionally active, and p300-dependent acetylation additionally enhances their activity (14, 23). Atrogin-1 blocks Akt-dependent Foxo1 and Foxo3a phosphorylation and enforces their localization in the nucleus, which may account in part for the transcriptional effects of atrogin-1 on these proteins. However, atrogin-1 also enhances the transcriptional activity of phosphorylation-defective constitutively active mutants of Foxo1 and Foxo3a that spontaneously localize to the nucleus, indicating that atrogin-1 modifies the activity of Forkhead proteins within the nucleus directly and not simply as a consequence of altered subcellular trafficking.
The coactivation of transcriptionally active proteins by ubiquitin ligases may seem paradoxical, but there is precedent for ubiquitin modifications regulating transcription. Monoubiquitylation of histones, transcription factors, and components of the core RNA polymerase machinery is a well-described mechanism for regulation of transcription (24–27). Indeed, the Forkhead protein Foxo4 was recently shown to be monoubiquitylated in response to hydrogen peroxide stimulation (28). However, the effects of atrogin-1 on Foxo1 and Foxo3a we observed here are distinctive in that atrogin-1 regulation is dependent on assembly of lysine 63–linked ubiquitin chains. Lysine 63–linked ubiquitin chains have typically been associated with intracellular signaling events by serving as a signal for recruitment of accessory proteins (15), but a clear role for this noncanonical ubiquitylation event as a regulatory mechanism for mammalian transcription factors has not been established. The promiscuity of atrogin-1 with respect to chain linkage topology is not unprecedented for a ubiquitin ligase (29, 30) and provides an elegant means for a ubiquitin ligase to mediate activities that are both dependent on and independent of proteasomes. In the case of atrogin-1, the ability to assemble ubiquitin chains of different linkages permits the inactivation of calcineurin through targeting to the proteasome at the same time that transcriptional activation of Forkhead transcription factors is favored. The assembly of noncanonical ubiquitin chains by atrogin-1 provides what we believe to be a new muscle-specific regulatory modification that regulates nuclear functions to suppress hypertrophic signaling within the heart.
Methods
Plasmids and antibodies.
The WT and mutants of atrogin-1, His- or HA-ubiquitin, Flag-Foxo1, and HA-Foxo1A3 have been described previously (8, 14). Flag-Foxo1 mutants (1–593, 1–517, 1–417, 1–261, and S256A) were gifts from D.J. Tindall (Mayo Clinic, Rochester, Minnesota, USA). HA-Foxo3a and Foxo3aA3, 6xDBE- and Bim-luciferase reporters were gifts from L. Guarente (Massachusetts Institute of Technology, Cambridge, Massachusetts, USA), and pGL2-p27kip1 luciferase reporter was a gift from T. Sakai (Kyoto Prefectural University of Medicine, Kyoto, Japan). siRNA sequences for atrogin-1 were previously described (8). Antibodies used were as follows: anti-Flag (Sigma-Aldrich); anti-Myc (clone 9E10; Santa Cruz Biotechnology Inc.); anti-HA (clone 12CA5; Roche Diagnostics); anti-Akt, anti–phospho-Akt (Ser473), anti–phospho-Akt (308), anti-mTOR, anti–phospho-mTOR (Ser2448), anti-p70S6K, phospho-p70S6K (Thr389), anti–GSK-3β, anti–phospho-GSK-3β (Ser9), anti-Foxo1, anti–phospho-Foxo1 (Ser256), and anti–phospho-Foxo3 (Thr32; all from Cell Signaling Technology); anti-Foxo3a (06-951; Upstate); anti-ubiquitin and anti-GAPDH (Chemicon International Inc.); and anti-mouse– or anti-rabbit–conjugated antibodies (Invitrogen).
Cell culture and adenovirus infection.
We cultured 293 cells in DMEM supplemented with 10% FBS at 37°C. Neonatal rat cardiomyocytes were isolated by enzymatic disassociation of 1- to 2-d-old neonatal rat hearts and infected with adenoviruses expressing vectors as previously described (8). Recombinant adenoviruses expressing GFP alone (Ad-GFP), Myc-tagged atrogin-1 and GFP (Ad–atrogin-1–GFP), siRNA-control, or siRNA–atrogin-1 driven by the cytomegalovirus promoter were generated using the AdEasy system (MP Biomedicals Inc.) as described previously (31). To induce the hypertrophic response, IGF-1, insulin, or PE was added to cultures at concentrations indicated in serum-free media for 24 h.
IP, Western blotting, and GST pulldown assays.
IP and IB were performed as described previously (8). Briefly, 293 cells were cotransfected with expression vectors for HA- or Myc-tagged atrogin-1, Flag-tagged Foxo1, or HA-tagged Foxo3 using FuGENE 6 (Roche Diagnostics). Tagged proteins were immunoprecipitated for 2 h at 4°C with the appropriate antibody. Beads were washed and analyzed by IB. GST pulldown assays were performed as described previously (8). Briefly, 293 cells were transfected with a Flag-Foxo1 or HA-Foxo3a expression plasmid for 36 h, and cells were lysed for 30 min in lysis buffer. Lysates were precleared with GST beads for 1 h and incubated with GST or GST–atrogin-1 fusion proteins for 1 h at 4°C. The bound beads were washed 4 times with lysis buffer and analyzed by IB.
Luciferase assays.
Luciferase activity assays were performed using the Luciferase Assay System (Promega) as described previously (8). Neonatal cardiomyocytes were transiently cotransfected with expression vectors carrying atrogin-1 or ΔF-box mutant, Foxo1 or Foxo3a, atrogin-1 siRNA or control vector, and luciferase reporter constructs using FuGENE 6 (Roche Diagnostics) or Lipofectamine 2000 (Invitrogen). Cells were lysed and assayed for luciferase activity 24–36 h after transfection. Data represent mean ± SEM of 3 independent experiments performed in duplicate and normalization for β-gal activity.
Immunofluorescence.
Cardiomyocytes were processed for immunofluorescence as described previously (8). Visualization of cardiomyocyte size was performed by immunostaining with anti–α-actinin antibodies. Fluorescent images were collected on an epifluorescence microscope (Eclipse E800; Nikon Inc.). Quantitation of cardiomyocyte cell surface area was performed on digitized images using NIH Image software. We examined 100 cardiomyocytes in 15–20 fields in 3 independent experiments.
In vivo ubiquitylation assays.
To assess ubiquitylation in vivo, 293 cells were transfected with expression vectors containing His-ubiquitin or HA-ubiquitin, Flag-Foxo1, HA-Foxo3a, and Myc–atrogin-1 using FuGENE 6. Lysate proteins were precipitated and analyzed by IB using appreciate antibodies as previously described (8).
In vitro ubiquitylation reactions.
For atrogin-1–mediated ubiquitylation, Skp1, Cul1, and Roc1 immunocomplexes were precipitated from transfected 293 cells with 2 μg anti-Myc antibody as described previously (8). Purified Foxo1 or Foxo3a and immunocomplexes were incubated in 30 μl ubiquitin reaction buffer, 60 ng E1, 600 ng Ubc13, and 10 μg ubiquitin in the presence of bacterially purified GST only, GST–atrogin-1, or GST–atrogin-1 ΔF-box (1 μg). After incubation at 30°C for 2 h, reaction products were analyzed by IB using the indicated antibodies.
Pulse-chase analysis.
Pulse-chase analysis was performed as described previously (25). Briefly, 293 cells were transfected with the indicated plasmid and then incubated for 2 h at 37°C in methionine- and cysteine-free media, followed by pulse-labeling for 15 min with 1 mCi/ml–1 35S-methionine-cysteine (Trans label; ICN Biochemicals). At the indicated times, cells were washed twice in PBS and lysed in IP lysis buffer. Lysates were subjected to IP using the anti-Flag or anti-HA antibodies. Immunoprecipitates were analyzed by SDS-PAGE followed by autoradiography.
Unloaded voluntary wheel exercise–induced cardiac hypertrophy.
Atrogin-1–/– mice backcrossed approximately 7 generations onto C57BL/6 mice were used as previously described (9). Heterozygous mice were used to generate Atrogin-1–/– and littermate Atrogin-1+/+ mice for the experiments described. All animal experiments were approved by the Institutional Animal Care and Use Committee of the University of North Carolina at Chapel Hill. Physiologic hypertrophy was induced by unloaded voluntary wheel running in female mice as previously described (32). Briefly, Atrogin-1–/– mice were assigned to either no-resistance wheel running (n = 6) or sedentary control (n = 3). WT littermates (n = 5) were assigned to no-resistance wheel running. Exercise wheels were equipped with a Mity 8 Cyclocomputer (model CC-MT400), which recorded distance, average speed, running time, and maximum speed. Voluntary running began at an average age of about 12 wk for all groups. Mice underwent echocardiography at baseline, ran voluntarily for 21 d, and underwent a final echocardiography before harvest. After 21 d of nonloaded, voluntary wheel running, mice underwent echocardiography followed by sacrifice via isoflurane inhalation overdose and subsequent cervical dislocation.
IGF-1/GH-induced cardiac hypertrophy.
The IGF-1/GH infusion model of hypertrophy was previously described (17). GH (Nutropin AQ; Genentech) and Long R3-IGF-1 (JRH Biosciences) were infused at 4 mg/kg each twice daily via subcutaneous injection. Echocardiography was performed with a Vevo 660 ultrasound system (VisualSonics Inc.) equipped with a 30-MHz transducer. All procedures were approved by and performed in accordance with the University of North Carolina Institutional Animal Care and Use Committee. Histologic analyses of heart tissues were performed according to standard protocols (8). Samples were stained with H&E for routine histologic examination and wheat germ agglutinin–TRITC conjugate to identify sarcolemmal membranes so that myofiber diameter could be quantified. Total RNA was purified from cultured cells or intact hearts with TRI zol (Invitrogen). For RT-PCR, the expression levels of Bim, p27kip1, GADD45, SOD2, and GAPDH (as a control) were measured with primers designed to detect mouse gene products as described previously (33).
Statistics.
Data are presented as mean ± SEM. Differences between groups were evaluated for statistical significance using 2-tailed Student’s t test. A P value less than 0.05 was considered significant. A 1-way analysis of variance was used to determine significant differences between groups in the voluntary running data. A pairwise multiple comparison was employed using the Holm-Sidak method to determine statistical significance between groups (Sigma Stat 3.5; Systat Inc.).
Supplementary Material
Acknowledgments
This study was supported by grants from the NIH (HL65619 and HL072347; to C. Patterson) and the China Natural Science Foundation (30570721; to H.-H. Li). C. Patterson is an Established Investigator of the American Heart Association and a Burroughs Wellcome Fund Clinical Scientist in Translational Research.
Footnotes
Nonstandard abbreviations used: Ad, adenovirus; GH, growth hormone; GSK-3β, glycogen synthase kinase–3β; GST, glutathione-S-transferase.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 117:3211–3223 (2007). doi:10.1172/JCI31757
Hui-Hua Li and Monte S. Willis contributed equally to this work.
References
- 1.Heineke J., Molkentin J.D. Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat. Rev. Mol. Cell Biol. 2006;7:589–600. doi: 10.1038/nrm1983. [DOI] [PubMed] [Google Scholar]
- 2.Hardt S.E., Sadoshima J. Negative regulators of cardiac hypertrophy. Cardiovasc. Res. 2004;63:500–509. doi: 10.1016/j.cardiores.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 3.Wilkins B.J., et al. Calcineurin/NFAT coupling participates in pathological, but not physiological, cardiac hypertrophy. Circ. Res. 2004;94:110–118. doi: 10.1161/01.RES.0000109415.17511.18. [DOI] [PubMed] [Google Scholar]
- 4.Taniyama Y., et al. Akt3 overexpression in the heart results in progression from adaptive to maladaptive hypertrophy. J. Mol. Cell. Cardiol. 2005;38:375–385. doi: 10.1016/j.yjmcc.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 5.Matsui T., Nagoshi T., Rosenzweig A. Akt and PI 3-kinase signaling in cardiomyocyte hypertrophy and survival. Cell Cycle. 2003;2:220–223. [PubMed] [Google Scholar]
- 6.Hardt S.E., Sadoshima J. Glycogen synthase kinase-3beta: a novel regulator of cardiac hypertrophy and development. Circ. Res. 2002;90:1055–1063. doi: 10.1161/01.res.0000018952.70505.f1. [DOI] [PubMed] [Google Scholar]
- 7.Kim Y.B., Peroni O.D., Franke T.F., Kahn B.B. Divergent regulation of Akt1 and Akt2 isoforms in insulin target tissues of obese Zucker rats. Diabetes. 2000;49:847–856. doi: 10.2337/diabetes.49.5.847. [DOI] [PubMed] [Google Scholar]
- 8.Li H.H., et al. Atrogin-1/muscle atrophy F-box inhibits calcineurin-dependent cardiac hypertrophy by participating in an SCF ubiquitin ligase complex. J. Clin. Invest. 2004;114:1058–1071. doi: 10.1172/JCI200422220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bodine S.C., et al. Identification of ubiquitin ligases required for skeletal muscle atrophy. Science. 2001;294:1704–1708. doi: 10.1126/science.1065874. [DOI] [PubMed] [Google Scholar]
- 10.Gomes M.D., Lecker S.H., Jagoe R.T., Navon A., Goldberg A.L. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc. Natl. Acad. Sci. U. S. A. 2001;98:14440–14445. doi: 10.1073/pnas.251541198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sandri M., et al. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117:399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ni Y.G., et al. Foxo transcription factors blunt cardiac hypertrophy by inhibiting calcineurin signaling. Circulation. 2006;114:1159–1168. doi: 10.1161/CIRCULATIONAHA.106.637124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skurk C., et al. The FOXO3a transcription factor regulates cardiac myocyte size downstream of AKT signaling. J. Biol. Chem. 2005;280:20814–20823. doi: 10.1074/jbc.M500528200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Motta M.C., et al. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116:551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- 15.Wu C.J., Conze D.B., Li T., Srinivasula S.M., Ashwell J.D. Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-kappaB activation [corrected]. Nat. Cell Biol. 2006;8:398–406. doi: 10.1038/ncb1384. [DOI] [PubMed] [Google Scholar]
- 16.Kirkpatrick D.S., et al. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat. Cell Biol. 2006;8:700–710. doi: 10.1038/ncb1436. [DOI] [PubMed] [Google Scholar]
- 17.Tanaka N., et al. Effects of growth hormone and IGF-I on cardiac hypertrophy and gene expression in mice. Am. J. Physiol. 1998;275:H393–H399. doi: 10.1152/ajpheart.1998.275.2.H393. [DOI] [PubMed] [Google Scholar]
- 18.DeBosch B., et al. Akt1 is required for physiological cardiac growth. Circulation. 2006;113:2097–2104. doi: 10.1161/CIRCULATIONAHA.105.595231. [DOI] [PubMed] [Google Scholar]
- 19.Muslin A.J., DeBosch B.2006Role of Akt in cardiac growth and metabolism. Novartis Found. Symp. 274118–126; discussion 126–131, 152–115, 272–116. . [PubMed] [Google Scholar]
- 20.Razeghi P., et al. Atrophic remodeling of the heart in vivo simultaneously activates pathways of protein synthesis and degradation. Circulation. 2003;108:2536–2541. doi: 10.1161/01.CIR.0000096481.45105.13. [DOI] [PubMed] [Google Scholar]
- 21.Matsumoto M., Han S., Kitamura T., Accili D. Dual role of transcription factor FoxO1 in controlling hepatic insulin sensitivity and lipid metabolism. J. Clin. Invest. 2006;116:2464–2472. doi: 10.1172/JCI27047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang H., et al. Skp2 inhibits FOXO1 in tumor suppression through ubiquitin-mediated degradation. Proc. Natl. Acad. Sci. U. S. A. 2005;102:1649–1654. doi: 10.1073/pnas.0406789102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuzaki H., et al. Acetylation of Foxo1 alters its DNA-binding ability and sensitivity to phosphorylation. Proc. Natl. Acad. Sci. U. S. A. 2005;102:11278–11283. doi: 10.1073/pnas.0502738102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Osley M.A., Fleming A.B., Kao C.F. Histone ubiquitylation and the regulation of transcription. Results Probl. Cell Differ. 2006;41:47–75. doi: 10.1007/400_006. [DOI] [PubMed] [Google Scholar]
- 25.Bres V., et al. A non-proteolytic role for ubiquitin in Tat-mediated transactivation of the HIV-1 promoter. Nat. Cell Biol. 2003;5:754–761. doi: 10.1038/ncb1023. [DOI] [PubMed] [Google Scholar]
- 26.Muratani M., Kung C., Shokat K.M., Tansey W.P. The F box protein Dsg1/Mdm30 is a transcriptional coactivator that stimulates Gal4 turnover and cotranscriptional mRNA processing. Cell. 2005;120:887–899. doi: 10.1016/j.cell.2004.12.025. [DOI] [PubMed] [Google Scholar]
- 27.Muratani M., Tansey W.P. How the ubiquitin-proteasome system controls transcription. Nat. Rev. Mol. Cell Biol. 2003;4:192–201. doi: 10.1038/nrm1049. [DOI] [PubMed] [Google Scholar]
- 28.van der Horst A., et al. FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat. Cell Biol. 2006;8:1064–1073. doi: 10.1038/ncb1469. [DOI] [PubMed] [Google Scholar]
- 29.Zhang B.H., et al. Serum- and glucocorticoid-inducible kinase SGK phosphorylates and negatively regulates B-Raf. J. Biol. Chem. 2001;276:31620–31626. doi: 10.1074/jbc.M102808200. [DOI] [PubMed] [Google Scholar]
- 30.Jiang J., et al. CHIP is a U-box-dependent E3 ubiquitin ligase: identification of Hsc70 as a target for ubiquitylation. J. Biol. Chem. 2001;276:42938–42944. doi: 10.1074/jbc.M101968200. [DOI] [PubMed] [Google Scholar]
- 31.Dai Q., et al. CHIP activates HSF1 and confers protection against apoptosis and cellular stress. EMBO J. 2003;22:5446–5458. doi: 10.1093/emboj/cdg529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Konhilas J.P., et al. Loaded wheel running and muscle adaptation in the mouse. Am. J. Physiol. Heart Circ. Physiol. 2005;289:H455–H465. doi: 10.1152/ajpheart.00085.2005. [DOI] [PubMed] [Google Scholar]
- 33.Nemoto S., Fergusson M.M., Finkel T. Nutrient availability regulates SIRT1 through a forkhead-dependent pathway. Science. 2004;306:2105–2108.. doi: 10.1126/science.1101731. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.