Abstract
Histamine receptor H1 (H1R) is a susceptibility gene in both experimental autoimmune encephalomyelitis (EAE) and experimental autoimmune orchitis (EAO), 2 classical T cell–mediated models of organ-specific autoimmune disease. Here we showed that expression of H1R in naive CD4+ T cells was required for maximal IFN-γ production but was dispensable for proliferation. Moreover, H1R signaling at the time of TCR ligation was required for activation of p38 MAPK, a known regulator of IFN-γ expression. Importantly, selective reexpression of H1R in CD4+ T cells fully complemented both the IFN-γ production and the EAE susceptibility of H1R-deficient mice. These data suggest that the presence of H1R in CD4+ T cells and its interaction with histamine regulates early TCR signals that lead to Th1 differentiation and autoimmune disease.
Introduction
Histamine [2-(4-imidazole) ethylamine] is a ubiquitous mediator of diverse physiological processes including neurotransmission and brain functions, secretion of pituitary hormones, and regulation of gastrointestinal and circulatory functions (1). Additionally, histamine is a potent mediator of inflammation and a regulator of innate and adaptive immune responses (2). Histamine exerts its effect through 4 receptors that belong to the 7-transmembrane G protein–coupled receptor family and are designated histamine receptor H1 (H1R), H2R, H3R, and H4R, according to the chronological order of their discovery (1, 3).
H1R couples to second messenger signaling pathways via the activation of the heterotrimeric Gαq/11 family of G proteins (1). Generally, activation of H1R leads to stimulation of phospholipase C, resulting in the hydrolysis of phosphatidylinositol 4,5-bisphosphate (PIP2) to form inositol-1,4,5-trisphosphate (IP3) and diacylglycerol (DAG), which cause calcium mobilization from intracellular stores and activation of PKC, respectively (3). In addition, H1R signaling also mediates other signaling pathways such as the production of cGMP, arachidonic acid, and nitric oxide (4), and the activation of NF-κB (5), STAT1 (6), STAT4 (7), and MAPK pathways (8–10). However, even though the H1R was the first histamine receptor to be identified and a large number of studies on histamine and H1R have been published in the last decade (11), little is known about the cell type–specific H1R signaling pathways.
In the immune system, histamine has been reported to be a potent modulator of innate and adaptive immune responses. Histamine, acting through H1R, affects the maturation of dendritic cells and alters their T cell–polarizing activity (12, 13). It regulates antigen-specific T cell effector functions and the related antibody isotype response (14). H1R signaling in splenocytes has been reported to modulate cytokine secretion by these cells (14–17), but to our knowledge, no study has addressed the role of H1R in purified CD4+ T cells.
We have previously demonstrated that Hrh1 (encoding the mouse H1R protein) is a shared susceptibility gene in experimental autoimmune orchitis (EAO) and in EAE, the autoimmune model of MS (17). In both MS and EAE, CD4+ T cells secreting IFN-γ (Th1) (18) and/or IL-17 (Th17) (19) are necessary and sufficient for eliciting EAE pathology and clinical signs. The relative contributions of each of these cytokines to the development of EAE in vivo are debated, because conflicting evidence exists on the importance of IFN-γ versus IL-17. On the one hand, the importance of IL-17 has been established in studies showing that EAE is diminished in IL-23–deficient but not IL-12–deficient animals (with no expression of the Th17-promoting or Th1-promoting cytokines, respectively) (20), and severe EAE has been observed in IFN-γ knockout mice and IFN-γR knockout mice (21, 22). These findings contrast with studies showing that either CD4+ Th1 cells (18) or CD4+ Th17 cells (23) can transfer EAE to naive recipients. Recent studies reporting the predominant presence of a pre-Th1, IFN-γ+IL-17+CD4+ T cell subtype early after induction of EAE with encephalitogenic myelin oligodendrocyte glycoprotein peptide 35–55 (MOG35–55) (24) may help resolve these apparent inconsistencies. Nevertheless, IFN-γ, alone or in conjunction with IL-17, is well established as a cytokine of relevance in EAE immunopathology.
We have previously shown that H1R-deficient (H1RKO) mice exhibit a significant delay in the onset of EAE and a reduction in the severity of the clinical signs compared with WT mice (17). This phenotype is associated with an immune deviation of the elicited CD4+ T cell population from a Th1 response to a Th2 response with no detectable difference in IL-17 secretion, suggesting that the CD4+ Th1 cells and the IFN-γ produced by them play an important role in the pathology of the disease. In this report, we studied the mechanism underlying the immune deviation and demonstrate that it was directly caused by H1R regulation of cytokine responses in CD4+ T cells, not by H1R expression in APCs. We also show that H1R was expressed in unstimulated CD4+ T cells but was rapidly downregulated upon activation. H1R was required for the activation of the p38 MAPK signaling pathway and for IFN-γ production in response to TCR stimulation in CD4+ T cells. Finally, H1R-mediated signaling in CD4+ T cells, independent of APCs, regulated the encephalitogenic Th1 effector cell response in EAE.
Results
H1R expression is required for IFN-γ production by CD4+ T cells.
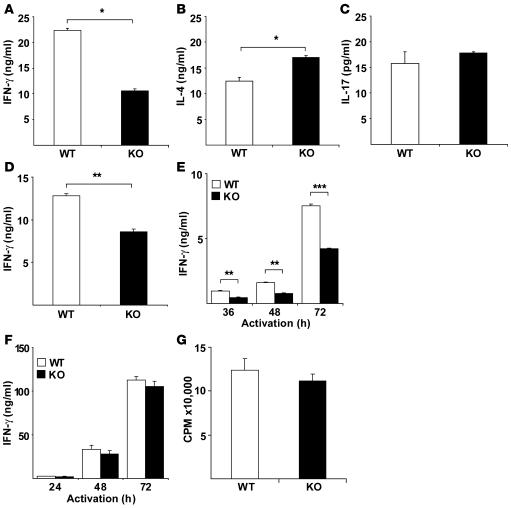
MOG35–55 peptide–immunized H1RKO splenocytes produce less IFN-γ and more IL-4 than do splenocytes from immunized WT mice (17). However, it is not clear whether this immune deviation is caused by lack of H1R signaling in CD4+ T cells or in APCs. To investigate the role of H1R in regulating IFN-γ production and Th1 differentiation, CD4+ T cells were purified from WT and H1RKO mice and activated with anti-CD3 and anti-CD28 mAbs in the presence of recombinant IL-12 and neutralizing anti–IL-4 mAb. After 4 days, Th1 effector cells were extensively washed and counted, and equal numbers of cells were restimulated with anti-CD3 mAb for 24 h. Th1 effector cells from H1RKO mice produced considerably less IFN-γ than did WT Th1 cells (Figure 1A). We also examined the production of IL-4 upon restimulation of Th2 effector cells generated in the presence of IL-4 and anti–IFN-γ mAb. A marginal increase in IL-4 production was observed in cells from H1RKO mice compared with cells from WT mice (Figure 1B). Recent studies have established IL-17 as an important cytokine in EAE (20). Consequently, we examined IL-17 production by Th17 cells generated in the presence of IL-6 and TGF-β and anti–IFN-γ and anti–IL-4 mAbs. There was no difference in IL-17 production between H1RKO and WT Th17 cells (Figure 1C). Moreover, we examined the role of H1R in nonpolarized effector cells, generated by stimulating cells in the absence of exogenous cytokines for 4 days. Effector cells were then restimulated with anti-CD3 mAb for 24 h. CD4+ T effector cells from H1RKO mice produced significantly less IFN-γ than did cells from WT mice (Figure 1D). Thus, under these conditions, IFN-γ production in H1RKO effector CD4+ T cells was impaired.
Figure 1. H1R is required for IFN-γ production by CD4+ T cells.
Purified CD4+ T cells from WT and H1RKO mice were activated with anti-CD3 (5 μg/ml) and anti-CD28 (1 μg/ml) mAbs either (A) in the presence of IL-12 (4 ng/ml) and anti–IL-4 mAb (10 μg/ml), (B) in the presence of IL-4 (30 ng/ml) and anti–IFN-γ mAb (10 μg/ml), or (C) in the presence of TGF-β (1 ng/ml), IL-6 (30 ng/ml), and anti–IFN-γ (10 μg/ml) and anti-IL4 mAbs (10 μg/ml). After 4 days, the cells were restimulated with anti-CD3 mAb (5 μg/ml) for 24 h. Production of (A) IFN-γ, (B) IL-4, and (C) IL-17 was determined by ELISA in triplicate. *P < 0.05, Student’s t test. (D) CD4+ T cells from WT and H1RKO mice were activated with anti-CD3 (5 μg/ml) and anti-CD28 (1 μg/ml) mAbs. After 4 days, cells were restimulated with anti-CD3 mAb (5 μg/ml) for 24 h, and IFN-γ production was determined by ELISA. **P = 0.002, Student’s t test. (E and F) WT and H1RKO CD4+ T cells were stimulated as in D for the indicated periods of time. Supernatants were analyzed for (E) IFN-γ (F = 168.8, P < 0.0001, 2-way ANOVA; **P < 0.01, ***P < 0.001, Bonferroni corrected post-hoc comparison) and (F) IL-2 by ELISA. (G) CD4+ T cells from WT and H1RKO mice were stimulated as in E, and 18 h [3H]-thymidine incorporation was measured in total 72 h culture. Data are representative of at least 2 independent experiments.
IFN-γ production by CD4+ T cells contributes to their differentiation into Th1 effector cells (25). To examine the role of H1R signaling in this process, purified CD4+ T cells from H1RKO and WT mice were stimulated with anti-CD3 and anti-CD28 mAbs for different periods of time, and IFN-γ production was quantified. CD4+ T cells from H1RKO mice produced significantly lower IFN-γ than did those from WT mice at all time points examined (Figure 1E). In contrast, no difference in IL-2 production between WT and H1RKO CD4+ T cells was observed (Figure 1F). Furthermore, proliferation was comparable between WT and H1RKO CD4+ T cells (Figure 1G). Taken together, these results demonstrated that H1R expression in CD4+ T cells plays a critical role in regulating IFN-γ production during the activation and differentiation of these cells.
Hrh1 expression is downregulated early upon TCR activation.
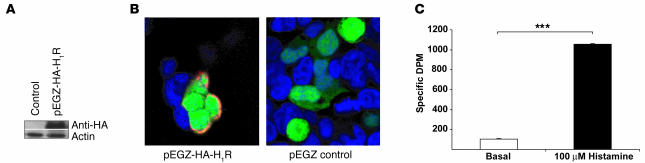
In order to demonstrate that the reduced secretion of IFN-γ by CD4+ T cells is caused by the absence of a functional H1R in these cells, we carried out H1R complementation in H1RKO CD4+ T cells by retroviral transduction. We generated a retroviral construct using the pEGZ-HA vector where H1R was subcloned downstream of a HA tag and upstream of IRES-EGFP. To confirm that the HA-H1R was properly expressed, we transiently transfected HEK293T cells with the pEGZ-HA-H1R construct and examined its expression by Western blot analysis using anti-HA mAb. A band corresponding to the HA-H1R size (~55 kDa) was present only in HA-H1R–transfected cells (Figure 2A). To demonstrate that the HA-H1R was expressed on the cytoplasmic membrane, the HA-H1R–transfected HEK293T cells were stained using anti-HA mAb and examined by confocal microscopy. HA-H1R was indeed found to be expressed on the cytoplasmic membrane only in HA-H1R–transfected cells (Figure 2B).
Figure 2. Expression and function of HA-H1R in HEK293T cells.
(A) HEK293T cells were transfected with empty pEGZ (control) and pEGZ-HA-H1R plasmids, and the expression of HA-H1R was determined by Western blot using an anti-HA mAb. Data are representative of at least 3 independent experiments. (B) HEK293T cells were transfected as in A, fixed, permeabilized, and stained with an anti-HA mAb (red) and Topro-nuclear dye (blue). EGFP expression (green) represents transfected cells. Cells were visualized by confocal microscopy. Data are representative of at least 3 independent experiments. (C) HEK293 cells were transfected with pHA-H1R-Gα11 fusion construct, and membrane fractions were isolated and used in [35S]GTPγS binding assays in the absence (basal) or presence of 10–4 M histamine. Samples were then used in immunoprecipitation using Gα11 antiserum, and the bound [35S]GTPγS was measured by liquid-scintillation spectrometry. ***P < 0.001, Student’s t test.
H1R coupling to second messenger pathways is primarily via Gαq/11 (1). The ability of the transfected HA-H1R to activate Gα11 was tested in a [35S]GTPγS binding assay. When membrane fractions from transfected HEK293 cells were used in the [35S]GTPγS binding assay, HA-H1R activated Gα11 in response to histamine (Figure 2C). Taken together, these results show that HA-H1R is properly expressed and is functional.
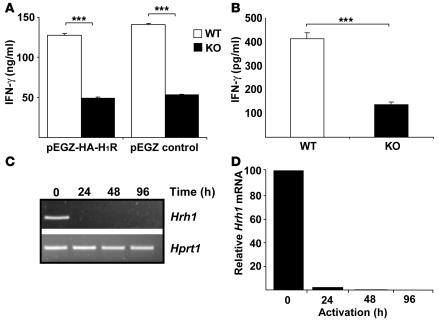
We next performed retroviral transduction: CD4+ T cells were isolated from H1RKO and WT mice, activated with anti-CD3 and anti-CD28 mAbs for 16 h, and transduced with either pEGZ or pEGZ-HA-H1R retroviruses. Expression of HA-H1R in transduced CD4+ T cells was confirmed by confocal microscopy and flow cytometry (data not shown). After 2 days, transduced CD4+ T cells were isolated by cell sorting based on EGFP expression, and equal numbers of cells were activated with anti-CD3 mAb for an additional 24 h. Both pEGZ- and pEGZ-HA-H1R–transduced CD4+ T cells from H1RKO mice produced significantly lower levels of IFN-γ than did those from WT mice (Figure 3A). These results indicate that the expression of H1R in activated CD4+ T cells does not restore IFN-γ production by H1R-deficient cells.
Figure 3. Hrh1 is downregulated upon activation in CD4+ T cells.
(A) CD4+ T cells from WT and H1RKO mice were stimulated in the presence of anti-CD3 and anti-CD28 mAbs for 16 h and then retrovirally transduced with pEGZ-HA-H1R or with empty pEGZ control plasmids. Transduced, sorted EGFP+ cells were then restimulated with anti-CD3 mAb, and 24 h later, the supernatants were harvested for determination of IFN-γ by ELISA in triplicate. Data are representative of 2 independent experiments. ***P < 0.001, Student’s t test. (B) Freshly isolated CD4+ T cells from WT and H1RKO mice were activated with anti-CD3 and anti-CD28 mAbs. After 24 h, IFN-γ production was determined by ELISA. Data are representative of at least 3 independent experiments. ***P < 0.001, Student’s t test. (C and D) CD4+ T cells were isolated from WT mice and stimulated with anti-CD3 and anti-CD28 mAbs. Cells were harvested at the indicated time points; total RNA was isolated and used to examine Hrh1 expression by conventional RT-PCR with Hprt1 as the endogenous control (C) and by quantitative real-time RT-PCR relative to B2m as the endogenous control (D). Data (representative of at least 3 independent experiments) are presented as expression relative to the unstimulated CD4+ T cells.
Retroviral transduction requires prior activation of CD4+ T cells for at least 16 h to induce cell cycling. Thus, if H1R is normally required during the early phase of activation concomitant with TCR engagement, the retroviral transduction would not rescue the H1R deficiency. The results presented above (Figure 1C) indicated that IFN-γ production was already reduced at 36 h in H1RKO CD4+ T cells compared with WT cells. We therefore examined IFN-γ production by H1RKO CD4+ T cells earlier during activation with anti-CD3 and anti-CD28 mAbs. Although lower levels of IFN-γ were present in WT CD4+ T cells at 24 h of activation, H1RKO CD4+ T cells still produced significantly less IFN-γ (Figure 3B), indicating that H1R plays a role early during the activation of CD4+ T cells.
To our knowledge, H1R expression during mouse T cell activation has not previously been investigated. We therefore analyzed Hrh1 expression in WT CD4+ T cells stimulated for different periods of time with anti-CD3 and anti-CD28 mAbs. Relative levels of Hrh1 were examined by conventional and quantitative real-time RT-PCR. CD4+ T cells markedly downregulated Hrh1 expression by 24 h after activation (Figure 3, C and D), further indicating that H1R plays a role early (i.e., less than 24 h) after TCR engagement and that it is not required for IFN-γ production by CD4+ T cells once the cells are activated.
Selective H1R expression in T cells in Tg mice restores IFN-γ production.
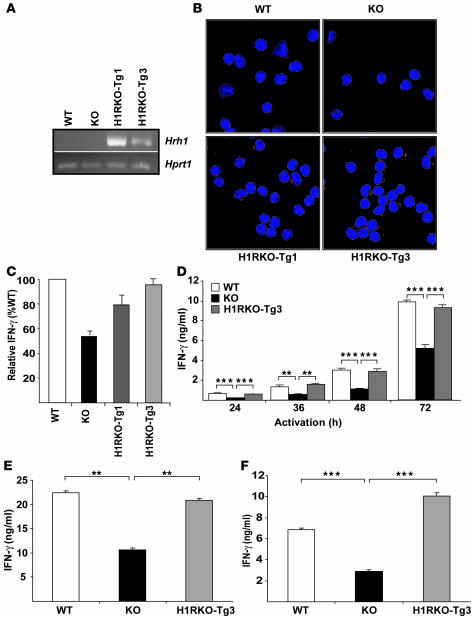
To examine the role of H1R during the initial activation of CD4+ T cells, we generated transgenic mice expressing H1R under the control of distal lymphocyte protein tyrosine kinase (dlck) promoter, which drives expression in T cells (26). Tg mice were generated directly on the C57BL/6J background. Two Tg founders were identified and crossed to H1RKO mice to obtain H1RKO mice expressing H1R selectively in T cells (H1RKO-Tg mice). The expression of the transgene in CD4+ T cells from 2 lines (H1RKO-Tg1 and H1RKO-Tg3) was confirmed by RT-PCR using transgene-specific primers (Figure 4A). We examined the surface expression of the transgene in CD4+ T cells by immunostaining using anti-HA mAb and confocal microscopy (Figure 4B). The transgene was expressed in CD4+ T cells from both transgenic lines. No differences in the total numbers or distribution of T cell subpopulations in the thymus and peripheral lymphoid tissues were observed among the WT, H1RKO, H1RKO-Tg1, and H1RKO-Tg3 lines (data not shown).
Figure 4. Transgenic expression of H1R in H1RKO CD4+ T cells complements IFN-γ production.
(A) Hrh1 transgene expression was analyzed by RT-PCR in CD4+ T cells from WT and H1RKO mice and the 2 independent lines of H1R transgenic mice crossed with H1RKO mice, HIRKO-Tg1 and HIRKO-Tg3. (B) CD4+ T cells were stained with anti-HA mAb (red) and visualized by confocal microscopy. Nuclear stain Topro (blue) is shown. (C) CD4+ T cells were activated with anti-CD3 and anti-CD28 mAbs for 72 h, and IFN-γ was determined by ELISA. Data are expressed as IFN-γ production relative to that by WT cells (set as 100%). (D) CD4+ T cells from WT, H1RKO, and HIRKO-Tg3 mice were stimulated as in C for the indicated periods of time, and IFN-γ was determined by ELISA. F = 55.1, P < 0.0001, 2-way ANOVA. **P < 0.01, ***P < 0.001, Bonferroni corrected post-hoc comparison. (E) CD4+ T cells were activated with anti-CD3 and anti-CD28 mAbs in the presence of IL-12 (4 ng/ml) and anti-IL-4 mAb (10 μg/ml). After 4 days, cells were restimulated and IFN-γ production was determined. F = 25.4, P < 0.001; 1-way ANOVA. **P < 0.01, Bonferroni corrected post-hoc comparison. (F) CD4+ T cells were activated with anti-CD3 and anti-CD28 mAbs. After 4 days, cells were restimulated with anti-CD3 mAb for 24 h, and IFN-γ production was determined by ELISA. F = 288.0, P < 0.0001; 1-way ANOVA. ***P < 0.001, Bonferroni corrected post-hoc comparison. Data are representative of at least 3 independent experiments.
We then examined whether the expression of H1R in H1RKO CD4+ T cells restored IFN-γ production. CD4+ T cells from WT, H1RKO, and H1RKO-Tg mice were stimulated with anti-CD3 and anti-CD28 mAbs, and IFN-γ levels were quantified. The levels of IFN-γ in CD4+ T cells from HIRKO-Tg3 mice were comparable to those of WT CD4+ T cells, whereas IFN-γ production by HIRKO-Tg1 CD4+ T cells remained slightly lower than that in WT CD4+ T cells but nevertheless significantly higher than that in H1RKO CD4+ T cells (Figure 4C). Analyses at different periods of time after activation confirmed that the transgenic expression of H1R in H1RKO-Tg3 CD4+ T cells fully restored IFN-γ production (Figure 4D).
We also studied IFN-γ production by Th1 polarized and nonpolarized effector cells from H1RKO-Tg mice. CD4+ T cells from WT, H1RKO, and H1RKO-Tg mice were differentiated in the absence of exogenous cytokines (nonpolarized) or in the presence of recombinant IL-12 and anti–IL-4 mAb (Th1). After 4 days, effector cells were restimulated with anti-CD3 mAb for 24 h, and IFN-γ production was measured. Compared with H1RKO effectors, both Th1 polarized (Figure 4E) and nonpolarized CD4+ effector T cells (Figure 4F) from H1RKO-Tg3 mice produced significantly more IFN-γ. Furthermore, the levels of IFN-γ in H1RKO-Tg cells were comparable to those in WT CD4+ T cells. Together, these data demonstrate that the presence of H1R at the time of activation of CD4+ T cells under both polarizing and nonpolarizing conditions regulates IFN-γ production and Th1 differentiation.
Impaired activation of p38 MAPK by TCR ligation in H1RKO CD4+ T cells.
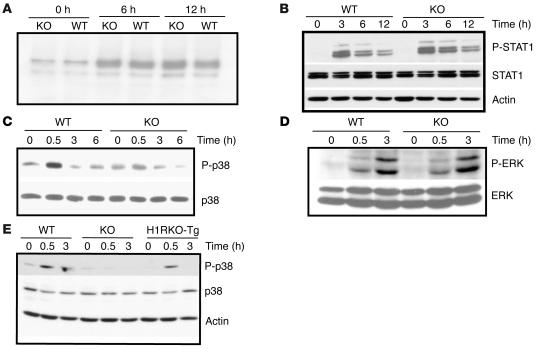
In order to dissect the molecular mechanism of H1R signaling in regulating IFN-γ production by CD4+ T cells, we examined the signaling pathways that have been previously associated with H1R in other cell types. NF-κB has been shown to be activated through H1R in green monkey kidney cells (5) and has been associated with regulation of IFN-γ expression in CD4+ T cells (27). Therefore, we performed EMSA to examine NF-κB DNA binding. CD4+ T cells from WT and H1RKO mice were stimulated with anti-CD3 and anti-CD28 mAbs for different periods of time. A significant difference in NF-κB activation between WT and H1RKO CD4+ T cells was not detected (Figure 5A). STAT1 is also known to regulate IFN-γ expression in CD4+ T cells (28), and it has recently been shown that H1R signaling regulates STAT1 phosphorylation in splenocytes (6). Therefore we examined activation of STAT1 by Western blot analysis in stimulated CD4+ T cells. Prior to 3 h of activation, STAT1 phosphorylation was not detected in WT or H1RKO cells (data not shown); however, phospho-STAT1 was detected after 3 h of activation, but there was no difference in STAT-1 phosphorylation between WT and H1RKO CD4+ T cells (Figure 5B). Although H1R signaling has also been reported to regulate STAT4 phosphorylation in splenocytes (7), phospho-STAT4 was not detected in WT and H1RKO CD4+ T cells after activation with anti-CD3 and anti-CD28 mAbs (data not shown).
Figure 5. Activation of p38 MAPK by TCR ligation requires H1R signals.
(A) Purified CD4+ T cells from WT and H1RKO mice were stimulated with anti-CD3 and anti-CD28 mAbs for the indicated periods of time, and nuclear extracts were prepared and analyzed for NF-κB DNA binding by EMSA. (B) CD4+ T cells from WT and H1RKO mice were stimulated with anti-CD3 and anti-CD28 mAbs for the indicated periods of time, and whole-cell lysates were prepared and analyzed for phospho-STAT1 (P-STAT1) and total STAT1 by Western blot analysis. Actin was used as loading control. (C) CD4+ T cells from WT and H1RKO mice were treated with anti-CD3 and anti-CD28 mAbs for the indicated periods of time, and whole-cell lysates were prepared and analyzed for phospho-p38 MAPK and total p38 by Western blot analysis. (D) CD4+ T cells from WT and H1RKO mice were activated with anti-CD3 and anti-CD28 mAbs for the indicated periods of time, and whole-cell lysates were prepared and analyzed for phospho-ERK and total ERK by Western blot analysis. (E) CD4+ T cells from WT, H1RKO, and HIRKO-Tg3 mice were stimulated with anti-CD3 and anti-CD28 mAbs for the indicated periods of time, and whole-cell lysates were analyzed for phospho-p38, total p38, and actin by Western blotting. Data are representative of at least 2 independent experiments.
H1R ligation has recently been shown to lead to the phosphorylation of p38 MAPK in DDT1MF-2 cells (10) and in human aortic endothelial cells (29). Activation of the p38 MAPK pathway is required for IFN-γ production and Th1 differentiation (30). We therefore examined the activation of p38 MAPK by Western blot analysis. CD4+ T cells from WT and H1RKO mice were stimulated with anti-CD3 and anti-CD28 mAbs for different periods of time. p38 MAPK was activated in WT CD4+ T cells but was markedly impaired in H1RKO CD4+ T cells (Figure 5C). In contrast, no difference in ERK MAPK activation was observed between WT and H1RKO CD4+ T cells (Figure 5D). As we reported previously (31), activation of JNK MAPK was not detected at earlier time points in both WT and H1RKO CD4+ T cells stimulated with anti-CD3 and anti-CD28 mAbs (data not shown). We further examined the activation of p38 MAPK by TCR ligation in H1RKO-Tg CD4+ T cells. Unlike H1RKO CD4+ T cells, the levels of phospho-p38 MAPK in H1RKO-Tg CD4+ T cells were equivalent to those in the WT CD4+ T cells (Figure 5E). Thus, TCR-mediated activation of p38 MAPK required the presence of H1R in CD4+ T cells.
Activation of p38 MAPK by TCR is mediated by histamine/H1R binding.
To understand the mechanism by which H1R may regulate TCR-mediated p38 MAPK activation, we examined whether histamine itself could activate the p38 MAPK in CD4+ T cells. Histamine was already present at low concentrations (about 10–7 M) in the serum used for the culture medium. Therefore, we assessed p38 MAPK phosphorylation in response to histamine using medium prepared with histamine-depleted serum (14). CD4+ T cells from WT and H1RKO mice were resuspended in the histamine-free medium and subsequently treated with histamine. p38 MAPK was activated by histamine in WT CD4+ T cells but not in H1RKO CD4+ T cells (Figure 6A), indicating that histamine activates this pathway in CD4+ T cells through H1R.
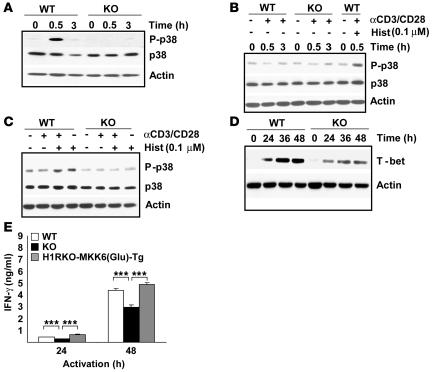
Figure 6. Activation of p38 MAPK by TCR ligation is mediated by histamine/H1R binding.
(A) CD4+ T cells from WT and H1RKO mice were treated with histamine (10–7 M) for the indicated periods of time in the histamine-free medium. Whole-cell extracts were used to analyze phospho-p38, total p38, and actin by Western blotting. (B) CD4+ T cells were isolated from WT and H1RKO mice and stimulated with anti-CD3 and anti-CD28 mAbs in the histamine free-medium for the indicated periods of time. CD4+ T cells stimulated in medium containing 10–7 M histamine (Hist) are shown as positive control for p38 MAPK activation. Phospho-p38, total p38, and actin are shown. (C) CD4+ T cells from WT and H1RKO mice were incubated with anti-CD3 and anti-CD28 mAbs, 10–7 M histamine, or both in the histamine-free medium for 30 minutes, and whole-cell lysates were analyzed for phospho-p38, total p-38, and actin by Western blotting. (D) CD4+ T cells from WT and H1RKO mice were stimulated with anti-CD3 and anti-CD28 mAbs for the indicated periods of time, and whole-cell lysates were analyzed for T-bet expression by Western blot. Actin is shown as loading control. (E) Purified CD4+ T cells from WT, H1RKO, and H1RKO-MKK6Glu Tg mice were stimulated with anti-CD3 and anti-CD28 mAbs for the indicated periods of time, and supernatants were analyzed for IFN-γ production by ELISA in triplicate. F = 21.7, P < 0.0001, 2-way ANOVA. ***P < 0.001, Bonferroni corrected post-hoc comparison. Data are representative of at least 2 independent experiments.
Because histamine was present in the normal medium used to activate CD4+ T cells with anti-CD3 and anti-CD28 mAbs (Figure 5, C and E), it was possible that the activation of p38 MAPK by TCR ligation was codependent upon histamine signaling through the H1R. To test this possibility, we examined p38 MAPK activation upon anti-CD3 and anti-CD28 mAb stimulation in histamine-free media. TCR ligation failed to activate p38 MAPK in both WT and H1RKO CD4+ T cells in histamine-free media (Figure 6B). In contrast, the absence of histamine did not affect TCR-mediated ERK activation (Supplemental Figure 1; supplemental material available online with this article; doi:10.1172/JCI32792DS1) or intracellular calcium mobilization (data not shown) in WT CD4+ T cells. To further demonstrate the selective requirement for histamine in TCR-mediated p38 MAPK activation, WT and H1RKO CD4+ T cells were stimulated in histamine-free media with anti-CD3 and anti-CD28 mAbs in the presence of histamine. TCR-mediated p38 MAPK activation was restored by histamine in WT CD4+ T cells but not in H1RKO CD4+ T cells (Figure 6C), indicating that binding of histamine to H1R was required for activation of p38 MAPK upon TCR ligation. Interestingly, the levels of phospho-p38 MAPK in WT CD4+ T cells treated with anti-CD3 and anti-CD28 mAbs and histamine were similar to the levels obtained when the cells were treated with histamine alone (Figure 6C). The inability of TCR stimulation to activate p38 MAPK in H1R-deficient cells in normal histamine-containing media (Figure 5C), activate p38 MAPK in the histamine-free media (Figure 6B), or further increase p38 MAPK activation when histamine was added back to the histamine-free media strongly suggest that the activation of p38 MAPK observed upon TCR ligation is dependent upon concomitant H1R signaling.
Although the precise mechanism by which p38 MAPK regulates IFN-γ production in CD4+ T cells remains unclear, recent studies have suggested that the activation of the this MAPK pathway is required for T-bet expression (32, 33), and T-bet regulates IFN-γ production (34). We therefore examined T-bet expression by Western blot analysis during activation of WT and H1RKO CD4+ T cells. T-bet levels were lower in activated H1RKO CD4+ T cells than in WT CD4+ T cells (Figure 6D). Thus, the impairment in p38 MAPK activation in the absence of H1R reduces the T-bet expression and thereby IFN-γ production by CD4+ T cells during TCR activation.
In order to demonstrate that reduced p38 MAPK activation in H1RKO CD4+ T cells is responsible for decreased IFN-γ production by these cells, we crossed H1RKO mice with the previously described dlck-MKK6Glu Tg mice (30). These mice express a constitutively active form of MKK6, a specific upstream activator of p38 MAPK, under the control of dlck promoter. Thus p38 MAPK is constitutively and selectively active in T cells in these mice. Anti-CD3 and anti-CD28 mAb–stimulated CD4+ T cells from H1RKO-MKK6Glu Tg mice produced significantly more IFN-γ than did CD4+ T cells from littermate H1RKO mice (Figure 6E), indicating that the diminished activation of p38 MAPK in H1RKO CD4+ T cells is responsible for the reduced IFN-γ production by these cells.
H1R signaling directly in CD4+ T cells regulates encephalitogenic Th1 effector responses.
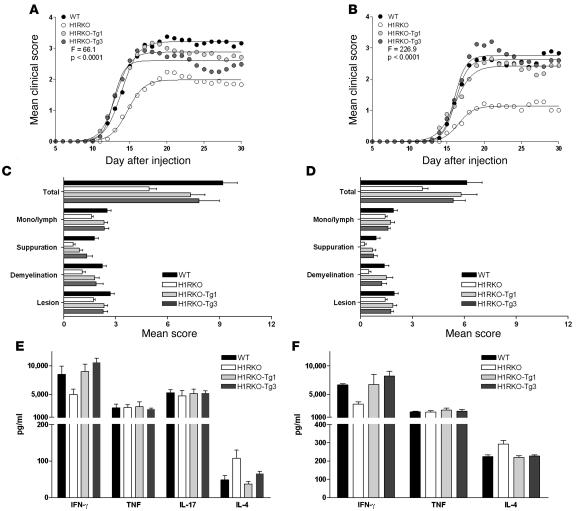
As a shared autoimmune disease susceptibility gene, Hrh1 has been shown to control numerous disease associated subphenotypes, including blood-brain barrier permeability, antigen presentation, and delayed-type hypersensitivity responses (13, 35). To assess whether H1R signaling in CD4+ T cells influences EAE by regulating encephalitogenic Th1 responses, we examined the susceptibility of H1RKO and H1RKO-Tg mice to EAE using the classical MOG35–55-CFA plus pertussis toxin (PTX) model and the 2× MOG35–55-CFA model (36), which does not use PTX as an ancillary adjuvant. Regression analysis (36) revealed that the clinical disease courses elicited with the MOG35–55-CFA plus PTX protocol (Figure 7A) was significantly more severe in WT and H1RKO-Tg mice compared with H1RKO mice (WT, F = 132.1, P < 0.0001; HIRKO-Tg1, F = 127.5, P < 0.0001; HIRKO-Tg3, F = 83.8, P < 0.0001). Identical results were obtained with the 2× MOG35–55 protocol (Figure 7B; WT, F = 226.9, P < 0.0001; HIRKO-Tg1, F = 134.0, P < 0.0001; HIRKO-Tg3, F = 215.8, P < 0.0001).
Figure 7. H1R signaling directly in CD4+ T cells regulates encephalitogenic Th1 effector responses.
Clinical EAE course (A and B), severity of CNS pathology (C and D), and ex vivo cytokine responses (E and F) of WT, H1RKO, and H1RKO-Tg mice were compared following immunization with MOG35–55-CFA plus PTX (A, C, and E) or 2× MOG35–55 and CFA (B, D, and F). Cytokine production was assessed by stimulating splenocytes with MOG35–55 on day 10 after injection, and supernatants were collected and quantified by ELISA in triplicate. The significance of differences in the course of clinical disease, CNS pathology indices, and cytokine responses were assessed by regression analysis (63), χ2 test, or ANOVA followed by Bonferroni corrected post-hoc comparisons. With the exception of TNF-α and IL-17 production, significant differences among the strains were detected for all parameters at P < 0.0001 — WT, H1RKO-Tg1, and H1RKO-Tg3 groups were equivalent and all significantly different from the H1RKO group.
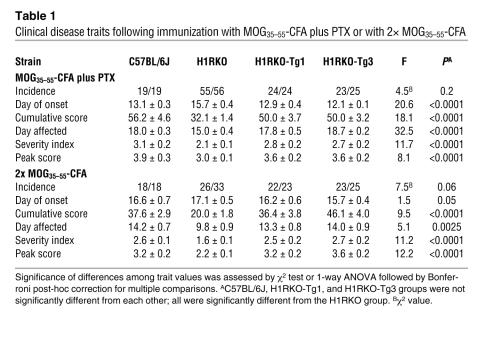
Analysis of EAE-associated clinical quantitative trait variables (37) revealed that the mean day of onset, cumulative disease score, number of days affected, overall severity index, and peak score were significantly different among the strains immunized with either MOG35–55-CFA plus PTX or 2× MOG35–55-CFA (Table 1). Bonferroni corrected post-hoc multiple comparisons for each trait variable revealed that the values were not significantly different among WT, HIRKO-Tg1, and HIRKO-Tg3 mice, all of which were significantly greater than those in H1RKO mice. Additionally, compared with H1RKO mice, both HIRKO-Tg1 and HIRKO-Tg3 mice immunized with MOG35–55-CFA plus PTX (Figure 7C) and 2× MOG35–55-CFA (Figure 7D) exhibited significantly more severe overall CNS pathology (38), which was equivalent in severity to that seen in WT mice. Therefore, H1R expression in CD4+ T cells alone was capable of complementing EAE susceptibility in H1R-deficient animals.
Table 1 .
Clinical disease traits following immunization with MOG35–55-CFA plus PTX or with 2× MOG35–55-CFA
We also examined cytokine production following ex vivo stimulation of splenocytes from mice immunized with MOG35–55-CFA plus PTX and 2× MOG35–55-CFA. The H1R transgene fully complemented IFN-γ production by H1RKO CD4+ T cells and restored IL-4 production to WT levels (Figure 7, E and F). In contrast, no significant differences in TNF-α or IL-17 production were detected among WT, H1RKO, and H1RKO-Tg mice. Together, these data indicate that H1R signaling in CD4+ T cells complements EAE severity independently of TNF-α and IL-17 production.
Discussion
Although H1R has been previously shown to play a role in regulating encephalitogenic Th1 immune response in EAE (17), it was unclear whether this was caused by the deficiency of H1R in CD4+ T cells or in APCs. In this study, we showed that the presence of H1R in CD4+ T cells was essential for the activation of p38 MAPK and IFN-γ production by these cells and that the lack of H1R in CD4+ T cells was responsible for the increased EAE resistance of H1RKO mice. These findings also explain the likely cause of the Th2 deviation and aberrant IL-4 production seen in the H1RKO mice (17), a result we confirmed in the present study. This deviation could logically be caused by the impairment in p38 MAPK activation that reduces T-bet expression and thereby IFN-γ production by CD4+ T cells during TCR activation. Without H1R, naive T cells cannot be driven into the full Th1 developmental pathway, and the result is an unbalanced immune repertoire that is generally thought to be protected from signs of EAE (39).
Even though the expression of H1R in CD4+ T cells has been reported previously (40), it was unknown how H1R is regulated during the activation phase of CD4+ T cells. Here we show, for the first time to our knowledge, that H1R gene expression is silenced early after the activation of CD4+ T cells. Modulation of H1R signaling, like that of other G protein–coupled receptors, is complex and includes receptor desensitization, internalization, and subsequent downregulation (41, 42). Desensitization of H1R is induced by both agonist-specific (homologous) and agonist nonspecific (heterologous) pathways, mainly involving PKC-mediated phosphorylation of H1R (43, 44). PKC activation has previously been shown to inhibit H1R both at the protein level and at the gene expression level (45–48). Because TCR ligation leads to potent activation of PKC (49), silencing of H1R expression in activated CD4+ T cells may be a consequence of PKC activation. Although the transcriptional regulation of H1R promoter is not well understood, H1R-mediated signaling has been shown to be necessary for continued H1R expression (47, 48). Thus, the loss of H1R gene expression in activated CD4+ T cells in mice may be a mechanism to turn off possible subsequent histamine signals in these cells. In humans, H1R expression is reported to increase in Th1 differentiated cells (15). However, Hrh1 was rapidly downregulated even during the Th1 differentiation of mouse CD4+ T cells (data not shown). These apparently contradictory results may be explained by the different origin of the T cells (mouse versus human) or by other differences in the culture conditions used.
H1R has been previously implicated in the regulation of IFN-γ production. H1R-deficient splenocytes have been shown to produce lower IFN-γ when activated by anti-CD3 and-CD28 mAbs or by specific antigen (14–17), but to our knowledge, no previous studies addressed the role of H1R in isolated CD4+ T cells. Here we show that H1R expression in CD4 cells was essential specifically for IFN-γ production by these cells, but not for IL-2 production or proliferation. A previous report showed hypoproliferation of total splenocytes from H1R-deficient mice in response to anti-CD3 mAb (14). However, the low proliferative response could be because of the H1R deficiency in cells other than CD4+ T cells, such as APCs (e.g., macrophages or dendritic cells), that also express H1R. Although CD4+ T cells also express H2R and H4R in addition to H1R, the selective restoration of the IFN-γ response in CD4+ T cells from H1RKO-Tg mice clearly demonstrates that signaling through H1R was necessary for regulation of IFN-γ production in these cells. However, we have not formally excluded the possibility that differences in H2R and H4R expression caused by loss of H1R-dependent cross regulation may also influence IFN-γ production.
Several studies have shown that p38 MAPK is activated in CD4+ T cells or total T cells upon TCR activation. Costimulatory molecules (such as CD28, 4-1BB, ICOS, and CD30) also contribute to the activation of p38 MAPK during activation (50). While most studies agree on the role of p38 MAPK on IFN-γ production and Th1 differentiation, recent studies have questioned the requirement of TCR-mediated p38 MAPK activation. Instead, they propose that activation of this pathway by cytokines such as IL-12 or IL-18 is probably more relevant (51). To date, the effect of other components also present in the milieu during TCR activation has not been addressed. Here we show, for the first time to our knowledge, that activation of p38 MAPK by TCR/CD28 ligation is dependent on the presence of histamine and its binding to H1R. A previous study has shown the requirement of H1R for ZAP-70 activation in H1RKO total splenocytes in conjunction with the hypoproliferative defect in these cells (14). However, here we show that in CD4+ T cells, H1R is not required for other key signaling pathways such as ERK activation (Supplemental Figure 1), NF-κB activation (Figure 5A), or calcium mobilization (data not shown) as well as for IL-2 production and proliferation. Thus, deficiency of H1R in CD4+ T cells appears to selectively impair the activation of the p38 MAPK pathway, but the mechanism remains to be investigated further. p38 MAPK is normally activated through the upstream MAPKK, MKK3 and MKK6 (and MKK4 in response to some stimuli) (52). It has previously been shown that GADD45 proteins interact with MEKK4, an upstream kinase of MKK3 and MKK6, and thus activate p38 MAPK (53). An alternative pathway for activation of p38 MAPK through its autophosphorylation has also been recently proposed (54). H1R signaling is mediated by Gαq/11 protein, which is also associated with TCR signaling through CD3ε (55). Thus, it is possible the H1R through Gαq/11 could regulate GADD45 members (α, β, and γ) and lead to p38 MAPK activation through either the classical or the alternative pathway in CD4+ T cells.
Epidemiological data indicate that the use of sedating H1R antagonists is associated with decreased MS risk (56), and in a small pilot study, patients with relapsing-remitting or relapsing-progressive MS given the H1R antagonist hydroxyzine remained stable or improved neurologically (57). Additionally, microarray analysis revealed that the H1R is overexpressed in the chronic plaques of MS patients (58). Historically, the role of histamine in autoimmune inflammatory disease of the CNS has been viewed as a mediator of the effector or inflammatory phase of the disease (59). However, recent data showing that EAE and neuroantigen-specific T effector cell responses are significantly different in histamine- and histamine receptor–deficient mice compared with WT mice revealed that histamine plays a role during the induction phase and priming of autoreactive effector T cells (17, 60, 61). In this regard, our results show that H1R signaling in T cells regulates Th1 effector functions, but not Th17 effector functions, as well as EAE severity independent of APCs and other hematopoietically derived cells. Moreover, our results demonstrate that H1R signaling in CD4+ T cells regulates the encephalitogenic Th1 effector responses during the priming of naive antigen-specific CD4+ T cells. Taken together, our findings suggest that pharmacological targeting of the H1R may be useful early in the treatment of MS and other autoimmune inflammatory diseases in which molecular mimicry, bystander activation (with or without epitope spreading), and viral persistence play a role in perpetuating immunopathology as a consequence of continual priming of pathogenic adaptive immune responses (62).
Methods
Mice.
C57BL/6J mice were purchased from The Jackson Laboratory. B6.129P-Hrh1tm1Wat(H1RKO) (14) mice were maintained in the animal facility at the University of Vermont. The experimental procedures used in this study were approved by the Animal Care and Use Committee of the University of Vermont.
For transgenic mouse generation, an HA-H1R construct was made by deleting the methionine of the Bphs-susceptible H1R allele (17) and adding an HA tag at the N terminus using TOPO cloning vector (Invitrogen). The HA-H1R was then subcloned downstream of the dlck promoter (26). The linear DNA fragment containing the dlck promoter, the HA-H1R gene, and the human growth hormone (hGH) intron and polyadenylation signal was injected directly into fertilized C57BL/6J eggs at the University of Vermont transgenic/knockout facility. Mice were screened by DNA slot blot using a BamHI–SacI 0.5-kb fragment from the hGH gene as a probe. Two founders were generated and were crossed to H1RKO mice to establish H1RKO-Tg mice. dlck MKK6Glu transgenic mice (30) were crossed to H1RKO mice to generate H1RKO-MKK6Glu transgenic mice.
Cell preparation and culture conditions.
CD4+ T cells were isolated from spleen and lymph nodes by negative selection for CD8+, MHC class II+, NK1.1+ and CD11b+ cells using magnetic beads from Qiagen as previously described (30). Purified CD4+ T cells were stimulated with plate-bound anti-CD3 (5 μg/ml) and soluble anti-CD28 (1 μg/ml) mAbs from BD Biosciences — Pharmingen. Th1 polarized CD4+ effector T cells were generated by culturing the CD4+ T cells (1 × 106 cells/ml) with anti-CD3 and anti-CD28 mAbs in the presence of 4 ng/ml recombinant IL-12 (R&D Systems) and 10 μg/ml anti–IL-4 mAb (BD Biosciences — Pharmingen). Th2 polarized CD4+ effector T cells were generated by activating cells (1 × 106/ml) with anti-CD3 and anti-CD28 mAbs in the presence of 30 ng/ml recombinant IL-4 (R&D Systems) and 10 μg/ml anti–IFN-γ mAb. Effector Th17 CD4+ T cells were generated by activating CD4+ T cells (1 × 106 cells/ml) with anti-CD3 and anti-CD28 mAbs in the presence of 1 ng/ml TGF-β (Peprotech Inc.), 30 ng/ml IL-6 (R&D Systems), 10 μg/ml anti–IFN-γ mAb, and 10 μg/ml anti–IL-4 mAb. After 4 days, the cells were extensively washed and counted, and equal numbers of cells were restimulated with anti-CD3 mAb. After 24 h, the supernatants were collected and IFN-γ, IL-4, and IL-17 were analyzed by ELISA. Nonpolarized effector cells were generated by culturing CD4+ T cells with anti-CD3 and anti-CD28 mAbs in the absence of exogenous cytokines for 4 days. The cells were then extensively washed and counted, and equal numbers of cells were restimulated with anti-CD3 mAb. After 24 h, the supernatants were collected, and IFN-γ was analyzed by ELISA.
Histamine dihydrochloride was obtained from Sigma-Aldrich. RPMI prepared with 10% Fetalclone bovine serum (Hyclone), serum dialyzed twice with 10,000-kDa molecular cutoff, was used as histamine-free medium.
Cytokine production.
ELISAs were performed on the cell culture supernatants as described previously (61), using the primary mAbs anti–IFN-γ, anti–IL-2, anti–IL-4, and anti–IL-17 and their corresponding biotinylated mAbs (BD Biosciences — Pharmingen). Other ELISA reagents included horseradish peroxidase–conjugated avidin D (Vector Laboratories), TMB microwell peroxidase substrate and stop solution (Kirkegaard and Perry Laboratories) and recombinant IFN-γ, IL-4, and IL-2 (R&D Systems) used as standards.
For ex vivo cytokine analysis, spleens were obtained from mice immunized 10 days earlier with either MOG35–55-CFA plus PTX or 2× MOG35–55-CFA, and single-cell suspensions were prepared at 1 × 106 cells/ml in RPMI medium and stimulated with 50 μg/ml MOG35–55. Cell culture supernatants were recovered at 72 h, and cytokine levels were measured by ELISA using anti–IFN-γ, anti–IL-4, and anti–IL-17 mAbs and their corresponding biotinylated mAbs (BD Biosciences — Pharmingen). TNF-α was quantified using an ELISA kit from BD Biosciences — Pharmingen.
Proliferation assays.
CD4+ T cells (2.5 × 105 cells/well) were activated with anti-CD3 and anti-CD28 mAbs for 72 h, and proliferation was determined by [3H]-thymidine incorporation during the last 18 h of culture.
Conventional and quantitative real-time RT-PCR.
Total RNA was extracted from CD4+ T cells using RNeasy RNA isolation reagent (Qiagen) as recommended by the manufacturer. cDNA generated from 1 μg total RNA was used in quantitative real-time RT-PCR using the SYBR green method. The sequences of Hrh1 primers used were as follows: forward, 5′-CCAGAGCTTCGGGAAGATAA-3′; reverse, 5′-ACCACAGCATGAGCAAAGTG-3′. B2m was used as reference gene, and relative mRNA levels were calculated using comparative threshold cycle (CT) method. For conventional RT-PCR, the cDNA was amplified by PCR and visualized on 1% gel. The primers described above were used for Hrh1, and the primers used for Hprt1 were as follows: forward, 5′-GTTGGATACAGGCCAGACTTTGTTG-3′; reverse, 5′-GAGGGTAGGCTGCCTATAGGCT-3′. To study the transgene expression in H1RKO-Tg mice, the cDNA prepared as described above was amplified using a forward primer in Hrh1 (5′-CTCCCGGACCACAGACTCAGA-3′) and a reverse primer in the third exon of hGH (5′-GACGGAGGTCTGGGGGTTCTG-3′), and the PCR product was visualized on 1% agarose gel.
Retroviral transduction experiments.
The retroviral vector plasmid pEGZ-HA was a generous gift from I. Berberich (University of Wuerzburg, Wuerzburg, Germany), and packaging vectors pHIT123 and pHIT60 were generous gifts from A. Klingsman (Oxford University, Oxford, United Kingdom). Two restriction sites, BamHI and EcoRI, were inserted into the mouse H1R cDNA by PCR and cloned such that the second codon was in frame with the HA tag of pEGZ, generating an HA-H1R fusion protein. pEGZ is a bicistronic system with IRES-EGFP. EGFP served as a marker for transfected cells.
The retroviral vector plasmids, pEGZ-HA-H1R or the empty pEGZ and the packaging vectors pHIT60 and pHIT123, were transiently transfected into HEK293T cells using the calcium phosphate method. After 2 days, the retrovirus containing supernatants were used to transduce (by centrifugation at 800 g for 3 h at 32°C) CD4+ T cells previously activated with anti-CD3 and anti-CD28 mAbs for 16 h. The transduced CD4+ T cells were cultured in the presence of 50 U/ml IL-2 for 2 days and were sorted using a FACSAria instrument (BD Biosciences — Pharmingen) based on their EGFP expression. Equal numbers of EGFP-positive cells were restimulated with anti-CD3 mAb, and 24 h later, IFN-γ was measured in the supernatant by ELISA.
Confocal microscopy.
HEK293T cells were transfected with pEGA-HA-H1R or empty pEGZ control vector (5 μg total DNA) using the calcium phosphate method. Cells were fixed, permeabilized, and stained using an anti-HA mAb (Cell Signaling Technologies) followed by incubation with Alexa Fluor 568–conjugated anti-mouse antibody (Invitrogen). TOPRO-3 nuclear stain (Invitrogen) was used as a nuclear marker. Cells were examined by confocal microscopy using a Zeiss LSM 510 META Confocal Laser Scanning Imaging System (Carl Zeiss Microimaging Inc.)
Cell lysates and Western blotting.
Whole-cell lysates were prepared from 1 × 106–5 × 106 cells in Triton lysis buffer and were then separated via SDS-PAGE and transferred to nitrocellulose membranes as described previously (63). Primary antibodies used for Western blot analysis include anti-HA (Abcam Inc.), anti-p38, anti–phospho-p38, anti–phospho-STAT1, anti–phospho-STAT4, anti–phospho-ERK, anti-ERK, anti–phospho-JNK, anti-JNK (Cell Signaling Technologies), anti–T-bet (a gift from L. Glimcher, Harvard University School of Public Health, Boston, Massachusetts, USA), and anti-actin (Santa Cruz Biotechnology Inc.).
[35S]GTPγS binding assay.
The HA-H1R cDNA was subcloned into pcDNA3 using restriction sites EcoRI and BamHI. The [35S]GTPγS binding experiments were initiated by the addition of 50 fmol receptor to an assay buffer (20 mM HEPES, pH 7.4; 3 mM MgCl2; 100 mM NaCl; 1 μM GDP; 0.2 mM ascorbic acid; and 100 nCi [35S]GTPγS) containing 100 μM histamine. Nonspecific binding was determined in the above condition with the addition of 100 μM GTPγS. Reactions were incubated for 15 min at 30°C and were terminated by the addition of 500 μl ice-cold buffer containing 20 mM HEPES (pH 7.4), 3 mM MgCl2, 100 mM NaCl, and 0.2 mM ascorbic acid. The samples were centrifuged at 16,000 g for 10 minutes at 4°C. The resulting pellets were resuspended in solubilization buffer (100 mM Tris, 200 mM NaCl, 1 mM EDTA, and 1.25% NonidetP-40) plus 0.2% SDS. Samples were precleared with Pansorbin for 1 h, followed by immunoprecipitation with C-terminal Gα11 antiserum. Finally, the immunocomplexes were washed with solubilization buffer, and bound [35S]GTPγS was estimated by liquid scintillation-spectrometry.
EMSA.
Nuclear extracts were prepared from anti-CD3 and anti-CD28 mAb–treated CD4+ T cells as previously described (64). Binding reactions for EMSA were carried out at room temperature using 2 μg nuclear proteins and [32P]dCTP-end labeled double-stranded oligonucleotide probes containing NF-κB binding site from the mouse κ intron enhancer (sense, 5′-GATCAGAGGGGACTTTCCGAGGGAT-3′; antisense, 5′-GATCCCTCGGAAAGTCCCCTCTGAT-3′). Samples were separated by electrophoresis under nondenaturing conditions and exposed to film for autoradiography.
Induction and evaluation of EAE.
Mice were immunized for the induction of EAE using either the MOG35–55-CFA double-inoculation (37) or the MOG35–55-CFA plus PTX single-inoculation protocols (36). For the double-injection protocol, mice were injected subcutaneously with a sonicated emulsion of 100 μg MOG35–55 and an equal volume of CFA containing 200 μg Mycobacterium tuberculosis H37RA (Difco Laboratories) in the posterior right and left flank; 1 week later all mice were similarly injected at 2 sites on the right and left flank anterior of the initial injection sites. Animals immunized using the MOG35–55-CFA plus PTX single-inoculation protocol received a sonicated emulsion of 200 μg MOG35–55 and equal volume of CFA containing 200 μg Mycobacterium tuberculosis H37RA by subcutaneous injections distributed equally in the posterior right and left flank and scruff of the neck. Immediately thereafter, each animal received 200 ng PTX (List Biological Laboratories) by intravenous injection. Mice were scored daily starting at day 5 after injection as previously described (36). Clinical quantitative trait variables, including disease incidence, mean day of onset, cumulative disease score, number of days affected, overall severity index, and peak score, were generated as previously described (37).
Brains and spinal cords were dissected from calvaria and vertebral columns, respectively, and fixed by immersion in 10% phosphate-buffered formalin (pH 7.2). Following adequate fixation, brains and spinal cords were trimmed, and representative transverse sections were embedded in paraffin, sectioned at 5 μm, and mounted on glass slides. Sections were stained with hematoxylin and eosin for routine evaluation and Luxol fast blue–periodic acid Schiff for demyelination. Sections from representative areas of the brain and spinal cords were scored in a semiquantitative fashion for the various lesions as previously described (38). An overall CNS pathology index for each lesions was obtained by calculating the average scores for the lesions observed in the brain and spinal cord.
Statistics.
Statistical analyses, as outlined in the figure legends, were performed using GraphPad Prism 4 software (GraphPad Software). A P value of 0.05 or less was considered significant.
Supplementary Material
Acknowledgments
This work was supported by NIH grants AI041747, AI058052, AI045666, NS036526, and AI051454 and by National Multiple Sclerosis Society grant RG-3575.
Footnotes
Nonstandard abbreviations used: EAO, experimental autoimmune orchitis; H1R, histamine receptor H1; H1RKO, H1R-deficient (mouse); MOG35–55, myelin oligodendrocyte glycoprotein peptide 35–55; PTX, pertussis toxin.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 117:3507–3518 (2007). doi:10.1172/JCI32792
References
- 1.Parsons M.E., Ganellin C.R. Histamine and its receptors. Br. J. Pharmacol. 2006;147(Suppl. 1):S127–S135. doi: 10.1038/sj.bjp.0706440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akdis C.A., Simons F.E. Histamine receptors are hot in immunopharmacology. Eur. J. Pharmacol. 2006;533:69–76. doi: 10.1016/j.ejphar.2005.12.044. [DOI] [PubMed] [Google Scholar]
- 3.Hill S.J., et al. International Union of Pharmacology. XIII. Classification of histamine receptors. Pharmacol. Rev. 1997;49:253–278. [PubMed] [Google Scholar]
- 4.Leurs R., Smit M.J., Timmerman H. Molecular pharmacological aspects of histamine receptors. Pharmacol. Ther. 1995;66:413–463. doi: 10.1016/0163-7258(95)00006-3. [DOI] [PubMed] [Google Scholar]
- 5.Bakker R.A., Schoonus S.B., Smit M.J., Timmerman H., Leurs R. Histamine H(1)-receptor activation of nuclear factor-kappa B: roles for G beta gamma- and G alpha(q/11)-subunits in constitutive and agonist-mediated signaling. Mol. Pharmacol. 2001;60:1133–1142. doi: 10.1124/mol.60.5.1133. [DOI] [PubMed] [Google Scholar]
- 6.Sakhalkar S.P., Patterson E.B., Khan M.M. Involvement of histamine H1 and H2 receptors in the regulation of STAT-1 phosphorylation: inverse agonism exhibited by the receptor antagonists. Int. Immunopharmacol. 2005;5:1299–1309. doi: 10.1016/j.intimp.2005.03.019. [DOI] [PubMed] [Google Scholar]
- 7.Liu Z., Kharmate G., Patterson E., Khan M.M. Role of H(1) receptors in histamine-mediated up-regulation of STAT4 phosphorylation. Int. Immunopharmacol. 2006;6:485–493. doi: 10.1016/j.intimp.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 8.Megson A.C., Walker E.M., Hill S.J. Role of protein kinase Calpha in signaling from the histamine H(1) receptor to the nucleus. Mol. Pharmacol. 2001;59:1012–1021. doi: 10.1124/mol.59.5.1012. [DOI] [PubMed] [Google Scholar]
- 9.Lipnik-Stangelj M., Carman-Krzan M. Histamine-stimulated nerve growth factor secretion from cultured astrocyctes is blocked by protein kinase C inhibitors. Inflamm. Res. 2004;53(Suppl. 1):S57–S58. doi: 10.1007/s00011-003-0327-0. [DOI] [PubMed] [Google Scholar]
- 10.Robinson A.J., Dickenson J.M. Activation of the p38 and p42/p44 mitogen-activated protein kinase families by the histamine H(1) receptor in DDT(1)MF-2 cells. Br. J. Pharmacol. 2001;133:1378–1386. doi: 10.1038/sj.bjp.0704200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simons F.E. Advances in H1-antihistamines. N. Engl. J. Med. 2004;351:2203–2217. doi: 10.1056/NEJMra033121. [DOI] [PubMed] [Google Scholar]
- 12.Caron G., et al. Histamine polarizes human dendritic cells into Th2 cell-promoting effector dendritic cells. J. Immunol. 2001;167:3682–3686. doi: 10.4049/jimmunol.167.7.3682. [DOI] [PubMed] [Google Scholar]
- 13.Caron G., et al. Histamine induces CD86 expression and chemokine production by human immature dendritic cells. J. Immunol. 2001;166:6000–6006. doi: 10.4049/jimmunol.166.10.6000. [DOI] [PubMed] [Google Scholar]
- 14.Banu Y., Watanabe T. Augmentation of antigen receptor-mediated responses by histamine H1 receptor signaling. J. Exp. Med. 1999;189:673–682. doi: 10.1084/jem.189.4.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jutel M., et al. Histamine regulates T-cell and antibody responses by differential expression of H1 and H2 receptors. Nature. 2001;413:420–425. doi: 10.1038/35096564. [DOI] [PubMed] [Google Scholar]
- 16.Bryce P.J., et al. The H1 histamine receptor regulates allergic lung responses. J. Clin. Invest. 2006;116:1624–1632. doi: 10.1172/JCI26150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma R.Z., et al. Identification of Bphs, an autoimmune disease locus, as histamine receptor H1. Science. 2002;297:620–623. doi: 10.1126/science.1072810. [DOI] [PubMed] [Google Scholar]
- 18.Baron J.L., et al. Surface expression of alpha 4 integrin by CD4 T cells is required for their entry into brain parenchyma. J. Exp. Med. 1993;177:57–68. doi: 10.1084/jem.177.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Park H., et al. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat. Immunol. 2005;6:1133–1141. doi: 10.1038/ni1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cua D.J., et al. Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature. 2003;421:744–748. doi: 10.1038/nature01355. [DOI] [PubMed] [Google Scholar]
- 21.Ferber I.A., et al. Mice with a disrupted IFN-gamma gene are susceptible to the induction of experimental autoimmune encephalomyelitis (EAE). J. Immunol. 1996;156:5–7. [PubMed] [Google Scholar]
- 22.Willenborg D.O., Fordham S., Bernard C.C., Cowden W.B., Ramshaw I.A. IFN-gamma plays a critical down-regulatory role in the induction and effector phase of myelin oligodendrocyte glycoprotein-induced autoimmune encephalomyelitis. J. Immunol. 1996;157:3223–3227. [PubMed] [Google Scholar]
- 23.Langrish C.L., et al. IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J. Exp. Med. 2005;201:233–240. doi: 10.1084/jem.20041257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suryani S., Sutton I. An interferon-gamma-producing Th1 subset is the major source of IL-17 in experimental autoimmune encephalitis. J. Neuroimmunol. 2007;183:96–103. doi: 10.1016/j.jneuroim.2006.11.023. [DOI] [PubMed] [Google Scholar]
- 25.Robinson D.S., O’Garra A. Further checkpoints in Th1 development. Immunity. 2002;16:755–758. doi: 10.1016/s1074-7613(02)00331-x. [DOI] [PubMed] [Google Scholar]
- 26.Wildin R.S., et al. Developmental regulation of lck gene expression in T lymphocytes. J. Exp. Med. 1991;173:383–393. doi: 10.1084/jem.173.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aronica M.A., et al. Preferential role for NF-kappa B/Rel signaling in the type 1 but not type 2 T cell-dependent immune response in vivo. J. Immunol. 1999;163:5116–5124. [PubMed] [Google Scholar]
- 28.Ramana C.V., Gil M.P., Schreiber R.D., Stark G.R. Stat1-dependent and -independent pathways in IFN-gamma-dependent signaling. Trends Immunol. 2002;23:96–101. doi: 10.1016/s1471-4906(01)02118-4. [DOI] [PubMed] [Google Scholar]
- 29.Steffel J., Akhmedov A., Greutert H., Luscher T.F., Tanner F.C. Histamine induces tissue factor expression: implications for acute coronary syndromes. Circulation. 2005;112:341–349. doi: 10.1161/CIRCULATIONAHA.105.553735. [DOI] [PubMed] [Google Scholar]
- 30.Rincon M., et al. Interferon-gamma expression by Th1 effector T cells mediated by the p38 MAP kinase signaling pathway. EMBO J. 1998;17:2817–2829. doi: 10.1093/emboj/17.10.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weiss L., et al. Regulation of c-Jun NH(2)-terminal kinase (Jnk) gene expression during T cell activation. J. Exp. Med. 2000;191:139–146. doi: 10.1084/jem.191.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koch A., et al. IL-12-induced T-bet expression and IFNgamma release in lymphocytes from asthmatics — Role of MAPkinases ERK-1/-2, p38(MAPK) and effect of dexamethasone. Respir. Med. 2006;101:1321–1330. doi: 10.1016/j.rmed.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 33.Jones D.C., Ding X., Zhang T.Y., Daynes R.A. Peroxisome proliferator-activated receptor alpha negatively regulates T-bet transcription through suppression of p38 mitogen-activated protein kinase activation. J. Immunol. 2003;171:196–203. doi: 10.4049/jimmunol.171.1.196. [DOI] [PubMed] [Google Scholar]
- 34.Szabo S.J., et al. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–669. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- 35.Gao J.F., et al. Analysis of the role of Bphs/Hrh1 in the genetic control of responsiveness to pertussis toxin. Infect. Immun. 2003;71:1281–1287. doi: 10.1128/IAI.71.3.1281-1287.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Teuscher C., et al. Evidence that the Y chromosome influences autoimmune disease in male and female mice. Proc. Natl. Acad. Sci. U. S. A. 2006;103:8024–8029. doi: 10.1073/pnas.0600536103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Butterfield R.J., et al. New genetic loci that control susceptibility and symptoms of experimental allergic encephalomyelitis in inbred mice. J. Immunol. 1998;161:1860–1867. [PubMed] [Google Scholar]
- 38.Butterfield R.J., et al. Identification of genetic loci controlling the characteristics and severity of brain and spinal cord lesions in experimental allergic encephalomyelitis. Am. J. Pathol. 2000;157:637–645. doi: 10.1016/S0002-9440(10)64574-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shaw M.K., et al. Local delivery of interleukin 4 by retrovirus-transduced T lymphocytes ameliorates experimental autoimmune encephalomyelitis. J. Exp. Med. 1997;185:1711–1714. doi: 10.1084/jem.185.9.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sachs B., Hertl M., Merk H.F. Histamine receptors on lymphocytes: distribution and functional significance. Skin Pharmacol. Appl. Skin Physiol. 2000;13:313–323. doi: 10.1159/000029939. [DOI] [PubMed] [Google Scholar]
- 41.McCreath G., Hall I.P., Hill S.J. Agonist-induced desensitization of histamine H1 receptor-mediated inositol phospholipid hydrolysis in human umbilical vein endothelial cells. Br. J. Pharmacol. 1994;113:823–830. doi: 10.1111/j.1476-5381.1994.tb17067.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smit M.J., Timmerman H., Hijzelendoorn J.C., Fukui H., Leurs R. Regulation of the human histamine H1 receptor stably expressed in Chinese hamster ovary cells. Br. J. Pharmacol. 1996;117:1071–1080. doi: 10.1111/j.1476-5381.1996.tb16699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fujimoto K., et al. Identification of protein kinase C phosphorylation sites involved in phorbol ester-induced desensitization of the histamine H1 receptor. Mol. Pharmacol. 1999;55:735–742. [PubMed] [Google Scholar]
- 44.Miyoshi K., Kawakami N., Horio S., Fukui H. Homologous and heterologous phosphorylations of human histamine H1 receptor in intact cells. J. Pharmacol. Sci. 2004;96:474–482. doi: 10.1254/jphs.fpj04031x. [DOI] [PubMed] [Google Scholar]
- 45.Pype J.L., Dupont L.J., Mak J.C., Barnes P.J., Verleden G.M. Regulation of H1-receptor coupling and H1-receptor mRNA by histamine in bovine tracheal smooth muscle. Br. J. Pharmacol. 1998;123:984–990. doi: 10.1038/sj.bjp.0701697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pype J.L., Mak J.C., Dupont L.J., Verleden G.M., Barnes P.J. Desensitization of the histamine H1-receptor and transcriptional down-regulation of histamine H1-receptor gene expression in bovine tracheal smooth muscle. Br. J. Pharmacol. 1998;125:1477–1484. doi: 10.1038/sj.bjp.0702222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miyoshi K., Das A.K., Fujimoto K., Horio S., Fukui H. Recent advances in molecular pharmacology of the histamine systems: regulation of histamine H1 receptor signaling by changing its expression level. J. Pharmacol. Sci. 2006;101:3–6. doi: 10.1254/jphs.fmj06001x2. [DOI] [PubMed] [Google Scholar]
- 48.Yoshimura S., et al. Histamine H1 receptor-mediated histamine H1 receptor gene expression. Inflamm. Res. 2005;54(Suppl. 1):S42–S43. doi: 10.1007/s00011-004-0419-5. [DOI] [PubMed] [Google Scholar]
- 49.Acuto O., Cantrell D. T cell activation and the cytoskeleton. Annu. Rev. Immunol. 2000;18:165–184. doi: 10.1146/annurev.immunol.18.1.165. [DOI] [PubMed] [Google Scholar]
- 50.Dodeller F., Schulze-Koops H. The p38 mitogen-activated protein kinase signaling cascade in CD4 T cells. Arthritis Res. Ther. 2006;8:205. doi: 10.1186/ar1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berenson L.S., Yang J., Sleckman B.P., Murphy T.L., Murphy K.M. Selective requirement of p38alpha MAPK in cytokine-dependent, but not antigen receptor-dependent, Th1 responses. J. Immunol. 2006;176:4616–4621. doi: 10.4049/jimmunol.176.8.4616. [DOI] [PubMed] [Google Scholar]
- 52.Kyriakis J.M., Avruch J. Mammalian mitogen-activated protein kinase signal transduction pathways activated by stress and inflammation. Physiol. Rev. 2001;81:807–869. doi: 10.1152/physrev.2001.81.2.807. [DOI] [PubMed] [Google Scholar]
- 53.Takekawa M., Saito H. A family of stress-inducible GADD45-like proteins mediate activation of the stress-responsive MTK1/MEKK4 MAPKKK. Cell. 1998;95:521–530. doi: 10.1016/s0092-8674(00)81619-0. [DOI] [PubMed] [Google Scholar]
- 54.Salvador J.M., et al. Alternative p38 activation pathway mediated by T cell receptor-proximal tyrosine kinases. Nat. Immunol. 2005;6:390–395. doi: 10.1038/ni1177. [DOI] [PubMed] [Google Scholar]
- 55.Stanners J., Kabouridis P.S., McGuire K.L., Tsoukas C.D. Interaction between G proteins and tyrosine kinases upon T cell receptor.CD3-mediated signaling. J. Biol. Chem. 1995;270:30635–30642. doi: 10.1074/jbc.270.51.30635. [DOI] [PubMed] [Google Scholar]
- 56.Alonso A., Jick S.S., Hernan M.A. Allergy, histamine 1 receptor blockers, and the risk of multiple sclerosis. Neurology. 2006;66:572–575. doi: 10.1212/01.wnl.0000198507.13597.45. [DOI] [PubMed] [Google Scholar]
- 57.Logothetis L., et al. A pilot, open label, clinical trial using hydroxyzine in multiple sclerosis. Int. J. Immunopathol. Pharmacol. 2005;18:771–778. doi: 10.1177/039463200501800421. [DOI] [PubMed] [Google Scholar]
- 58.Lock C., et al. Gene-microarray analysis of multiple sclerosis lesions yields new targets validated in autoimmune encephalomyelitis. Nat. Med. 2002;8:500–508. doi: 10.1038/nm0502-500. [DOI] [PubMed] [Google Scholar]
- 59.Bebo B.F., Jr., Yong T., Orr E.L., Linthicum D.S. Hypothesis: a possible role for mast cells and their inflammatory mediators in the pathogenesis of autoimmune encephalomyelitis. J. Neurosci. Res. 1996;45:340–348. doi: 10.1002/(SICI)1097-4547(19960815)45:4<340::AID-JNR3>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 60.Musio S., et al. A key regulatory role for histamine in experimental autoimmune encephalomyelitis: disease exacerbation in histidine decarboxylase-deficient mice. J. Immunol. 2006;176:17–26. doi: 10.4049/jimmunol.176.1.17. [DOI] [PubMed] [Google Scholar]
- 61.Teuscher C., et al. Attenuation of Th1 effector cell responses and susceptibility to experimental allergic encephalomyelitis in histamine H2 receptor knockout mice is due to dysregulation of cytokine production by antigen-presenting cells. Am. J. Pathol. 2004;164:883–892. doi: 10.1016/S0002-9440(10)63176-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fujinami R.S., von Herrath M.G., Christen U., Whitton J.L. Molecular mimicry, bystander activation, or viral persistence: infections and autoimmune disease. Clin. Microbiol. Rev. 2006;19:80–94. doi: 10.1128/CMR.19.1.80-94.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Farley N., et al. p38 mitogen-activated protein kinase mediates the Fas-induced mitochondrial death pathway in CD8 + T cells. Mol. Cell. Biol. 2006;26:2118–2129. doi: 10.1128/MCB.26.6.2118-2129.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang T.T., Ung P.M., Rincon M., Chow C.W. Role of the CCAAT/enhancer-binding protein NFATc2 transcription factor cascade in the induction of secretory phospholipase A2. J. Biol. Chem. 2006;281:11541–11552. doi: 10.1074/jbc.M511214200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.