Abstract
This review briefly summarizes recent progress in fundamental understanding and analytical profiling of the L-arginine/nitric oxide (NO) pathway. It focuses on key analytical references of NO actions and on the experimental acquisition of these references in vivo, with capillary electrophoresis (CE) and high-performance capillary electrophoresis (HPCE) comprising one of the most flexible and technologically promising analytical platform for comprehensive high-resolution profiling of NO-related metabolites. Second aim of this review is to express demands and bridge efforts of experimental biologists, medical professionals and chemical analysis-oriented scientists who strive to understand evolution and physiological roles of NO and to develop analytical methods for use in biology and medicine.
Keywords: Reviews, Nitrite, Peroxynitrite, 3-Nitrotyrosine, N-Nitrosotryptophan, ADMA
1. Introduction
Avalanche of findings triggered by the discovery of nitric oxide (NO) signaling in the mammalian circulation [1] exposes a fundamental role of this versatile free-radical as essential intermediate of intra-extra-cellular signaling and a key factor of endogenous toxicity [2,3], apoptosis [4,5], and pathogen-induced cellular defense [6–9]. It has been recognized that NO missions in signaling and toxicity have emerged with earlier life [10] and were greatly diversified upon acquisition of aerobic metabolism. NO has been recruited as a primary and auxiliary signal messenger through virtually all life forms and a variety of biological mechanisms ranging from quorum sensing and metabolic suicide in bacteria [11,12] to immune responses [13] and memory formation in human [14–16]. Remarkable progress has been made to understand structure and chemistry of NO-mediated reactions. However, concentrations, gradients, time of life and spatial profile of NO messengers, substrates, and products in biological systems and biological processes often remain vague. These values remain to be difficult to obtain directly with acceptable temporal and spatial resolution.
Detection and quantification of NO are especially complex tasks. It has small active concentrations, moderate stability and high reactivity with biogenic free radicals, metal coordinating molecules, and other biogenic active species in vivo. These biological factors together result in short time of life and complex analytical signature of this molecule. Nevertheless, important roles of NO in human health and animal physiology have generated a strong persistent effort to determine NO in biological matrices leading to the development of a variety of direct and indirect analytical assays. Many of these approaches require resources which exceed analytical expertise and instrumental capacity of a universal biological laboratory accumulating systematic understanding of mechanisms of action of NO. It is essential to consider that after 19 years eradiation of NO research the availability of an easy accessible and comprehensive analytical tool for evaluation of substrates and products of NO metabolism in biological matrices remains to be a key limitation.
NO is a short-lived species; however, evident synergy of several reciprocally coupled redox cascades in a circulating NO pool revealed over the last decade suggests the existence of a coordinated network of enzymatic, non-enzymatic, and membrane transport events which together integrate NO recycling/buffering system and lead to essential homeostasis of a NO pool in spatial and temporal dimension. This system remains to be resolved theoretically and practically upon acquisition of new data on reversible and irreversible interactions of NO and proximal NO metabolites with its molecular environment. Several new ideas about reactive nitrogen species (RNS)-mediated NO actions in biological systems have also demand for new analytical parameters. These analytes are becoming increasingly important to refine understanding of NO-mediated mechanisms and perhaps they would be absolutely necessary for understanding exact scenarios of NO actions. An expanding set of such biochemical parameters includes NO, nitrite, nitrate, higher nitrogen oxides, L-arginine and its dimethylarginine analogs, L-citrulline, small and peptide-incorporated nitrosothiols, and a variety of nitrated aromatic motives tightly interacting in biological matrices. Several CE applications addressing each of these analytes in biological matrices have been recently developed. However, primary effort and progress has been made to determine nitrite and nitrate which are currently considered as the most important members of the L-arginine/NO pathway analyzed by CE. The determination of the NO metabolites nitrite and nitrate in relevant biological matrices is emphasized in the present review. Other members of the L-arginine/NO family which are not derived from NO, such as L-arginine and its dimethyl analogs, will also be addressed in this work. For simplicity, these analytes, nitrite and nitrate will be collectively named NO metabolites.
2. Biological properties of NO pathways
NO is a free radical gas which is produced and circulated in biological systems as an average result of NO-generating and NO-scavenging pathways (Fig. 1). NO diffuses freely through cellular membranes and cytoplasm and interacts with redox components of biological matrices. It triggers specific signaling cascades and mediates translational and posttranslational changes of enzymatic activity directly or via RNS. It also may interact with biogenic free radicals as a potent protective free radical scavenger/terminator and antioxidant or as a radical-induced toxicity-amplifying precursor. It may escape rapid oxidative fate by binding to metalloporphyrins and thiol and aromatic moieties of proteins and small molecules which together level, extend, and buffer NO activity in spatial and temporal dimension. NO autooxidizes to nitrite. NO and nitrite are enzymatically oxidized in erythrocytes to nitrate which may undergo ion transport and renal excretion. Understanding oxygen-induced and redox-dependent reactions of NO along with a kinetic and capacity of NO-producing cascades are essential for the quantification of L-arginine/NO pathway and validation of analytical results.
Fig. 1.
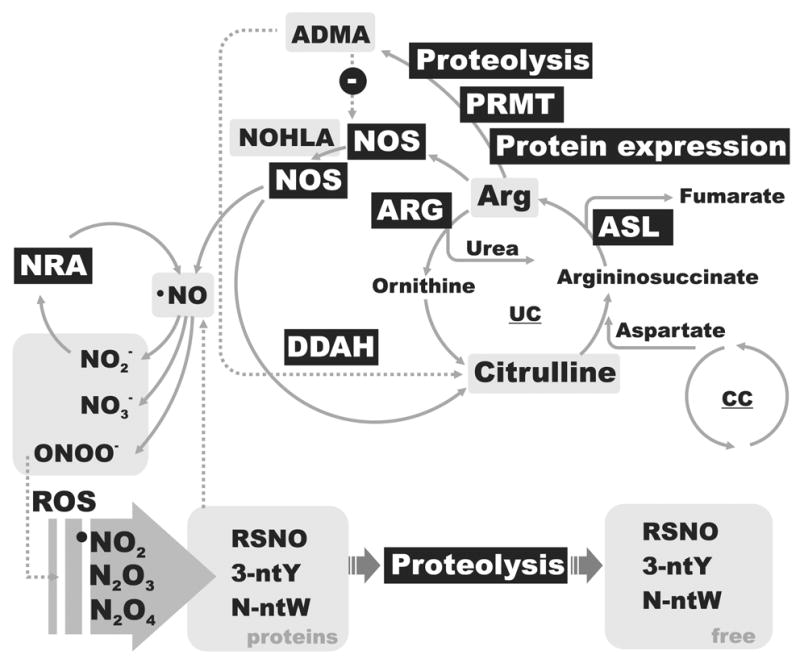
Simplified schematic of the L-arginine/NO pathway along with key analytical references. Enzymatic components are shown as a black box with white text. NO metabolic references are indicated by gray background boxes. Abbreviations are: ASL, arginine succinat lyase; DDAH, dimethylarginine dimethylaminohydrolase; NRA, nitrite reductase activity; CC, citrate cycle; UC, urea cycle; 3-NtY, 3-nitrotyrosine; N-ntW, N-nitrosotryptophan; ADMA, asymmetric dimethylarginine; RSNO, S-nitrosothiols; PRMT, protein-arginine methyltransferase.
2.1. NO production
High-capacity NO production in eukaryotic systems has two dominant but not unique routes: 1) the primary production via enzymatic oxygenation of the guanidine group of L-arginine to form NO and L-citrulline by a specific enzyme, NO synthase (NOS) [17]; and 2) the secondary production by enzymatic reduction of nitrite [18,19] via nitrite reductase activity (NRA) of xanthine oxidase [20,21], mitochondrial cytochrome complexes [22,23], deoxyhemoglobin [24–26], and some NOS isozymes under anoxic conditions [27]. It is generally assumed that the primary NO synthesis from L-arginine oxidation by NOS and following retrograde diffusion of the gaseous messenger is responsible for focal “neurotransmitter-like” actions of NO [28], as well as for pathogen-induced escalation of cytotoxic defense [29]. In contrast, the secondary NO formation route from nitrite anion is believed to be a basis of tonic circulation and retrograde “hormone-like” action of NO in organisms [30]. The latter also appears to be a fundamental mechanism for recycling of NO in cell, tissue, and whole organism [18,19]. Since NO synthesis from L-arginine oxidation and nitrite reduction varies in time and space, the contribution of both pathways should be considered in each particular bioanalytical scenario.
The capacity of intracellular NO synthesis depends on multiple factors (reviewed by Nathan and Xue in ref. [31]). It includes transcriptional [32], translational and posttranslational [33–35] regulations of NO-synthesis mechanisms, negative feedback regulation of NOS by NO [36–39], membrane transport of substrates [40–43], intracellular metabolic recycling of substrates [43–51], availability of molecular oxygen and cell respiration [52–57], and intracellular redox state [58–60].
2.1.1. Enzymatic NO synthesis by NOS
The primary enzymatic NO production occurs in two consecutive monooxygenations of L-arginine to L-citrulline with a toll of 1 L-arginine, 1.5 NADPH, 2 O2, and typically 5 electron transfer events [61] (Fig. 2). However stoichiometry of entire process may vary depending on the enzyme type, status, and redox conditions. It may involve either two-electron or one-electron oxidation in the terminal reactions resulting distinct nitroxy products [62,63]. Subsequent monooxygenation is catalyzed by an axially symmetrical homodimmeric NOS complex in which each subunit collaborates with five essential cofactors - prosthetic groups: FAD, FMN, iron coordinated haem, (6R)-5,6,7,8-tetrahydro-L-biopterin (BH4) and Ca2+-calmodulin [64,65].
Fig. 2.
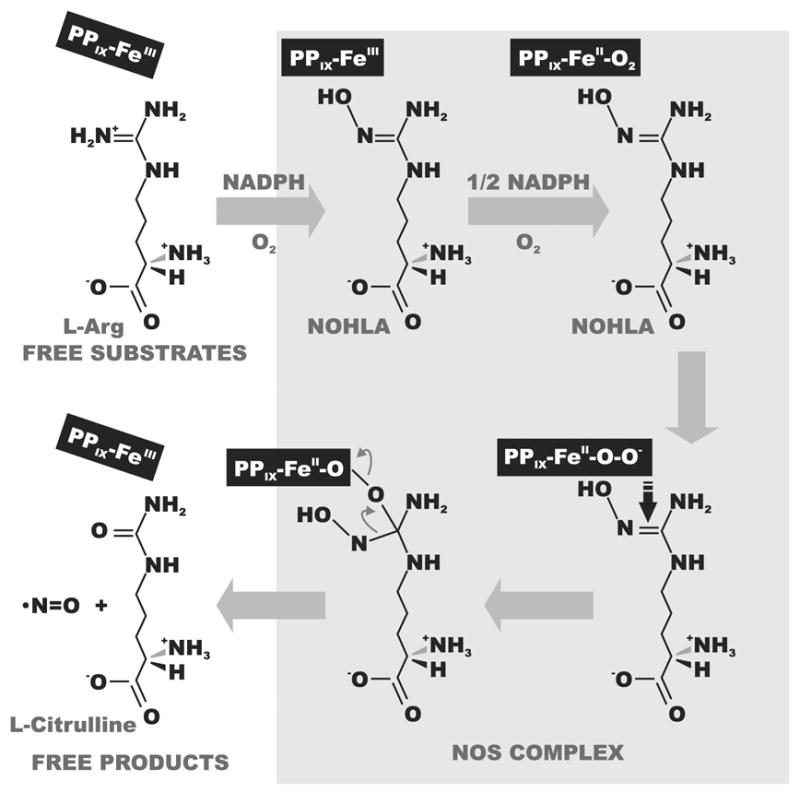
Enzymatic NO production by NOS. In this reaction one guanidino nitrogen of L-arginine undergoes a five-electron oxidation via NOHLA (NG-hydroxy-L-arginine) as the intermediate to yield one molecule of NO. PPIX-Fe, iron coordinated proto-porphyrin XI, a prosthetic group which function in the turnover of L-arginine.
There are three well characterized and functionally, morphologically, and structurally distinct NOS isoforms encoding by three specific genes in mammals; these NOS isozymes are members of the P450-containing monooxygenase system which core belongs to a subfamily in the P450 enzyme superfamily [66,67]. Earlier metazoan lineages obviously utilize a smaller number of separate genes which, however, may expand to large isozymes number through an alternative splicing of a pair (e.g. in genomic fishes and birds) or single (e.g. in genomic insects and some other invertebrates) genes per genome. NOS expansion in mammals is consistent with functional specialization of different isoforms. Mammalian NOS I (neuronal, nNOS) and NOS-III (endothelial, eNOS or universal, uNOS) are constitutively expressed. The constitutive isoforms (cNOS) generate NO in the nanomolar range (Table 1). In contrast, NOS II (hepatocyte, HE-NOS or inducible, iNOS) is transiently inducible upon cellular responses to some exogenous factors [32] and usually represents a terminal unit of a Ca2+-driven up-regulation cascade. Inducible isoforms generate a local NO pool in a micromolar range [68]. Names of NOS retrospect a location and action of particular isozymes from earlier reports. Consequently, essentially broader roles and distributions as well as synergetic activity of different NOS isozymes were established. All NOS isoforms act under tight transcriptional, translational, and posttranslational control, and likely participate in poly-enzymatic protein-protein interaction domains [32,69,70]. A putative mitochondrial form of NOS, mtNOS, has been recently reported ([34,71]; see also reviews [72–74]). Apparently, mtNOS acts in tight collaboration with mitochondrial electron transport pathways; however, a unique molecular identity and organelle assembling of such enzyme has remained under consideration [75].
Table 1.
Summary of experimentally determined NOS kinetic parameters
| Type | Alias | Forma | Cama | Ca++a | KMArg (μM) | KMO-O b (μM) | VmaxNO (Mol s−1 g−1) |
|---|---|---|---|---|---|---|---|
| NOS I | nNOS | c, m | + | + | 1.5–3 | 400 [36] | 1.6*10−7 [76] |
| NOS II | (HE)iNOS | i, s | + | − | 3–32 | 135 [36,78] | 2.7*10−5 |
| NOS III | (u)eNOS | c, m | + | + | 3 | 4 [36] | 2.5*10−7 |
| Type I – like. | mtNOS | c, m | + | + | 7 [79] | 7 [79] | 1.1*10−7 [34] |
| AtNOS1 | plantNOS | c, m | + | + | 12.5 | N.A. | 8.3*10−8 [77] |
| Type I-like | insectNOS | c, m | + | + | 2 | N.A. | 2 |
c, constitutive; i, inducible, m, membrane bound; s, soluble; Cam, calmodulin-dependent; Ca++, calcium-dependent.
Km for purified enzymes in vitro; close values were reported in vivo, e.g. KMO-O for NO synthase in the intact human lung was 190 μM [78].
N.A., not available
2.1.2. Enzymatic reduction of nitrogen oxides
The secondary NO production results from enzymatic reduction of nitrite which is catalyzed by a specific nitrite reductase (NIR; presently identified in bacteria, fungi and plants [79]) or/and auxiliary nitrite reductase activity (NRA) of multiple metalloenzymes in interaction with reducing environments and electron transport cascades, e.g. cytochrome oxidase in mitochondria, cytochrome P-450 in EPR, and deoxy-hemoglobin in erythrocytes [19,80–82]. Enzymatic reduction of nitrite typically occurs with participation of the electron donors NADH, NADPH, flavoproteins, and some other heme-containing proteins [19,23].
Enzymatic production of NO via NOS and NIR/NRA pathways are most capacitive and universal, but perhaps a very small fraction in a variety of NO synthesis cascades. Alternative mechanisms of NO production exist in eukaryotic and prokaryotic systems and may occasionally become a dominant mechanism of NO production under particular physiological conditions. For example, a large pool of NO and nitrogen oxides may be generated upon symbiotic and pathogenic interactions of organisms with prokaryotes which utilize specific nitrate/nitrite reductase mechanisms [20,83]. Intriguing diversity of orphan multi-copper proteins, which share a common ancestor with prokaryotic NIR, also exists in metazoan organisms [79], but the potential role of those enzymes in nitrite reduction catalysis remains to be analyzed. Bacterial nitrate/nitrite reductase mechanisms also provide a “side-door” for bioavailable nitrogen in nitrite/nitrate form escaping a dentification cascade and being recycled instead. In mammals, nitrate is generated upon oxidation of NO and nitrite. In general, nitrate is formed upon interaction NO and nitrite with oxyhemoglobin or some other oxidation mechanisms. Nitrate is considered as irreversible lost for NO recycling cascades and subsequently removed via active transport and excretion in organisms [30]. In bacteria intracellular nitrate concentration is regulated by active membrane transport and dentification cascades [84]. However, in hypoxic condition of upper alimentary canal bacterial conversion of dietary nitrate to nitrite may be a prevalent source of nitrite and consequently of NO for proximal tissues and entire organisms [85,86]. For example, eNOS itself may effectively reduce nitrite to NO under anoxic conditions [27], and similar biases are quite possible in other tissues.
2.1.3. Non-enzymatic production of NO
In addition to the enzymatic NO production routes from L-arginine and nitrite several important non-enzymatic routes which may account for burst of NO production in vivo have been identified. These routes include spontaneous reduction of tissue nitrite to NO under acidotic conditions, e.g. in the ischaemic heart [87,88], and reaction between L- or D-arginine and some arginine derivatives with hydrogen peroxide to form NO [89,90]. There are other “non-enzymatic” sources which may contribute to the local levels of NO, although in less extent and some times in conquer to the enzymatic production. These include spatial buffering and redistribution of NO and nitrite in organisms via reversible interaction with circulatory proteins and cells [30,91]. For example, NO may interact with haem-binding proteins in erythrocytes [26,92,93], other metal-binding proteins of cells and plasma [94,95], as well as with sulfhydryl (R-SH) and aromatic (R, indole or phenol) [96–98] groups of proteins, biopolymers and free amino acids (see also above).
2.2. NO precursors, metabolites and analytical references
Intracellular L-arginine and nitrite are major and essential precursors of endogenous NO. Governing evidence suggests that both NOS- and NIR/NRA-coupled NO production mechanisms undergo substrate- and product-dependent modulations. The fact that enzymatic availability of primary substrates is limited by biological membranes, while gaseous product NO diffuses freely in biological matrices, has important consequences for intra- and extra-cellular NO concentration and activity. It suggests that both NO synthesis pathways may rapidly consume substrates which are available in a frame of substrate diffusion domain and respectively may release NO only in equilibrium with a passive or, most likely active delivery of the substrates, e.g. via specific membrane transport of nitrite, L-arginine or it metabolic precursors. Depletion of NO substrates at the cellular level is partly compensated by substrate recycling cascades. For example, L-citrulline conversion to L-arginine in the urea cycle may act as substrate recycling mechanism for NOS. Enzymatic conversion of intracellular NO to nitrite anion provides substrate for NRA pathways and also reduces lost of NO upon diffusion of this molecule out of cell. Both, nitrite generation and L-arginine synthesis de novo would stabilize an intracellular NO pool. Obviously specific transporters and substrate recycling mechanisms act in synergy as a NO-generated and NO-regulated metabolic network.
2.2.1. NOS substrate, L-arginine
L-Arginine is a primary substrate of the NOS-mediated biosynthesis of NO. It has higher affinity to NOS reactive centers and apparent selectivity to generate NO in comparison to other guanidine group-containing substrates [61]. It also participates in protein synthesis and detoxification of ammonia formed during the nitrogen catabolism of amino acids (urea cycle). In addition, L-arginine serves as a metabolic precursor in the formation of creatine, polyamines, the excitatory neurotransmitter L-glutamate, the neuromodulator L-proline, the putative neurotransmitter agmatine, and the putative immunomodulator, arginine-containing tetrapeptide tuftsin. Upon apparent bottleneck in the L-arginine anabolism, L-arginine is usually considered to be a conditionally essential amino acid in specific tissues of adult organisms and an essential amino acid in developing organisms. Such an essentiality profile at the level of individual cells is often associated with constitutive or inducible NOS activity.
Normal L-arginine concentrations in human and animals plasma vary from 60 to 250 μM [99]. It is present in endothelial and neuronal cells at relatively steady state high concentrations of up to 1–2 mM [100,101], which implies existence of an active and regulated transport system. The intracellular L-arginine pool is generated by mean activity of protein digestion/synthesis pathways, amino acid metabolism [102], and secondary plasma membrane transporters. First two sources have been reported to be often insufficient to satisfy NO production demands [103]. L-Arginine has been shown to be a key or preferable substrate in several reminiscent mammalian transport systems, including system y+ and in the fewer capacity systems b0,+, B0,+, B0, and y+,L [40,104]. Most of these transport systems have been identified in a tight alliance with elevated synthesis of NO belong to SLC7 family (HUGO; phylogenetically-driven Solute Carrier Family nomenclature) [105], except for the sodium-dependent B0,+ [106,107] which represents a unique mammalian member A14 of the sodium neurotransmitter symporter family (SNF, SLC6A14). Three transporters comprising a cationic amino acid transporters subfamily of SLC7, CAT (member SLC7A1-A4), are considered as high-capacitance essential substrate providers for normal NOS activity in different tissues [42]. CAT-1 and CAT-3 are associated with constitutive eNOS and nNOS activity, respectively, while both low- and high-affinity isoforms of CAT-2 (A and B isoforms respectively) were found to be co-induced with iNOS in macrophage-pathogen interaction responses and some other tissues [41,105]. A protein-protein-interaction metabolon involving NOS and L-arginine transporters have been identified to facilitate NO production [45,108].
Intracellular L-arginine is substrate and product in the urea cycle [109]. This cycle is important for the waist disposal of ammonia, high-capacity L-citrulline-to-L-arginine recycling and a TCA-bridged L-arginine synthesis pathways in which L-arginine is synthesized de novo in the second part of the cycle via argininosuccinate synthetase (ASS)- and argininosuccinate lyase (ASL)-catalyzed reactions. ASS catalyzes the formation of argininosuccinate from L-citrulline and aspartate, whereas ASL cleaves argininosuccinate to yield fumarate and L-arginine [110]. Therefore, the transport and subsequent metabolism of various amino acids could benefit L-arginine synthesis indirectly.
2.2.2. NO oxidation product and nitrite reductase substrate, nitrite anion
Circulating nitrite pool in organisms is primarily a result of combined activity of NOS and NO oxidation pathways. In oxygenated biological environment NO may undergo a number of spontaneous and enzymatically facilitated oxidative and radical reactions [111], unequivocally leading to the relatively stable nitrite and to the inert nitrate. The half-life of NO under normoxic conditions in vitro can be up to 30 s, however it may differ in vivo [112]. The oxidation kinetics in vivo depends on local redox conditions as well as on nature and status of metalloenzymes which may interact with NO and oxygen. The half-life may increase at low NO concentrations and under hypoxic and reducing intracellular conditions up to 100~500 s [113]. The half-life of NO decreases in the presence of oxidized forms of haem-binding proteins participating in the cellular respiration and relevant electron transport systems [1,114–116]. NO oxidation represents a major source of intracellular nitrite since other capacitive sources are unknown and perhaps absent in organisms. In contrast, nitrate- and nitrite-specific transport coupled to nitrate reductases is most prominent and apparently essential for regulation of nitrite pool in prokaryotes [117] and plants [118]. Homologous components for nitrate/nitrite transport are largely unknown in organisms, but there is some evidence of nitrite transport via a Cl−/HCO3− exchanger [119] which is member of the bicarbonate transporter family (SLC4). Nevertheless, it has been shown that dietary nitrate [86,120,121] as well as symbiotic and/or pathogenic bacteria immobilized nitrate [88,122–124] may play an important role in the formation of nitrite at pathogenic and normal levels and consequently in regulating NO pools in higher organisms.
2.2.3. Peroxynitrite
NO undergoes a variety of oxidative and radical reactions generating a number of stable reaction products and intermediates with a wide range of stability and reactivity [125]. One such a product, the peroxynitrite anion (ONOO−), has attracted wide attention in the area of NO research. ONOO− is the dissociation product of peroxynitrous acid, ONOOH, pKa 6.5–6.8. Although it is a frequent key word in scientific literature, data on biogenesis, action, and role of this extremely reactive anion with a half-life of <1 s [126] remain very fragmented and often are controversial. Peroxynitrite is the primary product of the reaction of superoxide anion and NO, with other reactions also being capable of producing peroxynitrite (see Eqs. below and ref. [125]).
| (1) |
| (2) |
| (3) |
ONOO− has been identified as a universal factor of cellular toxicity, causing oxidative mispairing and scission of DNA strands [2,127–130], degenerative oxidation and nitration of proteins [131,132], and oxidative damage to lipids [133,134]. Peroxynitrite undergoes homolysis to the free radicals •NO2 and •OH (Eq. 2). Peroxynitrite also rapidly reacts with CO2 to yield nitrosoperoxycarbonate (ONOOCO2−), which is the supposed predominant fate of peroxynitrite undergoing homolysis to form carbonate and nitrogen dioxide radicals. These radicals can recombine to finally form carbon dioxide and nitrate. Carbonate and nitrogen dioxide free radicals are likely responsible for peroxynitrite-induced toxicity. Peroxynitrite can nitrosate thiols with low yield, however, there is some controversy about the putative mechanisms [135,136].
Another potent nitrogen oxide of the RNS family, i.e. dinitrogen trioxide (N2O3), is produced upon autooxidation of NO [137–139] (Eqs. 4 and 5).
| (4) |
| (5) |
It has been suggested that the majority of nitrosation occurs via a series of reactions between dinitrogen trioxide and thiols or aromatic moieties [137–139]. It was also proposed that a heterogeneous, NO-derived pool of higher nitrogen oxides (i.e., NO2, N2O3, and N2O4) and intermediate organic NO derivatives may be responsible for a given profile of NO-dependent modifications in vivo [140]. Recently, HPCE with UV excitation and an array diode detector was employed to identify peroxynitrite, nitrite, and nitrate in standard samples. The LOD of peroxynitrite reported in this work was suitable for detection of contaminations in peroxynitrite preparations from commercial sources but not in biological samples [141]. Authors mentioned that use of electrochemical detection may improve analysis of peroxynitrite in biological samples with CE.
2.2.4. S-Nitrosothiols (RSNO)
In biological systems, pools of thiols and S-nitrosothiols may act as scavenger of RNS [135], NO donors [94] and/or NO transport vehicles [91,139,142]. For example, concentrations of thiols in the forms of albumin, cysteine, homocysteine, and glutathione in the human vascular system are 680 μM [143], ~10 μM [144], 5 up to 15 μM in healthy vs diseases subjects [145], and 7 μM [146], respectively. While a small fraction of circulated thiols of < 1% is accessible to S-nitrosation, it may be sufficient to compensate a basal RNS level in healthy subjects. The S-transnitrosation of protein cysteinyl moieties is a reversible facile modification that plays an important role in posttranslational alterations of protein function and signal transduction [147]. S-Transnitrosation has been reported to be equally important like phosphorylation, myristylation, palmitylation and sialytation [147]. The S-nitrosation of proteins and peptides can produce guanylyl cyclase-independent signaling via reversible protein modification. It can also produce exchange and retrograde scattering of nitrosyl group in the network of cysteinyl and aromatic residuals of proteins [148–151].
A basal level of S-nitrosothiols is always present in biological systems [91,152,153], and it increases during inflammation [154–157]. It can be regulated via intracellular traffic and active plasma membrane transport of S-nitroso-L-cysteine by an L-type amino acid transport system (LATs of the SLC7 family) [158–161]. S-Nitrosothiols may be important component of the NO pathway, but this issue is problematic and controversially discussed with regard both to concentration and biological roles. Additional analytical effort would be necessary to justify S-nitrosothiols as a systemic biomarker of NO activity in biological systems.
2.2.5. Aromatic nitrosation, 3-nitrotyrosine and N-nitrosotryptophan
In addition to peroxynitrite, higher nitrogen oxides produced from the autooxidation of NO can interact with aromatic moieties of proteins and small biological molecules such as amino acids, monoamine neurotransmitters, and peptides. During the last decade, an increasing volume of evidence suggests the important physiological role of tyrosine and tryptophan nitrosation. It appears that the efficiency of such an aromatic nitrosation is comparable to that of S-nitrosation. At the same time, the cumulative frequency of these amino acids in protein sequences e.g. 1.47%W and 3.6%Y vs. 1.69%C as well as their good accessibility in a 3D protein pocket, except for tryptophan, result in a relatively high probability of modification by RNS attack,. The nitrosation of aromatic moieties may play important roles in buffering NO-RNS actions [139,140] conformational alteration of protein functions [168], and additional source providing NO and nitrosyl groups to thermodynamically favorable reactions (e.g. nitrosation of thiols [140]); however, a major effort has been made to understand the role of 3-nitrotyrosine [162–166], and to a lesser extent of N-nitrosotryptophan [138,139,167–169], in pathological conditions and diseases.
The experimentally determined extent of peroxynitrite-induced nitration of tyrosine has proved the concept that biogenic RNS can promote aromatic nitration [164–166,170]. However, the role of peroxynitrite as a nitration agent has been questioned [171,172], and an alternative has been suggested: the more potent nitrosation via NO2 interaction facilitated by the presence of transition metal centers [172–174]. In addition to nitration, inflammatory conditions can facilitate other oxidative modifications in tyrosine, including chlorination, bromination, and hydroxylation with efficiencies comparable to S-nitrosation, e.g. 10−8 – 10−7 mol per g of protein [175]. The protein nitration yield in vivo is low, e.g. 1 – 5 NO-Tyr per 104 Tyr residues [175]. Values from 6–80 μmol/mol (Tyr/NO-Tyr) were reported in reference [176], and ~ 10 ng NO-Tyr/mg protein in plasma and tissue up to 170 ng/mg in actin-enreched fractions of tissue proteins have been reported in reference [177]. It has been demonstrated recently that N-terminal-blocked tryptophan moieties can effectively be nitrosated by N2O3 [138,139], outlining the pivotal role of high nitrogen oxides in aromatic nitrosation.
Nitration and oxidation of aromatic moieties may impact enzymatic function [138,139,167–169]. On the other hand, these reactions may destabilize protein structure, making them more accessible to cellular proteolysis [175]. Presumably, nitrosated amino acids are less efficient substrates in metabolic and protein synthesis pathways, which may lead to increases of nitrosated and oxidized aromatic compounds in the cytoplasm, specific tissues, circulation and urinary excretion. From the analytical perspective, the detection of nitrosated small molecules can be used to qualify systemic as well as organ- and tissue-specific NO-induced toxicity [178–180].
2.2.6. Asymmetric dimethylarginine (ADMA)
ADMA is a biogenic analogue of L-arginine and a potent inhibitor of NOS-catalyzed NO production [181]. It is a regular sub-product of enzymatic methylation of polypeptides on the guanidino group of internal L-arginine residues. This methylation reaction is catalyzed by enzymes protein-arginine N-methyltransferases (PRMTs), which interacts with specific methyl acceptor motifs in peptide chain and yields NG-monomethylarginine (L-NMMA), NG,NG-dimethylarginine (asymmetric dimethylarginine, ADMA) and NG,N′G-dimethylarginine (symmetric dimethylarginine, SDMA) [182]. PRMTs utilize an intermediate of the homocysteine pathway, S-adenosylmethionine, as a donor of methyl groups. These enzymes are now categorized into two categories, type I and type II, which yield ADMA and SDMA, respectively. Both enzymes can form L-NMMA as an intermediate in the formation of dimethylated species. Subsequent degradation of irreversibly methylated arginine-containing peptides is responsible for the formation and accumulation of ADMA and SDMA which are present both in cells and in systemic circulation. ADMA but not SDMA produces local and general NOS inhibition. These inhibitory ADMA actions have been associated with a variety of NO-related disorders including atherosclerosis [183], other cardiovascular diseases [184,185], diabetes mellitus [186,187], erectile dysfunction [188], kidney disease [189,190], and aging [191–193].
The systemic clearance of ADMA occurs upon catalytic hydrolysis by the dimethylarginine dimethylaminohydrolase (DDAH) to L-citrulline and dimethylamine (DMA), membrane transport by SLC7 family members, and urinary excretion of ADMA as well as DMA [194,195]. A correlation between ADMA plasma levels and the severity of disease has been suggested as an analytical reference for disease risk factors [196]. An experimentally determined ADMA concentration of 3–10 μM was found to inhibit vascular NOS [197–200], while ADMA concentrations of 10–300 μM were found to cause endothelial collapse [201]. The effect of ADMA infusion in humans and analytical approaches used in the quantification of ADMA have recently been reviewed [202,203]. Effective HPCE techniques for the quantification of L-arginine derivatives, including ADMA, in biomedical samples have been reported [204,205]. Example of CE-LIF analysis of ADMA in serum samples from healthy and memorialized human subjects are shown in Fig. 3.
Fig. 3.
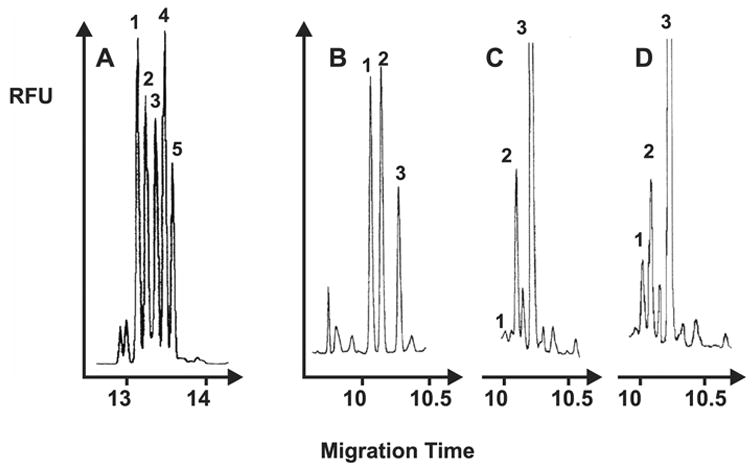
Example of HPCE-LIF detection of ADMA in standard and human subject. (A) Electropherogram of standard methylated arginines: (peak 1) SDMA, 6.25 nM; (peak 2) ADMA, 6.25 nM; (peak 3) MMA, 6.25 nM; (peak 4) L-homoarginine (internal standard), 6.25 nM; (5) LArg, 5.0 nM. Migration conditions: boric acid 50 mM, CAPS 20 mM at pH 10, +20 kV (35 μA). Electropherograms of artificial serum standards (B), healthy subject serum containing low (C) concentration (0.31 μM) of ADMA and hemodialysed subjects serum containing high (D) concentration (2.26 μM) of ADMA (peak 1), using L-homoarginine (LH, peak 2) as an internal standard and L-arginine (peak 3). Electrophoretic conditions: boric acid 50 mM, CAPS 20 mM, at pH 11.5, 30 kV. Serum samples were diluted 1000-fold in water prior to the analysis derivatized with FITC. Separation conditions were 50 mM boric acid and 20 mM 3-(cyclohexylamino)-1-propanesulfonic acid (CAPS) adjusted to high pH. A Zeta automatic CE instrument (Picometrics, Ramonville, France) equipped with a modular LIF detector and a 20 mW argon laser (wavelength nm) was used. Reprinted from [204], Journal of Chromatography B 741, E. Causse, N. Siri, J.F. Arnal, C. Bayle, P. Malatray, P. Valdiguie, R. Salvayre, F. Couderc. Determination of asymmetrical dimethylarginine by capillary electrophoresis-laser-induced fluorescence, 77–83, 2000, with permission from Elsevier.
2.3. NO targets
NO plays important biological roles as a signal messenger, protective agent, and cytotoxic factor without strict boundaries between those actions. A degree of its impact on cellular metabolism transiently depends on multiple and equally important components including generation, buffering, and degradation of the NO pool, local redox environment, as well as presence and accessibility of NO-specific molecular targets. One of the most important mechanisms of NO action leading to biological activity is the soluble guanylyl cyclase cascade. Other important mechanisms of action involve S-transnitrosylation, and disulfide formation in proteins and nitration of aromatic residues (see above).
2.3.1. Soluble guanylyl cyclase (sGC)
High-sensitivity and high-amplification of NO signaling involves sGC that converts guanosine-5′-triphosphate (GTP) into the second messenger, cyclic guanosine-3′,5′-monophosphate (cGMP) [206,207]. sGC enzyme comprises an heterodimer α1β1 and a single Fe2+-haem moiety with the haem-binding site located to the β-subunit, where histidine residue 105 forms a covalent link to the Fe2+-centre of the haem group [208–210]. Structures of several bacterial orthologs of sGC which now are solved reveal molecular basis of sGC/NO interaction [211,212]. NO forms a five-coordinated NO complex with haem cofactor of β-sGC subunit which leads to a hundred-fold more active conformation of the heterodimer, e.g. 10–100 nmol cGMP/mg×min vs 10–40 μmol cGMP/mg×min [213–215]. Assays of sGC/NO interaction have revealed complex kinetics with specific activation, deactivation, desensitization and resensitization indexes (reviewed in ref. [216]). Briefly, purified sGC in vitro has an EC50 of 85–250 nM of NO [217,218], is fully activated by NO within a few milliseconds [219], remains constantly active upon available substrate and ligand, and deactivates in a seconds-to-minutes range after NO disappears from the reaction scene [220,221]. sGC in vivo exhibits EC50 of 45 nM of NO and an approximately 25-fold faster deactivation [222]. Downstream cGMP signaling cascades include stimulation of G-family protein kinases, altering cGMP-gated ion channels, and modulation of cGMP-binding phosphodiesterases [206]. Termination of cGMP signaling occurs upon metabolic deprivation of the cGMP. This reaction is catalyzed by phosphodiesterase-5 which is often co-expressed and spatially colocalized with NOS [223].
3. Analytical approaches to study NO metabolites
The important role of NO in human physiology and health maintains a steady effort to determine NO and proximal metabolites in biological matrices leading to the development of an immense variety of analytical approaches which may be categorized as: (1) optical methods including spectral colorimetric and ultraviolet spectrophotometric methods, fluorometric assays, and chemiluminescence; (2) separation techniques including HPLC, HPCE, and GC coupled with various detectors; (3) electrochemical methods including cyclic voltammetry, amperometric sensors, and analyte-selective exchange potentiometric sensors; (4) MS-based methods; and (5) electron paramagnetic resonance spectroscopy EPR (for comprehensive reviews see refs. [224–226]). Many of these approaches require resources and expertise which exceed the potential of regular biological laboratories. Hence, after 19 years expansion of NO research the availability of simple and easily accessible analytical tools for evaluation of substrates and products of the L-arginine/NO pathway in relevant biological matrices remains to be a key limitation. HPLC and HPCE play increasingly important role in the NO research, primarily because it is most universal with respect to analytes. In addition, these separation techniques become increasingly accessible in regular biological and medical laboratories. HPLC analysis with focus on nitrite and nitrate in biological samples is subject of another review [227]. The present review focuses on the current status and trends in the development of HPCE applications for profiling the NO pathway with some emphasis on applications of various CE approaches in biomedical and life sciences.
3.1. High-performance capillary electrophoresis (HPCE) of the NO pathway
Modern HPCE comprises an interplay of analytical sampling, separation and detection techniques [228–230]. The sample is loaded and separated in a 10- to 100-μm bore of 0.1- to 1-m long fused silica capillary or anisotropicaly etched silicon micro-channel (microchip capillary electrophoresis, MCE, reviewed in ref. [231]). The spatial separation of analyte is a subject of analytical performance. It occurs upon application of high-voltage steady-state electric field as a result of differential electrophoretic motility (EM) of analytes and electro-osmotic flow (EOF) of separation media. Key advantages of HPCE over slab gel electrophoresis are efficient and uniform dispassion of current-induced heat and rapid separation with application of remarkable electrical field and current densities, typically 200–500 V/mm and 10 – 60 μA, respectively. In a variety of bioanalytical applications HPCE is comparable to HPLC. Major advantages of HPCE over HPLC are simplicity of separation framework in parallel with a wide variety of separation mode available, minor if any requirements for sample treatment, opportunity to measure analytes in sub-microliter volumes of samples, and several orders of magnitude better resolution compared to HPLC, e.g. >50,000–500,000 vs. 5,000 theoretical plates [232]. Resolution can be as high as 6.1 million theoretical plates upon separation with an ultrahigh voltage CE at 120 kV [233]. This unprecedented resolution results from virtually flat electrophoretic and electroosmotic velocity profiles across the capillary channel and absence of mass transfer which limits resolution in HPLC. In addition, the technological progress in the development of HPCE hardware follows the rapid progress in electronics and optics design including developing integrated high-voltage power supplies, miniaturizing and expanding range of light-emitting sources and improving high-resolution, low-noise analog electronic technologies. With reduction of price and increase of availability of such technologies, HPCE has becoming one of the most technological, cost-efficient and flexible analytical framework which can be easily modified at sample injection, separation, and detection sites. For the same reason, HPCE became one of the most potent techniques for future integration into high-throughput microanalytical devices and lab-on-the-chip platforms prepared in an integrated technological process and suitable for routine biomedical and biological laboratories with a convenient analytical expertise. Factors influencing EM and EOF include complex equilibrium of analytes with additives, ion pairing, pH, interaction of analytes and capillary surface modification and can be employed to improve separation properties.
There are six separation modalities in HPCE, three of which are proven to be very useful for analysis of the L-arginine/NO pathway. The capillary zone electrophoresis (CZE) is the most simple and convenient method which is widely used for separation of inorganic ions [234–237] and small biomolecules [238], and even both categories simultaneously [239]. Because nitrite and nitrate are easily accessible to CZE, this methodology became a generally employed HPCE technique to analyze these anions in biological samples. Isotachophoresis (ITP) can be used for preconcentration of charged analytes from diluted samples or from samples containing low amounts of analytes (for review see ref. [240]). Micellar electrokinetic chromatography (MEKC) is almost exclusively used for analysis of small molecules such as drug and their metabolites. MEKC is a very potent technique to determine small organic members of the NO pathway including L-arginine, ADMA and SDMA [204,205,241]. Other HPCE modes, including capillary isoelectric focusing (CIEF), capillary electrochromatography (CEC) and gel-sieving capillary electrophoresis generally used for separation of medium and large size molecules, and specific analytical tasks were not included in this review.
In HPCE sample can be loaded using hydrodynamic displacement (pressure or gravity enforced), electrokinetic (electrical field enforced, including electro migration and EOF) or mixed injection modes. Isoelectric focusing or staking of analyte in a pre-loaded, low conductivity plug is often used to improve detection of trace analytes, e.g. in highly diluted samples. Separation conditions depend on the concentration and pH of a separation buffer and additives which modify properties of inner silica surface of the capillary and impact EOF (summarized in Table 2). Optimization of loading, separation and detection may lead to remarkable improvement of separation and precision of the analysis. HPCE ability to snap-shot a set of analytes with similar electrophoretic properties, e.g. inorganic anions, is particularly important to obtain a comprehensive analytical profile of complex metabolic processes such as those of the L-arginine/NO pathway. Not surprisingly, HPCE analysis of NO-related metabolites is one of most rapidly progressing area of separation assays. To that date, there are over 200 reports describing different aspects of HPCE analysis of the L-arginine/NO pathway. Among those over 50 reports describe usage of different detector modalities and optimized HPCE procedures to analyze biological and biomedical samples.
Table 2.
Analytical characteristics of selected CE/HPCE methods for analysis of L-arginine/NO pathway metabolites in biological samples
| Analytes | Matrix | Sample dilution, derivatization |
Detection mode |
capillary | Separation buffer |
Determined concentrations, comments |
LOD or LOQa, comments |
Ref. |
|---|---|---|---|---|---|---|---|---|
| L-Arg, L-Cit, L-Arginino-succinate | isolated neurons, ganglia, and hemolymph from molluscs. | 1:104; fluorescamine | LIF 350–6 nm; PMT | FC | 30 mM borate buffer 20 mM SDS, pH 9.5 | L-Cit/L-Arg ratios 2.5 (NO-positive B2 cell) and 0.1–0.2 (NO-naive cells). | LODs Arg/Cit 11/15 nM | [101] |
| 3-ntY; nitrosated and chlorinated amino acids | standard mixture | native fluorescence | UVF 200 nm | FC | 50 mM borate buffer, variable pH, and EOFM spermine | large volume sample stacking | LOD 2.5–10 nM | [179] |
| ADMA, and other arginines | human serum control vs. hemodialysed subjects | FITC | LIF | FC | 50 mM boric acid 20 mM 3-(cyclohexylamino)1-propanesulfonic acid pH 9.5, 10 or 11.5; | ADMA control 0.3μM, coronary stenosis 1.2 μM | LOD 50 nM (Plasma) | [204] |
| ADMA, and other arginines | human blud plasma | 4-fluoro-7-nitrobenz ofurazan | LIF | FC | borate buffer at pH 9.4; MEKC | includes a MEKC optimization | LOQ 20 μM (UV) 100 nM (LIF); LOQ plasma 125 nM | [205] |
| NO3− | human urine | 1:50 | UV 254 nm | FC | chromate, Nice-Pak OFM Anion-BT, pH8.0 | ND | [244] | |
| NO2−, NO3− | human serum, urine, brain tissue homogenate | 1:10, 1:100 | UV 214 nm | FC | 10 mM sodium sulfate, 2.5% OFM-OH | LOD25 μg/l | [245] | |
| NO2−, NO3− | rat dorsal root ganglia, biopsy, tissue homogenate | 1:103; 1:104 SPME Cl− cleanup | CD | FC | 81.5 mM sodiumborate, 25 mM arginine, 0.5 mM TTAH, pH 9.5; ITP injection | NO2−/NO3− were 96-24/231±29 nM; | LOD = 8.9/3.54 nM | [247] |
| NO2−, NO3− | pheochromocy toma cell line PC12 | cell culture supernat ant | UV 214 nm | FC | 150 mM Tris-phosphate, 6 μm hexadecyltrimethy-ammonium chloride, pH 7.0 | synergistic stimulation of tumor necrosis | LOD NO2− ~25 pM | [254] |
| NO2−, NO3− | rat brain perfusate | microdia lysis 50 nl/min | UV 214 nm | FC | 20 mM phosphate buffer; 2 mM CTAC, pH 3.5 | Brain perfusate[NO2−/NO3−] = 3.4/15.5 μM; 1.5 min separation | LOD0.96/2.86μM | [256] |
| NO2−, NO3− | human serum | 1:10 | UV 214; LPA | MCE | 2 mM Na2HPO4, 12 mM KCL, 412 mM NaCl, 5.43 mM urea, 4,72 mM glucose | 6.5 sec separation | LOD NO2−/NO3− = 24/12 μM | [264] |
| 3-ntY | diabetic and control rat urine | 1:1 acetonirile | UV 214 nm | FC | 150 mM phosphoric acid, 0.5 mM CTAB, pH 6.4 | large volume sample stacking | LOD normal/stac King = 1.77/0.08μM | [292] |
| NO2−, NO3− | identified molluscan neurons, ganglia, and hemolymph | 1:1, 1:10, 1:50 | UV 214 nm | FC | 100 mM NaCI, 2 mM CTAC | highest NO2−/NO3− found in NO positive neurons 2–12 μM | LODs < 4μM | [387] |
| NO2−, NO3− | rat ASF, plasma | 1:1 | CD ConCap | FC | 100 mM CHES, 40 mM LiOH, pH 9.3, 80 mM spermine; 8% propan-2-ol | Repeatability range5.3–9.8% for various ions | ND | [400] |
| NO2−, NO3− | human serum | 1:10 | UV 214 nm | FC | 25 mM sodium sulfate, 5% Nice-Pak (OFM) Anion-BT | Repeatability, 6.4–10%; reproducibility, 9–11%; capillaries are stable for over 300 analyses | ND | [406] |
| NO2−, NO3− NOx | CSF from MS and control subjects, lumbar puncture | 1:10; | UV 214 nm | FC | 25 mM sodium sulfate 5% NICE-Pak (OFM) Anion-BT | SCF NO2−/NO3− level 304/82 and 34/134 % in active and inactive MS respectively vs. control | ND | [421] |
| NO2−, NO3− | human saliva | 1:100 | CD suppressed | CFS | 2 mM sodium tetraborate | repeatability, ≤5%; reproducibility, ≤10% | LOD=8–10 μg/l | [429] |
| NO3− | human serum, urine | 1:25, 1:50 | UV 250 nm | FC | 2.25 mM pyromellitic acid, 6.5 mM NaOH, 0.75 mM HMH, 1.6 mM triethanolamine, pH 7.7 | repeatability, 7–11%; reproducibility, 10–15% | LOD=1–2 mg/l | [430] |
| NO3− | rat urine | 1:40 | UV 214 nm | FC | 25 mM sodium phosphate, 0.5% DMMAPS, 1% Brij-35 | repeatability, <3%; no deterioration of capillaries was observed for over300 analyses | LOD=0.5 mg/l | [431] |
| NO2−, NO3− | human serum | NA | UV 214 nm | FC | 750 mM sodium chloride, 5% Nice-Pak OFM Anion-BT | repeatability, <5%; reproducibility, <8%; recovery, 97–114% | LOD=0.1 mg/l | [432] |
| NO3− | human serum, milk | ion-exchange | UV 235 nm | FC | 50 mM DoTAB, 18 mM sodium tetraborate, 30 mM Na2HPO4, 10% propan-2-ol, pH 7.0 | repeatability, 1.8–13.5% | LOD=55.8, 2.5 and 8.7 mg/l | [433] |
| NO2−, NO3− | human serum | NA | UV 200 nm | FC | 200 mM lithium chloride, 0.7 mM TTAH, 5 mM triethylamine | repeatability, ≤ 7.3%; reproducibility, ≤6.4% | ND | [434] |
| NO2−, NO3− | human serum, urine, CSF, human, plant tissue | 1:5 | UV 214 nm | FC | 8 mM Na2HPO4, 4 mM NaH2PO4, 1.4% NaCl, 0.1% poly-ethylene glycol | repeatability, <6%; urinary nitrate analysis was compared with enzymatic assay | LOD=0.3 mg/l | [435] |
| NO2−, NO3− | human serum, urine | 1:100 | UV 214 nm | FC | 15 mM sodium sulfate, 2.5% (OFM)-OH, pH 8.0 | repeatability, 3.0–3.3%; reproducibility, 4.6–5.0%; recovery, 92–113% | LOD=1 μM | [436] |
| NO2−, NO3− xanthine oxidase, NAD+, NADH | human plasma, serum | NA | UV 214 nm | FC | 150 mM sodium chloride, 5 mM Tris-HCl, 2 mM TTAB, pH 7.4 | linearity 0.2–1 mM; repeatability ≤3%RSD; reproducibility ≤1% | ND | [437] |
| GSH, GSSG, GSNO | human erythrocyte extract | 1:2 | UV 200 nm | CFS | 40 mM sodium phosphate, 0.1 mM diethylenetriaminepent aacetic acid, pH 2.2 | mobility GSSG, GSH and GSNO was reported | ND | [438] |
| NO2−, NO3− | rat ASF, plasma sub-microliter samples | 1:1 | UV 214 nm | FC | 50 mM sodium phosphate, 0.5 mM spermine, pH 3 | [NO2−/NO3−] = basal ASF 83/102, plasma undetectable/70 μM; lipopolysaccharide induced ASF=103/386 plasma 138/377 μM | LOD=30/10μM | [439] |
LOD and LOQ values listed here are presented as reported in the original publications cited.
Abbreviations: AA, amino acids; ADMA, asymmetric dimethylarginine; ASF, airway surface fluid; CSF, cerebrospinal fluid; CFS, coated fused silica; CTAC, cetyltrimethylammonium chloride; DMMAPS, 3-(N,N-dimethylmyristylammonio)propanesulfonate; FS, fused silica; LPA, linear photodiode array; MCE, microchip capillary electrophoresis; ND, not determined; PMT, photomultiplier.
3.2. HPCE detection
Micro-scale dimension and high separating voltage conditions associated with HPCE require special adjustments on the detector including electrical isolation of its hardware or “0” voltages clamp at the detector site plus adequate miniaturization of the detection cell. The detection modalities that were employed in HPCE separation of the NO metabolites include direct or indirect UV and visible light absorbance detection (UV/VIS), conventional UV source-induced fluorescence (CIF), laser-induced fluorescence (LIF), contact conductivity (CD) or contact-less conductivity (capacitively coupled conductivity detector, CCD), electrochemical amperometry, and electrospray mass spectrometry. The variety of HPCE detection techniques have been comprehensively reviewed recently [242,243]. Modification of basal electrolyte and physicochemical properties of analytes are essential to enable a particular detection mode and improve resolution. For example, reduced conductivity basal electrolyte would be necessary for direct conductivity detection; similarly, optically active additives or derivatization of non-fluorescent analytes with fluorogenic agents would be essential for UV/VIS-AE or CIF/LIF detection respectively.
3.2.1. Direct UV absorbance detection
Direct UV absorbance detection with UV-transparent carrier electrolytes and UV-absorbing analytes is the traditional way to quantify separated analytes and it is the most commonly used HPCE detection mode for the measurement of NO metabolites in biological samples. UV absorbance detection was first used with CZE to detect simultaneously nitrate and nitrite in biological samples [244,245] at about the same time as HPLC technique was used for similar analyses [246]. Minor detection interference with other anions was observed by using various electrophoretic and separation modalities and buffers, except for chloride which may interfere with electrokinetic sample loading and nitrite detection in biological liquid and tissue samples [247–249]. A range of 210–220 nm for UV absorbance detection with apparent efficiency maximum at 214 nm is commonly used for the detection of nitrite and nitrate. In general, CZE with direct UV absorbance detection does not require pretreatment of biological sample, and use of organic solvents makes this technique useful for “direct sampling”. However, selectivity may be dramatically improved after ultrafiltration and stacking of homogenized and diluted biological samples. An LOQ of ~25 ng/ml was achieved during earlier assays for both ions in biological samples when they were analyzed in ultrafiltrates of plasma, urine and brain tissue extracts [245]. This sensitivity was sufficient to quantify nitrate and nitrite in the mammalian brain extracts, but was insufficient to detect the nitrite ion in biological fluids with apparently low nitrite content. Other bioanalytical examples and improvements of this technique have been reported subsequently [250–261].
Sensitivity and number of detected NO metabolites in complex biological matrices with direct UV approach may be increased by extending excitation band in the short wavelength UV range and band-pass filtration of appropriate spectral windows in combination with a spectra-resolving devise such as array multianode PMT assembly, photodiode array, high-sensitivity line scan or square matrix scan CCD camera. Applicability of UV detection also expanded with developing new technologies such as light-emitting diodes and lasers with different spectral properties. For example HPCE/direct UV/diode array bundle have been used for the simultaneous detection of nitrite and nitrate as well as N-nitroso-L-arginine [262], peroxynitrite [141] and NG-hydroxy-L-arginine [263].
MCE in combination with diode array has also been employed for the ultra rapid separation detection of NO metabolites [264]. This approach showed excellent stability even within a very short 6.5-s separation interval. However, described MCE application demands further development because currently it provides LOD values of 24 μM for nitrite and 12 μM for nitrate, which are insufficient to quantify these ions in many biological samples.
3.2.2. Indirect UV absorbance detection
Indirect UV absorbance detection is a generic detection mode for the HPCE and HPLC analysis of inorganic ions as well as some organic species. It has been applied to the analysis of NO metabolites in biological samples, because many members of the L-arginine/NO family exhibit limited potential for UV absorbance detection. In the indirect UV absorbance detection analytes are detected as a UV transparent migration band in UV absorbing background electrolyte. This technique is simple and inexpensive. It provides convenient and reliable assay with a sufficient method detection limit, reproducibility and selectivity in biological matrices. However, it is mainly applicable when selectivity and LOQ are not critical issues and analytes of interest are present in the sample within the submicromolar-millimolar range (Table 2). Indirect UV absorbance detection requires the modification of background electrolyte (BGE) by an additive (probe) of a relatively high UV absorptivity compared to analytes, which results in reversed recorded peaks. At the same time, electrophoretic mobility close to that in analytes (for mobility matching rules see ref. [265]) and appropriate concentration are essential for separation performance (reviewed in refs. [266] and [250]). For example, inorganic anions including NO2− and NO3− can be efficiently separated in basal electrolytes with addition of chromate [267], pyromellitic acid [268] and salicylate [269]. Other useful probes for simultaneous separation of a range of inorganic anions and organic acids include p-aminobenzoic acid [270], phthalic acid [271], 5-sulfosalicylic acid [272], trimellitic acid (TMA) [273,274], and 1,3,5-benzenetricarboxylic acid [275]. Future development of HPCE/UV bundle technique may lead to a convenient approach for determination of nitrite, nitrate and some other NO-related metabolites in biological samples. LED-sources combined detectors can be especially useful for indirect detection in the UV/VIS region, e.g. detection of anions using a strongly absorbing electrolyte [276]. LED has excellent stability enabling very low instrumental noise and detector performance which in combination with low cost and inerrability comprises a very promising detection technology in terms of low cost and integration of disposable CE devises.
3.2.3. Direct/indirect fluorescence and laser-induced fluorescence (LIF) detection
The vast majority of the members of the L-arginine/NO pathway are not fluorescent in the easy achievable excitation range e.g. deuterium, tungsten, or xenon arc sources. However, because fluorescence detection provides exceptionally high sensitivity especially in combination with laser-induced excitation, a substantial ongoing effort aims at developing assays based on the derivatization of NO metabolites by fluorogenic agents. Alternatively, a chemically-inert fluorescent probe can be added to the BGE for indirect fluorescence detection of separated analytes. This approach may be useful if derivatization and other derivatization-induced changes in the separation efficiency are concern [277]. In addition, a series of laboratory techniques for post-column derivatization/detection has been developed (reviewed in ref. [278]). Detectors with conventional UV sources that may operate at wavelength above 185 nm allow a relatively wide selection of derivatising agents, whereas the selection of fluorophores for LIF detection is limited to a specific set with excitations corresponding to wavelengths of available lasers. Commonly used LIF devices utilize Ar ion (457, 488 nm), HeCd (325, 422 nm), pulsed diode (355, 405 nm), solid state UV (266 nm), and laser diode double-pumped solid-state (LD-DPSS, 473 nm [279]) lasers.
A variety of derivatizing agents and derivatization techniques was proposed (recently reviewed with respect to amino acid analysis in ref. [280]; see also refs. [281–283] for detailed derivatization protocols). Floyd with coworkers [284] have described CZE-LIF system and proposed a fluorescamine derivatization protocol suitable for separation and simultaneous quantification of L-arginine, L-citrulline, L-argininosuccinate, L-ornithine, and L-arginine phosphate in isolated neurons of the marine slugs, Pleurobranchaea californica and Aplysia californica. They report LOD in the range 5×10−17– 1.7×10−14 mol which were corresponded to 5×10−9 – 1.7×10−5 M in cells under study, hence, justifying sufficient sensitivity of the method to determine intracellular levels of these metabolites. The consequent progress in the single-neuron HPCE-LIF analysis of amino acids in particular showed this technique to be useful for analysis of L-arginine and L-citrulline with LOD values of 8.4×10−17 mol (1.1×10−8 M) and 1.1×10−16 mol (1.5×10−8 M), respectively, which may aid in determining NOS activity at the level of individual cells with high as well as low NOS activity [101,285]. Examples of inorganic and organic metabolites recordings from identified individual neurons shown in Fig. 4. In addition, it has been shown that L-arginine/L-citrulline molar ratio indeed changes upon inhibition of NOS by a specific NOS inhibitor, i.e. L-iminoethyl-N-ornithine, in the isolated nitrergic neurons. HPCE-LIF assay has been optimized for analysis of amino acids including L-arginine and its metabolites in human serum plasma [286,287], microdialysis samples from rat thalamus [288,289], and cerebrospinal fluids [290,291].
Fig. 4.
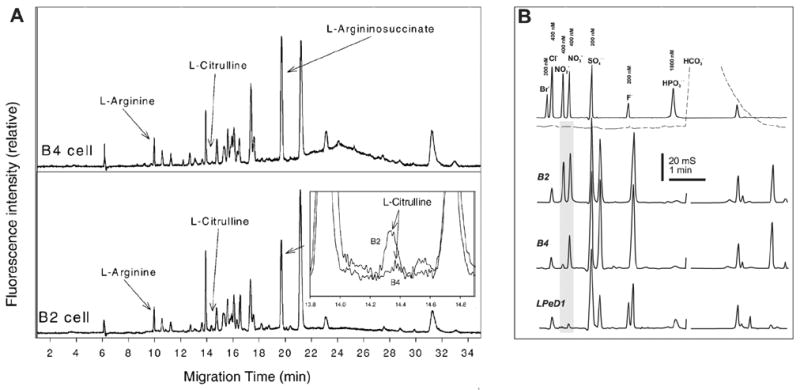
An example of HPCE analysis of NO metabolites in isolated neurons, putative NO-ergic B2, peptidergic B4, and LPeD1 cells examples are shown. (A) CZE-LIF separation and detection of arginine/citrulline ratio in the samples of isolated B2 and B4 neurons. Arginine and citrulline peaks are identifiable at electropherograms, but citrulline levels are very low in B4 neuron, see magnified superimposed traces in insertion. Sample was derivitised with fluorescamine and separated in 40 cm beyond the detector point long capillary using a borate/SDS BGE. LIF setup included 350–356 nm Ar/Kr ion laser microscope objective spatial filter and photomultiplier tube. (B) Electrophoretic separation/detection of NO2−/NO3− in selected identifiable molluscan neurons. CZE-contact CD was performed in arginine/borate BGE (pH 9.5) with addition of 0.5 mM TTAOH electro-osmotic modifier. Samples were clean up with SPME techniques and introduced using ITP staking injection. The separation was carried at −28 kV. Reprinted from [101], Journal of Inorganic Biochemistry 99, L.L. Moroz, R.L. Dahlgren, D. Boudko, J.V. Sweedler, P. Lovell, Direct single cell determination of nitric oxide synthase related metabolites in identified nitrergic neurons, 929–939, 2005, with permission from Elsevier.
HPCE-LIF was also used to analyze ADMA and SDMA labeled with fluorescein isothiocyanate (FITC) in plasma samples from patients with renal failure, peripheral arterial occlusive disease or clinically asymptomatic hypercholesterolemia [204]. More recently, Trapp et al. [203] have reported the development of efficient electrophoretic methods for the separation and quantification of L-arginine and several related metabolites including dimethylarginine, NG-monomethyl-L-arginine, L-homoarginine, L-ornithine, and L-citrulline. Amino acids were derivatized with 4-fluoro-7-nitrobenzofurazan, separated using MEKC with deoxycholic acid added as a surfactant and detected using LIF. This approach was successful to resolve ADMA and L-arginine in human plasma samples (LOQ for ADMA, 125 nM) and suggested to be useful in clinical diagnostic of cardiovascular diseases. Reverse EOF and large-volume sample stacking have been employed for the quantification of 3-nitrotyrosine in urine of diabetic rats [292] (Fig. 5). Authors have optimized separation using a 0.15 M phosphoric acid with 0.5 mM CTAB at pH 6.4 (NaOH) and reached LOD values for 3-nitrotyrosine of 177 nM for normal and 8 nM for large-volume sample stacking injection modes. These LOD values were sufficient to distinguish 3-nitrotyrosine in urine from diabetic and control rats [292]. Nitrosated aromatic compounds can be detected using native fluorescence-induced UV excitation. For example in the study on N-nitrosotryptophan-dependent nitrosation of thiols authors used a commercial CZE apparatus with UV absorbance detection at 280 nm to quantify N-acetyltryptophan and N-acetyl-N-nitrosotryptophan; LOQ value was ~15 μM at pH 7.4 [139]. Analysis of the decarboxylated L-arginine, agmatine, in human and rat tissues and urine samples has been achieved recently by using an optical fiber light emitting diodes (LED) induced fluorescence detector [293].
Fig. 5.
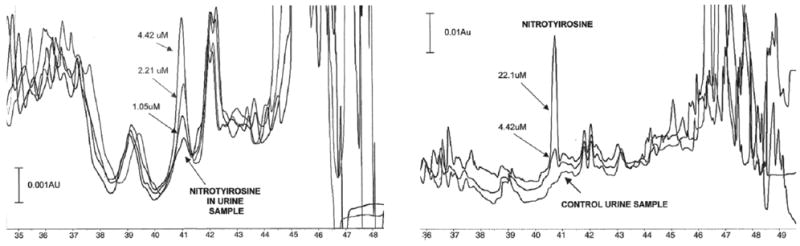
An example of HPCE/direct UV absorbance detection analysis of 3-nitrotyrosine in mammalian urine samples. Electrophoretic 3-nitrotyrosine peaks in urine samples from diabetic (left) and control (right) rats after large-volume sample stacking-CE. The samples were spiked with three or two levels of analyte 3-nitrotyrosine at 1.05, 2.21, 4.42 μM. BGE was 15 M phosphoric acid, 0.5 mM of CTAB at pH 6.4. Separations were performed in a 75-cm fused silica capillary, i.d. 50 μm, using −15 kV voltages. Pressure injection and direct UV detection (214 nm) was used. Reprinted from [292], Journal of Chromatography B 809, N. Maeso, A. Cifuentes, C. Barbas, Large-volume sample stacking-capillary electrophoresis used for the determination of 3-nitrotyrosine in rat urine, 147–152, 2004, with permission from Elsevier.
3.2.4. Contact and contactless conductivity detection (CD, CCD, C4D)
Conductivity detection comprises a set of well established and widely used techniques in combination with various HPCE separation modes. It is very attractive for developing integrated and microscale CE platforms since it is well suited for microfabrication technologies, does not require chemical modification of analytes, and its miniaturization is usually not compromised, but has positive effects on sensitivity and selectivity (for a comprehensive review see ref. [294]). Conductivity detectors fall into two major categories: contact and contact-less (CD, CCD or C4D, capacitively coupled contactless conductivity detection). CD design is more straightforward compared to CCD enabling higher sensitivity and selectivity, but at the same time the implementation of electrically coupled low signal electronic is far more challenging [295]. The recording framework must be organized to avoid electrical shock and interference from any HV sources or must be galvanically isolated using a battery-powered circuit with electrically uncoupled signal transfer, e.g. optical (up to 30 kV protection) or radiofrequency. In addition, CD requires precise alignment of the outlet of the separation capillary or the on-column electrode pair, and may be associated with significant thermal or temporal drifts as well as active surface contamination.
Several types of CD have been proposed [296–299]. Analytical grade CDs utilizing an AC impedance measurement allow the avoidance of artifacts of sensor surface polarization and interferences from the galvanically coupled HV source. Post-column contact CD with a cell-based on a fiber optic coupler was first commercialized in 1996 as a stand-alone unit (Crystal 1000, Thermobioanalysis Inc.; see also ref. [295]). This unit was designed for integration with modular HPCE (PrinCE Technologies, Netherlands) using a ConCap capillary assembly [300]. An example of ion analysis with Crystal 1000 is presented in Fig. 4B. The commercial CCD TraceDec® with a sensor head permitting use of fused silica capitally up to 5 μm has recently become available from Innovative Sensor Technologies (Strasshof, Austria).
After a prototype of a contactless conductivity detector was designed for capillary ITP [301] and a theoretical treatment of contactless conductivity measurement was proposed [302,303], it became increasingly employed in HPLC and HPCE analyses [294]. Slightly different CCD techniques were proposed for HPCE in 1998 [304,305] and subsequently improved (for reviews see refs. [306–310]). In CCD, the impedance is measured as a function of the cell capacitance value which depends on multiple factors, including the dielectric (conductivity) profile of environment and the geometry of cell and electrodes.
The optimal output of CCD depends nonlinearly on excitation frequency, whereas the maximum represents a narrow frequency range window. Placing a grounded shield between the CCD electrodes reduces the electrode coupling artifact and can substantially improve the signal-to-noise ratio (SNR) as well as the selectivity of the detector. However, it also introduces an additional technological challenge. Also, CCD will have reduced sensitivity on a capillary with high external diameter/aperture diameter values. The optimal measurement frequency of CCD depends on the BGE conductance; therefore, the optimization of an HPCE/CCD method may involve a frequency adjustment. In addition, such dependency is nonlinear and may be problematic in applications with dynamic changes in BGE conductivity. In spite of these difficulties, the popularity of CZE-CD and -CCD bundles is increasing. In detecting UV-silent inorganic ions, these techniques provide comparable or 10-fold better LOQ compared to UV absorbance detection [295,311].
The BGE composition for use in CZE in combination with CD has been previously investigated, focusing on the optimization of separation conditions and detector response [306,312,313]. Conductivity-based assays were also optimized in a number of practical works relevant to the analysis of the L-arginine/NO pathway [101,179,247–249,313–317]. The major challenge of CD/CCD in the determination of nitrite and nitrate in biological samples is the high concentration of Cl−. In vivo, the concentration of Cl− is 2–3 orders of magnitude higher, and its electrophoretic mobility is close to that of NO2− and NO3−. Therefore, chloride can interfere with sampling and detection of low quantity anions, especially of nitrite. To avoid this, several approaches were developed. Kaniansky and colleagues have used CZE/CD with integrated ITP cleanup framework, which allows a reliable detection of nitrite, fluoride, and phosphate in the presence of a molar excess of chloride, nitrate, and sulfate ions in the model mixture [314]. This study suggests that a similar technique could be used to determine macro- and micro- constituents at molar ratios <2×104. The type of combination of CZE and ITP sample treatment can be especially useful for planar chip CE technologies such as the one introduced by Kaniansky and colleagues [318]. Our group has optimized a CZE/CD technique to measure nitrite and nitrate concentrations in hemolymph, tissues, and individual neurons samples from model mollusks [101,315], subepithelial and epithelial tissue microsamples from the midgut of mosquito larvae [249], and submicroliter tissue samples from mammalian neuronal tissues [247]. To reduce chloride interference and improve NO2− detection, we use sample cleanup with a home-made solid-phase microextraction (SPME) cartridge filled with Dionex OnGuard-Ag cationic exchange resin. Samples diluted 100–10,000-fold were cleaned and analyzed using an ITP stacking procedure that yields LOD values of 8.9 nM (0.41 ppb) and 3.54 nM (0.22 ppb) for nitrite and nitrate relative to undiluted sample matrix [247]. A description of cartridge preparation and CZE/IPT has been published [248]. Up to 200 cartridges can be prepared from single DIONEX cartridge and used up to 3 times each with highly diluted samples (unpublished personal observation).
The application of CZE/CCD to the analysis of biomedical laboratory blood serum and urine samples was demonstrated by Wan et al. [316]. In this work, potassium, sodium, calcium, and magnesium cations as well as chloride anion were determined in human serum; ammonium, nitrate, and sulfate ions were also successfully measured in urine. The problem with the high molar excess of sodium and chloride in these samples was reduced by BGE optimization. Optimal separation was achieved in 6.5 mM maleic acid/7.5 mM L-arginine and in 13 mM maleic acid/15 mM L-arginine at pH 5.5 for the determination of cations and anions, respectively. However, in that study nitrite was not detectable in the biological samples analyzed using this technique.
Substantial effort was made to develop new CZE/CCD assays for organic conductivity-inert substances such as amino acids, peptides, and neurotransmitters that could be relevant to the future bioanalytical profiling of the L-arginine/NO pathway. For the separation of amphoteric amino acids, many of which are zwitterions at neutral pH, a low or high pH of the BGE is necessary. Similar conditions also enhance conductivity detection. The optimization of CZE/CCD for the comprehensive detection of proteinogenic amino acids was first described by Coufal et al. [319]. An acidic BGE based on 2.3 M acetic acid with 0.1% hydroxyethylcellulose (HEC) allows the separation and detection of 20 L-amino acids in cationic form with LOD values ranging between 9.1 and 29 μM. Biomedical samples of urine and saliva have been analyzed with this technique, which resulted in good separation and detection in general, including the best LOD value for L-arginine [319].
In 2003, Tanyanyiwa et al. reported a CZE technique with a high-voltage CCD especially useful for the detection of low amounts of non-derivatized amino acids [320]. Using low pH lactic acid-based BGE, these authors were able to optimize the detection for L-arginine, L-lysine, and L-histidine, reaching LOD values of ~20, 30 and 40 nM, respectively; the remaining proteinogenic L-amino acids showed LOD values in the range of 10−5–10−6 M. Basic conditions (pH 10–11) generally lead to a more stable baseline and more consistent sensitivity, which enabled sub-micromolar LOD values for all 20 amino acids. Tanyanyiwa et al. also showed that nine of 20 amino acids can be detected in urine samples, although the quantification of real samples was not reported [320]. More recently, CZE/CCD was used to analyze free amino acids and some other metabolites in amniotic fluid samples [317]. BGE with 1.7 M acetic acid and 0.1 wt% of HEC (pH 2.15) was supplemented with acetonitrile. This significantly improved separation and sample stacking, resulting in LOD improvement, e.g. 1.5 μM for L-arginine and 6.7 μM for aspartate.
The optimization of a biogenic amines assay with CZE/CCD was recently described by Kvasnicka and Voldrich [321]. Comparison of CCD for the analysis of small inorganic ions by CZE has been also performed recently [322]. In addition to the rapid progress of CZE/CCD technology for fused silica capillaries, several on-chip platforms have been proposed (for recent reviews see refs. [294,308–310,323]). In summary, CCD is a very potent and promising technique for both fused silica capillary- and micro-chip-based CZE platforms; however, applications focusing on profiling of L-arginine/NO pathway in biological samples have yet to be developed.
3.2.5. Electrochemical detection (ECD)
Electrochemical detection (ECD) comprises a series of techniques which quantify analytes through an electrical signal from electrolysis, redox, or phase transfer events at the interface between analytes (usually aqueous) and the surface of specific chemical sensors. This technique is usually classified by the modality of the recording conditions, such as cyclic voltammetry, amperometry, potentiometry, and conductivity. The last category does not involve an active electrochemical reaction and has been addressed above. The sensor of the electrochemical detector is usually electrically coupled to a capillary outlet, which implies similar requirements as contact conductivity detection. There is one repot of contact-less integrated ECD with independent battery power and optical communication interface [324].
Amperometry is the simplest ECD technique. It scales analyte concentrations through currents between working and reference electrodes. These currents are generated by an electrochemical redox reaction of the analyte at the surface of the electrode. Such a current is described by Faraday’s law.
Amperometric assays employ fixed voltage, whereas the techniques that utilize scan voltages are classified as voltametric (e.g., cyclic voltammetry and squire wave voltammetry). The chemical properties and appropriate electrical potential of the substrate-interacting surface are employed to reduce the kinetic barrier of electrochemical reaction and generate detectable currents. Nobel metals, glassy carbon, allotropes of carbon, and enzymatically modified surface sensors that increase substrate coordination efficiency are usually used. Cyclic voltammetry techniques use cyclically adjusted voltage to detect specified redox reaction currents and analytes. Scanning voltage provides a comprehensive current/voltage (IV) profile of the electrochemical reactions of the analyte or mixture of analytes [232]. This increases the informative capacity of the assay and eliminates interference between species. For example, with scanning voltammetry, the concentrations of all redox species in a sample, including NO, NO2− and NxOy or GSH, GSSG and GSNO, can be determined by the distribution and amplitude of anodic current peaks [325,326].
A variety of ECD methods were used as stand-alone techniques for in vivo monitoring of NOS activity [327]. A variety of amperometric sensors specifically developed for the analysis of NO recently has been reviewed [325,326,328,329]. In combination with CE, the ECD techniques (CEECD) expand the range of accessible analytes without derivatization. ECD offers an additional dimension to optimize the selectivity and sensitivity of detection, and it could provide additional information about the identity of analytes on the basis of their electrochemical properties. CEECD has rapidly expanded to an attractive alternative of technologically more changing UV/LIF and CD/CCD-based technique in both conventional fused silica capillary assay and microchip-based frameworks; see for example refs. [330] and [331–333], respectively.
The remarkable analytical potential of CEECD was reported by Olefirowicz and Ewing in their pioneering study that used CE with an amperometric detector to determine catecholamines in whole-cell and cytoplasmic fractions of identified molluscan neurons [334]. Microchip-integrated CEECD has been developed and tested with biomedical samples, showing excellent analytical performance and good technological potential for microfabrication [335]. The effort to develop CEECD applications focuses on the analysis of neurochemically and metabolically important small biogenic and essential molecules, including amino acids [336–342], monoamines [343–352], excitatory and inhibitory neurotransmitters with low electrochemical activity [342,353], and some vitamins and cofactors such as ascorbic acid and glutathione [354–364].
Recent improvements in CEECD technique include the development of lab-on-a-chip devices that integrate sample clean-up and preconcentration, electrochemical derivatization, and different types of electrodes for electrochemical detection [365]. Interesting design of in-channel ED for microchip HPCE which uses optically isolated RS-232 port has recently been published by Martin et al. [324,366]. Optimization of separation/recordings with CEECD yields overall LOD values for different analytes in the 10−6–10−7 M range. Hence, this technology could be valuable in the determination of NO-modified organic metabolites that have yet to be addressed. No HPCE applications with amperometric, voltammetric, or potentiometric detectors that directly analyzed nitrite and nitrate have yet been published.
3.2.6. Electrospray ionization mass spectrometry (ESI-MS) detection
The use of CE in tandem with mass spectroscopy (MS) is growing exponentially. This growth reflects the recent increase in research demand and laboratory accessibility of these techniques. It also reveals the enormous analytical potential of these approaches in solving some of today’s hottest biological problems, including identification and structural analysis of small metabolites, peptide sequencing, and detection of posttranslational modifications in proteins and nucleotides. Among the variety of MS techniques used in combination with CE (for a comprehensive review see ref. [367]) the electrospray ionization (ESI) interface is the most conventional for integration with HPCE (for a review see ref. [368]). One drawback of CE-ESI-MS, however, is the limited time frame for data-dependent mass determination. Another CE-MS detection framework without that limitation is matrix assisted lased desorption/ionization-time of flight (MALDI-TOF) MS (reviewed in ref. [369]). This technique may be especially valuable for the analysis of subminiature cellular and subcellular samples ([370,371]). Of the three most common ESI-MS interfaces in CE to become the first commercially available technique used coaxial liquid sheath flow (a.k.a. sheath-flow interface). This was followed by a sheathless technique with liquid junction and then a technique with conductively-coated tapped capillary tip. The latter two techniques appear to be especially suitable for low-flow electrospray or nanospray conditions of HPCE (see Fig. 1 in ref. [372] for the schematics). ESI-MS techniques are also commonly used to analyze biomedical samples in tandem with HPLC [373]. Both GC-MS and LC-MS were essential tools in establishing the fine mechanisms of NO production and became increasingly used to analyze intermediate and terminal metabolites of the L-arginine/NO pathway in a variety of biomedical samples. These MS-based techniques were recently historically and comprehensively reviewed in ref. [374]. In contrast, CE-ESI-MS appears to be just emerging as a bioanalytical approach for the evaluation of NO-related pathways. At present, there is a single published report on CE-ESI-MS analysis of NO-related metabolites [263]. In this work Meulemans used CE in combination with ESI-MS and photodiode array detectors to analyze products of electrochemically-induced oxidation of NG-hydroxy-L-arginine in vitro. It has been stated that this assay can be employed for the analysis of biological samples. Clearly, CE-MS has not yet been established in the field of NO research as GC-MS and LC-MS have been. However, the broad implication of this technique in parallel and as an alternative to chromatography-based approaches is already evident. In addition, apparent limitation of LC-MS to perform analysis in subcellular volumes and to determine traces of NO action may be relatively easily accessible by CE-MS with online sample handling such as ITP, and cleanup. This predicts a great niche for CE-MS in the analysis of products of nitrosylation at the nM-level with subcellular resolution.
4. HPCE profile of NO metabolites in biomedical samples
There are two analytical problems in evaluating NO functions. The first, which is probably more relevant to clinical assays, is the challenge of detecting and quantifying small amounts of specific metabolites in circulation and excretion samples. The second, which is relevant to biomedical assays dealing with NO role in biological processes, is the determination of analytes in small sample volumes, ideally using a single cell or even part of a cell and preferably nondestructive sampling approaches. For example, density of NO-ergic neurons varies dramatically across biological species, developmental stages, tissues, and neuronal structures. The study of local NO action demands measuring concentrations of NO and related metabolites with a single-cell resolution. With more cells of different metabolic characteristics to be included into the samples the heterogeneity would increase and the assay would be less precise. The accumulation of chemically stable members of the L-arginine/NO family such as L-citrulline, nitrite, or 3-nitrotyrosine in tissue, circulation, or excretion may be good reference for the evaluation of pathological NO elevation in the entire organism. It is critical to realize that alternative sources for some of those metabolites exist. Validation of the potential problems in the assay of NO metabolites as well as individual variation in the concentration of NO metabolites in cell, tissues, and body fluids in health and disease remain very important parts of HPCE analysis of the NO pathway and may substantially improve assessment of a risk or status in multiple diseases and disorders.
4.1. Cellular and subcellular resolution HPCE
The ability of HPCE to analyze extremely small and highly diluted samples (nanoliter to picoliter volumes in a capillary load; up to 100,000-fold dilution) with an excellent LOD (e.g., 10−16–10−20 mol corresponding to 10−9–10−6 M) has a “fatal attraction” for bioanalytical and biomedical use. For example, HPCE is the technique of choice for high resolution spatial and temporal profiling of certain biological phenomena. It is analytically attractive because it can quantify more than one analyte per run. For comparison, other single-cell techniques (e.g. electrochemical microelectrodes and fluorescence labelling) typically resolve a single analyte, with minor exception. Presently, several HPCE methods have been developed to analyze single-cell samples (reviewed in refs. [375–378]). A few important breakthroughs should be addressed: HPCE of subcellular metabolic domains including individual mitochondrion [379], nuclei [380], tiny cytoplasmic fractions withdrawn from a single cell [381,382], and subcellular environment [254,383,384], and exocytosis samples [254,383,384]. The possibility to perform HPCE analysis of NO-related metabolites with organelle resolution would be important for the validation and calibration of new analytical tools such as fluorescent NO markers (see for example refs. [385,386]). It also may contribute important information about intracellular distribution of proximal NO metabolites.
4.2. Single-neuron and neuronal tissue HPCE of NO metabolites
The brain is the most metabolically heterogeneous tissue in organisms. Hence, single-cell (single-neuron) assays can provide most realistic values. This is essential for the fundamental understanding of neuronal NO pathways and functions. Several single-cell HPCE assays have been developed specifically to identify NO metabolites within isolated neurons of gastropod mollusks, a group of classical neuronal model organisms. Some gastropod species that have large, easily accessible and identifiable NO-ergic neurons provide experimental advantages for such studies. The first work on determining nitrite and nitrate levels in individual neuronal cells has been published by Sweedler’s group [387]. Individual NO-positive buccal neurons from the sea slug Pleurobranchaea californica were electrophoretically separated, and NO2− and NO3− were measured by direct UV absorbance detection. For both ions the method provided LOD and LOQ values of <200 fmol and <4 μM, respectively. The NO2− and NO3− levels in individual neurons associated with NOS-specific NADPH-diaphorase staining were from 2 μM and 12 μM down to undetectable levels in NO-naïve cells. Neuronal tissue and hemolymph samples from other gastropod molluscs were also reported in this work.
Recently a more comprehensive evaluation was performed for the simultaneous determination of L-arginine, L-citrulline, NO2− and NO3− in a selected set of NO-ergic and NO-naïve molluscan neurons [101]. HPCE/LIF of OPF-derivatized samples was used for the amino acid assay, and the CZE-CD technique was used for inorganic anions (Fig. 4B). LOD and LOQ values were 84 amol and 11 nM for L-arginine, 110 amol and 15 nM for L-citrulline, and each <200 amol and <10 nM for NO2− and NO3−, respectively. It has been estimated that less than 1% of a single cell is required for such assays. A similar finding was reported in the analysis of isolated dorsal root ganglia from rat [248]. However, in that case, an integrated biopsy sample of mammalian tissue was analyzed. Such sampling procedure would probably be valuable for laboratory animal experiments and would provide important information about mammalian neuronal NO. However, the dissociation and analysis of individual cells is more problematic with a mammalian model. Both these reports used solid-phase extraction (SPE) clean-up following ITP stacking sample injection.
4.3. Microdialysis and push-pull sampling HPCE of NO metabolites
Microdialysis and push-pull sampling in combination with HPCE has been proposed as a versatile technique to determine the concentration of neurotransmitters and other biologically active substances under physiological conditions and upon drug administration in animal models [388–392]. Sakamoto et al. [393] used in vivo brain dialysis and CZE analysis of NO2− and NO3− to study the effects of mild hypothermia on trauma-induced synthesis of NO in the rat parietal cortex after traumatic brain injury. They showed a significant increase in NO end-products in samples from posttraumatic rats. The Kennedy group has employed a microdialysis-coupled rapid CE-LIF system which allows a 30-s frame scan of the amino acid profile in the free moving rat [394]. MECC mode allows the quantitative determination of 15 amino acids in brain perfusate. A similar technique was also used to identify neurotransmitter pools in rats with genetic absence epilepsy [395]. Analytical parameters quantified in both of these reports would be sufficient to identify key organic components of the L-arginine/NO pathway. Example of microdialysis systems that can be used in combination with CE-LIF for analysis of human cerebrospinal fluid samples with 1-min sampling have been recently described by Parrot et al. [396].
Certain improvement in the analysis of cerebrospinal fluid using HPCE on low-flow push-pull perfusion samples has been made in recent years. A 1.5-min approach was proposed by Gao et al. for the separation and analysis of NO2− and NO3− in rat striatum [256]. This method was especially optimized for NO2−. It utilized 20 mM phosphate, 2 mM CTAC BGE at pH 3.5, electrokinetic injection, and 214-nm direct UV absorbance detection. Reported LOD values were 0.96 μM for NO2− and 2.86 μM for NO3−.
The same group recently proposed an automated nanoliter-scale injection device which uses 5-nl volume injections and operates with <1-μl sample volume [397]. This system was employed for the analysis of NO2− and NO3−. In addition, Hadamard transform (HT) and sample overage was used to improve detection and resolution parameters [397]. Hadamard transform is a generalized class of Fourier transforms that is becoming increasingly employed to improve the SNR of analytical recordings. HPCE detection of NO2− in biological samples is a good target for such an improvement. For example, McReynolds et al. [251] utilized a pseudorandom alternating sample-buffer injection according to a Hadamard matrix, using conventional capillaries with an original “T” connector coupler. By applying an 83-element injection sequence, they were able to improve the SNR by a factor of 4.5 over a single injection mode, leading to an LOQ value of 0.56 μM for nitrite [251]. A detailed description of HPCE/HT applications with a focus on planar chip CE techniques has been recently published [398].
4.4. Other HPCE assays of NO metabolites
Infrequently but very effectively, HPCE has been used to identify NO metabolites beyond the analysis of neuronal samples. For example, the role of NO in cystic fibrosis and other lung diseases [399] can be analyzed in a variety of aspects using HPCE techniques. The first application of the technique for microanalysis of lung airway surface fluid (ASF) was with HPCE and CD, and was published in 1997 by Govindaraju et al. [400]. The use of CD offers an excellent analytical flexibility sufficient to determine important ions including cations (Na+, K+, Ca2+, Mg2+) and anions (Cl−, NO2−, NO3−, SO42−, PO43−, HCO3−) in the airway fluid microsamples. Four years later, Govindaraju et al. reported HPCE of NO2−/NO3− in sub-microliter quantities of ASF and plasma using a UV detector (214 nm). Reported basal concentrations were 102 ± 12 μM for NO3− and 83 ± 28 μM NO2− in rat ASF, and 70 ± 1.0 μM for NO3− and below the LOD value for NO2− in plasma samples. After lipopolysaccharide was intratracheally administered, the concentration of these NO metabolites increased significantly, e.g. 387 ±16/377 ± 88 and 103 ± 7.0/138 ± 17 (all values in μM) for ASF and plasma, respectively.
Another example of nitrite/nitrate detection in epithelial samples investigated the profile of these anions in the midgut epithelia of the tropical disease vector mosquito larvae, Aedes aegypti [249]. Similar to neuronal model molluscs, this simple arthropod organism offers remarkable advantages in the study of epithelial functions at the level of individual cells. Submicroliter fragments of epithelial tissue (~100 cells) as well as tiny apical and basal liquid samples were analyzed using CZE, SPE and ITP staking injection [249]. In addition to major biological anions present in the samples, nitrite and nitrate were detected. One order magnitude higher concentrations of NO2−/NO3− (e.g. 1.48/0.78 μM) were found in the anterior midgut in comparison to other parts of the larval alimentary canal. Anterior midgut is associated with high alkalinization (up to pH 11 [249]), high expression of NOS, and elevated NO production (unpublished). Despite that the role of NO in gastrointestinal function and infection [401–403] has been evident for a long time, so far, this is the sole report of nitrite and nitrate in intestinal tissue with HPCE. In addition, the accumulation of nitrite and nitrate has been determined by HPCE in microdialysis samples from the wall of the stomach and proximal colon during vagal nerve stimulation [404].
4.5. Clinical HPCE of NO metabolites
The high sensitivity and resolution of HPCE and the relative simplicity of various HPCE methods and equipment generate a strong interest in adapting such assays for the quantification of NO-related metabolites in human health and disease. In particular, a large number of methods were developed to assay nitrite and nitrate in human serum [255,257,258,264,405,406] and urine [253,407–409] as indicators of systemic and whole body NO synthesis, respectively. Other biochemical parameters of the L-arginine/NO pathway became increasingly used in assessing disease risk factors and in laboratory diagnostics of samples from patients and laboratory mammalian models. This set includes ADMA [203–205], S-nitrosothiols [410–416], and 3-nitrotyrosine [179,292,417,418].
NO-related HPCE assays have been reported in growing numbers relevant to particular human disease diagnostics and treatment. The severity of neurological symptoms associated with systemic lupus erythematosus (SLE), an inflammatory disease in which up to two thirds of patients present neurological symptoms, and with multiple sclerosis (MS) were extensively evaluated using HPCE analysis of nitrite and nitrate [419–422]. Svenungsson et al. have used HPCE to determine the concentration of NO metabolites in the CSF of patients with varying severity of neuropsychiatric lupus erythematosus (NPLE) [423]. The determination by HPCE of CSF and plasma concentrations of NO metabolites in postoperative patients with subarachnoid hemorrhage has been reported by Sadamitsu et al. [424].
Ischemia-reperfusion myocardial and endothelial damage after heart surgery was addressed in a clinical study by Carlucci et al. [425]. In addition to a comprehensive set of assays for the evaluation of oxidative stress and energy balance in the heart by analyzing biopsy samples before, during, and after cross-clamping, they employed HPCE for plasma samples from the coronary sinus and measured a decrease in the nitrate plus nitrite levels. Panossian et al. [426] have used CZE to determine plasma nitrite (as an indicator of inflammatory NO formation) in patients with family Mediterranean fever (FMF). They also studied its modulation by Immuno-Guard, a novel herbal preparation which relieves the severity and longevity of FMF attacks. These authors demonstrated a relationship between the beneficial effect of Immuno-Guard in reducing the severity of inflammatory attacks and the increase in nitrite plasma concentrations. The antioxidant effect of sinivastatin, associated or not associated with alpha-tocopherol, was studied in hypercholesterolemic subjects. The levels of electronegative low-density lipoprotein (LDL-), nitrotyrosine, thiols (homocysteine, glutathione, cysteine, and methionine), and lipid-soluble antioxidants in blood plasma were determined by using multiple techniques, including an HPCE-LIF assay of free thiols, as well as HPLC and ELISA [427]. Danilov et al. [428] compared the concentrations of nitrite and nitrate in CSF and plasma of 61 MS patients with distinct stages of multiple sclerosis. They found that the nitrite CSF levels correlated with clinical progression of the disease. They concluded that NO is involved in the pathogenesis of MS and that the determination of nitrite levels may be a useful marker for disease activity. Some other work related to the detection of organic NO metabolites is summarized in Table 2 and has been briefly discussed above, along with particular separation and detection technology.
5. Concluding remarks
Biological NO is one of the most complex and controversial phenomena in the life history and physiology of modern organisms. It is also one of the most crucial factors in the study of human health and disease, including diagnostics and therapeutic efforts. The physiological and pathological actions of NO involve a variety of direct and indirect biochemical parameters and targets, and these are briefly summarized in the first part of this review. An impressive effort has been made to better understand recognized mechanisms of action of NO, as well as to identify new potential of this exciting molecule with respect to interactions with the metabolic and physiological universe of organisms. Not surprisingly, this effort continues to increase. An especially critical part of NO actions, i.e. transcellular signaling messenger, neurotransmitter, and putative retrograde hormone with small active concentrations and enormous amplification and biological activity potential, remains to be comprehensively understood. It was recently recognized that human activity may generate NO in amounts that could impact the ozone layer of the planet [440,441], but the effect of this activity on the biosphere is difficult to assay since an infinite variety of possible interactions between NO and NO-related pathways within the biosphere exist. Recently, the vital role of NO in virtually all life forms has been recognized. This has generated a second wave in the expansion of NO research, with the involvement of new species from all life Kingdoms. The recent post-genomic transition of biological science and the emerging new high-throughput technologies for screening molecular and genetic references of NO action have played an important role in the recognition of novel and cross-specific NO actions (reviewed by Thum and Bauersachs in ref. [442]). Hence, incremental growth of NO studies in the post-genomic era is predictable.
Almost twenty years have passed since the discovery of L-arginine-derived NO. The greatest problem in the field remains the absence of a simple, reliable, and inexpensive technology to measure NO and a variety of auxiliary metabolites produced upon NO reaction and metabolism as well as of other members of the L-arginine/NO pathway. Most analytical approaches for the evaluation of NO production and action remain “state of the art,” with the apparent consensus that they likely will retain the “state of the art” status for some time. It is obvious therefore, that progress in the analytical chemistry of the NO pathway demands an analytical breakthrough. The second part of this review briefly summarizes recent progress in the development of HPCE as one of the most promising technologies to provide this essential breakthrough in the study of NO’s chemistry, biology, physiology and pathology.
The remarkable research progress that will supply the critical mass in the predictable expansion of HPCE in the near future is apparent: (1) there is a number of reports that addressed and greatly optimized separation and detection conditions for a variety of NO and NO-related metabolites; (2) there is a set of good work which assesses a diversity of detection technologies in combination with HPCE and specifically aimed practical approaches for the detection of NO metabolites, including organic and inorganic compoounds of the L-arginine/NO pathway; (3) there is a number of innovative work that utilizes the emerging lab-on-a-chip technology and is directly or indirectly applicable to high-efficiency and high-throughput profiling of the L-arginine/NO pathway. Apparently, there is only a single missing component: the absence of persistent industrial effort that would address the commercial development of analytical HPCE. Despite remarkable progress in microelectronic technologies and in CE “know-how,” most commercially available HPCE equipment utilizes decades-old electronics, software, sample load, and separation concepts. In addition, the commercial framework is over-complicated, bulky, unreasonably expensive, fragile, and difficult to service in a regular laboratory. There is also an apparent gap in the industrial deployment of analytical HPCE equipment for metabolic research in comparison to HPCE for genomic and proteomic research.
In summary, the market comprising experimental biology, analytical biochemistry, and clinical diagnostics is ready to acquire new CE technology for the study of NO. The current progress in HPCE of the NO pathway highlights two possible directions in the future development of this technology: (1) a flexible analytical framework based on a fused silica capillary with interchangeable UV/LIF, CCD, and ECD detectors, and a submicroliter automatic sampling device; and (2) disposable lab-on-a-chip platforms that will include a sample clean-up and separation micro-fluidic maze integrated with a CCD or contact CD/ED pattern. New algorithms for separation data-analysis, such as pseudorandom injection and Hadamard transform, will be integrated in computer interface to improve method sensitivity by lowering the SNR.
Fig. 6.
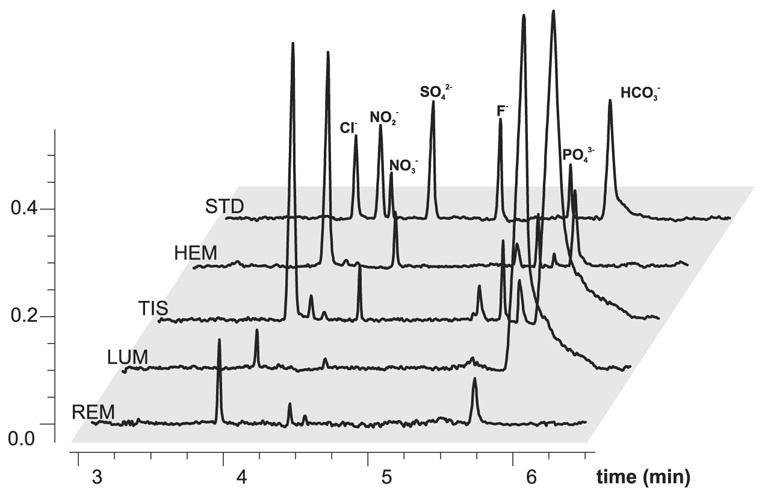
CZE-CD analysis of anions profile in epithelial tissue and subepithelial liquid samples from midgut of mosquito larvae. Orthogonal overlay of typical electrophoretic traces are shown. Samples were introduced by pressure. The 81.5/25 mM arginine/borate BGE9.5 with 0.5 mM of TTAOH was used. A commercial PrinCE HPCE unit with Crystal-1000 CD was used. A fused silica capillary (50 μm i.d., 70-cm length) was used and the separation voltage was −20 kV. Abbreviations are: REM, rearing media; LUM, ectoperitrophic fluid (lumen) sample from the anterior midgut area; TIS, anterior midgut tissue sample; HEM, hemolymph; STD, 2-ppm mixture of anion standards. Z-axis is a conductivity signal. Reproduced from [249], Proceedings of the National Academy of Sciences of the United States of America 98, D.Y. Boudko, L.L. Moroz, W.R. Harvey, P.J. Linser, Alkalinization by chloride/bicarbonate pathway in larval mosquito midgut,15354–15359, 2001, with permission from the National Academy of Sciences of the USA.
Acknowledgments
This work was supported by NIH-NIAID research grant AI030464.
Nomenclature
- ADMA
asymmetric dimethylarginine
- ASF
airway surface fluid
- ASL
argininosuccinate lyase
- ASS
argininosuccinate synthetase
- BGE
background electrolyte
- CAPS
3-cyclohexylamino-1-propanesulfonic acid
- CCD, C4D
contactless capacitively coupled conductivity detector
- CD
contact conductivity detector
- CE
capillary electrophoresis
- C/FS
coated/fused silica capillary
- CHES
2-(N-cyclohexylamino)ethanesulfonic acid
- CSF
cerebrospinal fluid
- CTAC
cetyltrimethylammonium
- CZE
capillary zone electrophoresis
- DMF
N,N-dimethylformamide
- DMMAPS
3-(N,N-dimethylmyristylammonio)propanesulfonate
- DoTAB
dodecyltrimethylammonium bromide
- ECD
electrochemical detection
- EM
electrophoretic motility
- EOF
electro-osmotic flow
- ESI
electrospray ionization
- HBED
N,N-di(2-hydroxybenzyl)ethylenediamine-N,N-diacetic acid
- HEC
hydroxyethylcellulose (HEC)
- HIBA
hydroxyisobutyric acid
- HMH
hexamethonium hydroxide
- HPCE
high-performance capillary electrophoresis
- ITP
isotachophoresis
- LOD
limit of detection
- LOQ
limit of quantification
- MALDI-TOF
matrix assisted lased desorption/ionization-time of flight
- MCE
microchip capillary electrophoresis
- MES
2-(N-morpholino)ethanesulfonic acid
- MS
mass spectrometry
- Nice-Pak OFM Anion-BT
trademark of EOF modifiers (Waters)
- NIR
nitrite reductase
- NO
nitric oxide
- NOS
nitric oxide synthase
- NRA
nitrite reductase activity
- OFM-OH
trademark of EOF modifiers (Waters)
- PC
polyacrylamide-coated capillary
- PRMTs
protein-arginine N-methyltransferases
- RNS
reactive nitrogen species
- RSNO
S-nitrosothiols
- SDMA
symmetric dimethylarginine
- SDS
sodium dodecyl sulfate
- SNR
signal-to-noise ratio
- SPE
solid-phase extraction
- SPME
solid-phase micro-extraction
- TCA
trichloroacetic acid
- TTAH
tetradecyltrimethylammonium hydroxide
Footnotes
This paper is part of a special issue entitled “Analysis of the L-arginine/NO pathway”, guest edited by D. Tsikas.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Ignarro LJ, Fukuto JM, Griscavage JM, Rogers NE, Byrns RE. Proc Natl Acad Sci USA. 1993;90:8103. doi: 10.1073/pnas.90.17.8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tannenbaum SR, Tamir S, Derojaswalker T, Wishnok JS. Amer Chem Soc, Washington. 1994. Nitrosamines and Related N-Nitroso Compounds; p. 120. [Google Scholar]
- 3.Langrehr JM, Hoffman RA, Lancaster JR, Simmons RL. Transplantation. 1993;55:1205. doi: 10.1097/00007890-199306000-00001. [DOI] [PubMed] [Google Scholar]
- 4.Fehsel K, Kroncke KD, Kolb-Bachofen V. Methods Enzymol. 1996;269:426. doi: 10.1016/s0076-6879(96)69043-0. [DOI] [PubMed] [Google Scholar]
- 5.Brune B, von Knethen A, Sandau KB. Eur J Pharm. 1998;351:261. doi: 10.1016/s0014-2999(98)00274-x. [DOI] [PubMed] [Google Scholar]
- 6.MacMicking J, Xie QW, Nathan C. Annu Rev Immunol. 1997;15:323. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- 7.Akaike T, Maeda H. Immunology. 2000;101:300. doi: 10.1046/j.1365-2567.2000.00142.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bogdan C. Nat Immunol. 2001;2:907. doi: 10.1038/ni1001-907. [DOI] [PubMed] [Google Scholar]
- 9.Stafford JL, Neumann NF, Belosevic M. Crit Rev Microbiol. 2002;28:187. doi: 10.1080/1040-840291046731. [DOI] [PubMed] [Google Scholar]
- 10.Aguirre J, Rios-Momberg M, Hewitt D, Hansberg W. Trends Microbiol. 2005;13:111. doi: 10.1016/j.tim.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 11.Yoon SS, Hennigan RF, Hilliard GM, Ochsner UA, Parvatiyar K, Kamani MC, Allen HL, DeKievit TR, Gardner PR, Schwab U, Rowe JJ, Iglewski BH, McDermott TR, Mason RP, Wozniak DJ, Hancock REW, Parsek MR, Noah TL, Boucher RC, Hassett DJ. Developmental Cell. 2002;3:593. doi: 10.1016/s1534-5807(02)00295-2. [DOI] [PubMed] [Google Scholar]
- 12.Hassett DJ, Cuppoletti J, Trapnell B, Lymar SV, Rowe JJ, Yoon SS, Hilliard GM, Parvatiyar K, Kamani MC, Wozniak DJ, Hwang SH, McDermott TR, Ochsner UA. Adv Drug Delivery Rev. 2002;54:1425. doi: 10.1016/s0169-409x(02)00152-7. [DOI] [PubMed] [Google Scholar]
- 13.Coleman JW. Int Immunopharmacol. 2001;1:1397. doi: 10.1016/s1567-5769(01)00086-8. [DOI] [PubMed] [Google Scholar]
- 14.Hawkins RD, Son H, Arancio O. Prog Brain Res. 1998;118:155. doi: 10.1016/s0079-6123(08)63206-9. [DOI] [PubMed] [Google Scholar]
- 15.Martin SJ, Grimwood PD, Morris RGM. Annu Rev Neurosci. 2000;23:649. doi: 10.1146/annurev.neuro.23.1.649. [DOI] [PubMed] [Google Scholar]
- 16.Yamada K, Mizuno M, Nabeshima T. Life Sci. 2002;70:735. doi: 10.1016/s0024-3205(01)01461-8. [DOI] [PubMed] [Google Scholar]
- 17.Palmer RMJ, Ashton DS, Moncada S. Nature. 1988;333:664. doi: 10.1038/333664a0. [DOI] [PubMed] [Google Scholar]
- 18.Reutov VP. Biochemistry (Mosc) 1999;64:528. [PubMed] [Google Scholar]
- 19.Reutov VP, Sorokina EG. Biochemistry (Mosc) 1998;63:874. [PubMed] [Google Scholar]
- 20.Zhang Z, Naughton D, Winyard PG, Benjamin N, Blake DR, Symons MCR. Biochem Biophys Res Commun. 1998;249:767. doi: 10.1006/bbrc.1998.9226. [DOI] [PubMed] [Google Scholar]
- 21.Godber BLJ, Doel JJ, Sapkota GP, Blake DR, Stevens CR, Eisenthal R, Harrison R. J Biol Chem. 2000;275:7757. doi: 10.1074/jbc.275.11.7757. [DOI] [PubMed] [Google Scholar]
- 22.Kozlov AV, Staniek K, Nohl H. FEBS Lett. 1999;454:127. doi: 10.1016/s0014-5793(99)00788-7. [DOI] [PubMed] [Google Scholar]
- 23.Nohl H, Staniek K, Kozlov AV. Redox Rep. 2005;10:281. doi: 10.1179/135100005X83707. [DOI] [PubMed] [Google Scholar]
- 24.Liu XP, Miller MJS, Joshi MS, Sadowska-Krowicka H, Clark DA, Lancaster JR. J Biol Chem. 1998;273:18709. doi: 10.1074/jbc.273.30.18709. [DOI] [PubMed] [Google Scholar]
- 25.Nagababu E, Ramasamy S, Abernethy DR, Rifkind JM. J Biol Chem. 2003;278:46349. doi: 10.1074/jbc.M307572200. [DOI] [PubMed] [Google Scholar]
- 26.Cosby K, Partovi KS, Crawford JH, Patel RP, Reiter CD, Martyr S, Yang BK, Waclawiw MA, Zalos G, Xu XL, Huang KT, Shields H, Kim-Shapiro DB, Schechter AN, Cannon RO, Gladwin MT. Nat Med. 2003;9:1498. doi: 10.1038/nm954. [DOI] [PubMed] [Google Scholar]
- 27.Gautier C, van Faassen E, Mikula I, Martasek P, Slama-Schwok A. Biochem Biophys Res Commun. 2006;341:816. doi: 10.1016/j.bbrc.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 28.Snyder SH. Science. 1992;257:494. doi: 10.1126/science.1353273. [DOI] [PubMed] [Google Scholar]
- 29.Wei XQ, Charles IG, Smith A, Ure J, Feng CJ, Huang FP, Xu DM, Muller W, Moncada S, Liew FY. Nature. 1995;375:408. doi: 10.1038/375408a0. [DOI] [PubMed] [Google Scholar]
- 30.Rassaf T, Kleinbongard P, Kelm M. Kidney Blood Press Res. 2005;28:341. doi: 10.1159/000090188. [DOI] [PubMed] [Google Scholar]
- 31.Nathan C, Xie QW. J Biol Chem. 1994;269:13725. [PubMed] [Google Scholar]
- 32.Forstermann U, Kleinert H. Naunyn Schmiedebergs Arch Pharmacol. 1995;352:351. doi: 10.1007/BF00172772. [DOI] [PubMed] [Google Scholar]
- 33.Searles CD, Miwa Y, Harrison DG, Ramasamy S. Circ Res. 1999;85:588. doi: 10.1161/01.res.85.7.588. [DOI] [PubMed] [Google Scholar]
- 34.Elfering SL, Sarkela TM, Giulivi C. J Biol Chem. 2002;277:38079. doi: 10.1074/jbc.M205256200. [DOI] [PubMed] [Google Scholar]
- 35.Aktan F. Life Sci. 2004;75:639. doi: 10.1016/j.lfs.2003.10.042. [DOI] [PubMed] [Google Scholar]
- 36.Santolini J, Meade AL, Stuehr DJ. J Biol Chem. 2001;276:48887. doi: 10.1074/jbc.M108666200. [DOI] [PubMed] [Google Scholar]
- 37.Mori M, Gotoh T, Nagasaki A, Takiguchi M, Sonoki T. J Inherit Metab Dis. 1998;21:59. doi: 10.1023/a:1005357608129. [DOI] [PubMed] [Google Scholar]
- 38.Li D, Hayden EY, Panda K, Stuehr DJ, Deng HT, Rousseau DL, Yeh SR. J Biol Chem. 2006;281:8197. doi: 10.1074/jbc.M507328200. [DOI] [PubMed] [Google Scholar]
- 39.Gautier C, Negrerie M, Wang ZQ, Lambry JC, Stuehr DJ, Collin F, Martin JL, Slama-Schwok A. J Biol Chem. 2004;279:4358. doi: 10.1074/jbc.M305048200. [DOI] [PubMed] [Google Scholar]
- 40.Bae SY, Xu Q, Hutchinson D, Colton CA. Biochim Biophys Acta. 2005;1745:65. doi: 10.1016/j.bbamcr.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 41.Nicholson B, Manner CK, Kleeman J, MacLeod CL. J Biol Chem. 2001;276:15881. doi: 10.1074/jbc.M010030200. [DOI] [PubMed] [Google Scholar]
- 42.Closs EI, Simon A, Vekony N, Rotmann A. J Nutr. 2004;134:2752S. doi: 10.1093/jn/134.10.2752S. [DOI] [PubMed] [Google Scholar]
- 43.Hallemeesch MM, Lamers WH, Deutz NEP. Clin Nutr. 2002;21:273. doi: 10.1054/clnu.2002.0571. [DOI] [PubMed] [Google Scholar]
- 44.Selamnia M, Robert V, Mayeur C, Delpal S, Blachier F. Biochim Biophys Acta. 1998;1425:93. doi: 10.1016/s0304-4165(98)00056-7. [DOI] [PubMed] [Google Scholar]
- 45.McDonald KK, Zharikov S, Block ER, Kilberg MS. J Biol Chem. 1997;272:31213. doi: 10.1074/jbc.272.50.31213. [DOI] [PubMed] [Google Scholar]
- 46.Flam BR, Hartmann PJ, Harrell-Booth M, Solomonson LP, Eichler DC. Nitric Oxide. 2001;5:187. doi: 10.1006/niox.2001.0340. [DOI] [PubMed] [Google Scholar]
- 47.Mori M, Gotoh T. Biochem Biophys Res Commun. 2000;275:715. doi: 10.1006/bbrc.2000.3169. [DOI] [PubMed] [Google Scholar]
- 48.Lee TJF, Yu JG. Nitric Oxide: Novel Actions, Deleterious Effects and Clinical Potential. New York Acad. Sci.; New York: 2002. p. 73. [Google Scholar]
- 49.Goodwin BL, Solomonson LP, Eichler DC. J Biol Chem. 2004;279:18353. doi: 10.1074/jbc.M308160200. [DOI] [PubMed] [Google Scholar]
- 50.Stuehr DJ. J Nutr. 2004;134:2748S. doi: 10.1093/jn/134.10.2748S. [DOI] [PubMed] [Google Scholar]
- 51.Mori M, Gotoh T. J Nutr. 2004;134:2820S. doi: 10.1093/jn/134.10.2820S. [DOI] [PubMed] [Google Scholar]
- 52.Whorton AR, Simonds DB, Piantadosi CA. Am J Physiol Lung Cell Mol Physiol. 1997;16:L1161. doi: 10.1152/ajplung.1997.272.6.L1161. [DOI] [PubMed] [Google Scholar]
- 53.Dweik RA, Laskowski D, Abu-Soud HM, Kaneko FT, Hutte R, Stuehr DJ, Erzurum SC. J Clin Invest. 1998;101:660. doi: 10.1172/JCI1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brown GC. Biochim Biophys Acta. 1999;1411:351. doi: 10.1016/s0005-2728(99)00025-0. [DOI] [PubMed] [Google Scholar]
- 55.Gardner PR, Martin LA, Hall D, Gardner AM. Free Radic Biol Med. 2001;31:191. doi: 10.1016/s0891-5849(01)00569-x. [DOI] [PubMed] [Google Scholar]
- 56.Griffiths C, Yamini B, Hall C, Garthwaite J. Biochemical J. 2002;362:459. doi: 10.1042/0264-6021:3620459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nalwaya N, Deen WM. J Theor Biol. 2004;226:409. doi: 10.1016/j.jtbi.2003.09.015. [DOI] [PubMed] [Google Scholar]
- 58.Gorren ACF, Bec N, Lange R, Mayer B. Redox Cell Biology and Genetics, Pt B. Academic Press Inc.; San Diego: 2002. p. 114. [Google Scholar]
- 59.Thomas DD, Miranda KM, Espey MG, Citrin D, Jourd’Heuil D, Paolocci N, Hewett SJ, Colton CA, Grisham MB, Feelisch M, Wink DA. Nitric Oxide. Pt. D, Acad. Press Inc.; San Diego: 2002. p. 84. [DOI] [PubMed] [Google Scholar]
- 60.Reynaert NL, Ckless K, Wouters EFM, Van Der Vliet A, Janssen-Heininger YMW. Antioxid Redox Signaling. 2005;7:129. doi: 10.1089/ars.2005.7.129. [DOI] [PubMed] [Google Scholar]
- 61.Mansuy D, Boucher JL. Free Radic Biol Med. 2004;37:1105. doi: 10.1016/j.freeradbiomed.2004.06.031. [DOI] [PubMed] [Google Scholar]
- 62.Miller RT, Martasek P, Omura T, Masters BSS. Biochem Biophys Res Commun. 1999;265:184. doi: 10.1006/bbrc.1999.1643. [DOI] [PubMed] [Google Scholar]
- 63.Wink DA, Feelisch N, Fukuto J, Chistodoulou D, Jourd’heuil D, Grisham MB, Vodovotz Y, Cook JA, Krishna M, DeGraff WG, Kim S, Gamson J, Mitchell JB. Arch Biochem Biophys. 1998;351:66. doi: 10.1006/abbi.1997.0565. [DOI] [PubMed] [Google Scholar]
- 64.Alderton WK, Cooper CE, Knowles RG. Biochemical J. 2001;357:593. doi: 10.1042/0264-6021:3570593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ghosh DK, Salerno JC. Front Biosci. 2003;8:D193. doi: 10.2741/959. [DOI] [PubMed] [Google Scholar]
- 66.Degtyarenko KN, Archakov AI. FEBS Lett. 1993;332:1. doi: 10.1016/0014-5793(93)80470-f. [DOI] [PubMed] [Google Scholar]
- 67.Nelson DR, Koymans L, Kamataki T, Stegeman JJ, Feyereisen R, Waxman DJ, Waterman MR, Gotoh O, Coon MJ, Estabrook RW, Gunsalus IC, Nebert DW. Pharmacogenetics. 1996;6:1. doi: 10.1097/00008571-199602000-00002. [DOI] [PubMed] [Google Scholar]
- 68.Moncada S, Palmer RMJ, Higgs EA. Pharmacol Rev. 1991;43:109. [PubMed] [Google Scholar]
- 69.Kone BC, Kuncewicz T, Zhang WZ, Yu ZY. Am J Physiol Renal Physiol. 2003;285:F178. doi: 10.1152/ajprenal.00048.2003. [DOI] [PubMed] [Google Scholar]
- 70.Stuehr DJ. Annu Rev Pharmacol Toxicol. 1997;37:339. doi: 10.1146/annurev.pharmtox.37.1.339. [DOI] [PubMed] [Google Scholar]
- 71.Tatoyan A, Giulivi C. J Biol Chem. 1998;273:11044. doi: 10.1074/jbc.273.18.11044. [DOI] [PubMed] [Google Scholar]
- 72.Brookes PS. Mitochondrion. 2004;3:187. doi: 10.1016/j.mito.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 73.Kato K, Giulivi C. Front Biosci. 2006;11:2725. doi: 10.2741/2002. [DOI] [PubMed] [Google Scholar]
- 74.Ghafourifar P, Cadenas E. Trends Pharmacol Sci. 2005;26:190. doi: 10.1016/j.tips.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 75.Lacza Z, Pankotai E, Csordas A, Geroo D, Kiss L, Horvath EM, Kollai M, Busija DW, Szabo C. Nitric Oxide. 2006;14:162. doi: 10.1016/j.niox.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 76.Bredt D, Snyder S. Proc Natl Acad Sci USA. 1990;87:682. doi: 10.1073/pnas.87.2.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Guo FQ, Okamoto M, Crawford NM. Science. 2003;302:100. doi: 10.1126/science.1086770. [DOI] [PubMed] [Google Scholar]
- 78.Dweik RA, Laskowski D, Abu-Soud HM, Kaneko F, Hutte R, Stuehr DJ, Erzurum SC. J Clin Invest. 1998;101:660. doi: 10.1172/JCI1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nakamura K, Go N. Cell Mol Life Sci. 2005;62:2050. doi: 10.1007/s00018-004-5076-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dejam A, Hunter CJ, Schechter AN, Gladwin MT. Blood Cells Mol Dis. 2004;32:423. doi: 10.1016/j.bcmd.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 81.Huang Z, Wong AL, Partovi KS, Schechter AN, Gladwin MT. Blood. 2003;102:758A. [Google Scholar]
- 82.Gladwin MT, Cosby K, Partovi KS, Patel RP, Crawford JH, Kim-Shapiro DB, Schechter AN, Cannon RO. Blood. 2003;102:255A. [Google Scholar]
- 83.Payne WJ, Liu MY, Bursakov SA, Le Gall J. Biofactors. 1997;6:47. doi: 10.1002/biof.5520060106. [DOI] [PubMed] [Google Scholar]
- 84.Philippot L. Trends Microbiol. 2005;13:191. doi: 10.1016/j.tim.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 85.Mowat C, McColl KEL. Best Pract Res Clin Gastroenterol. 2001;15:523. doi: 10.1053/bega.2000.0196. [DOI] [PubMed] [Google Scholar]
- 86.Tsuchiya K, Kanematsu Y, Yoshizumi M, Ohnishi H, Kirima K, Izawa Y, Shikishima M, Ishida T, Kondo S, Kagami S, Takiguchi Y, Tamaki T. Am J Physiol Heart Circ Physiol. 2005;288:H2163. doi: 10.1152/ajpheart.00525.2004. [DOI] [PubMed] [Google Scholar]
- 87.Brown GC. Eur J Biochemistry. 1995;232:188. doi: 10.1111/j.1432-1033.1995.tb20798.x. [DOI] [PubMed] [Google Scholar]
- 88.Duncan C, Dougall H, Johnston P, Green S, Brogan R, Leifert C, Smith L, Golden M, Benjamin N. Nat Med. 1995;1:546. doi: 10.1038/nm0695-546. [DOI] [PubMed] [Google Scholar]
- 89.Nagase S, Takemura K, Ueda A, Hirayama A, Aoyagi K, Kondoh M, Koyama A. Biochem Biophys Res Commun. 1997;233:150. doi: 10.1006/bbrc.1997.6428. [DOI] [PubMed] [Google Scholar]
- 90.Moroz LL, Norby SW, Cruz L, Sweedler JV, Gillette R, Clarkson RB. Biochem Biophys Res Commun. 1998;253:571. doi: 10.1006/bbrc.1998.9810. [DOI] [PubMed] [Google Scholar]
- 91.Rassaf T, Feelisch M, Kelm M. Free Radic Biol Med. 2004;36:413. doi: 10.1016/j.freeradbiomed.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 92.Dejam A, Hunter CJ, Pelletier MM, Hsu LL, Machado RF, Shiva S, Power GG, Kelm M, Gladwin MT, Schechter AN. Blood. 2005;106:734. doi: 10.1182/blood-2005-02-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gladwin MT, Schechter AN, Kim-Shapiro DB, Patel RP, Hogg N, Shiva S, Cannon RO, 3rd, Kelm M, Wink DA, Espey MG, Oldfield EH, Pluta RM, Freeman BA, Lancaster JR, Jr, Feelisch M, Lundberg JO. Nat Chem Biol. 2005;1:308. doi: 10.1038/nchembio1105-308. [DOI] [PubMed] [Google Scholar]
- 94.Gaston B. Biochim Biophys Acta. 1999;1411:323. doi: 10.1016/s0005-2728(99)00023-7. [DOI] [PubMed] [Google Scholar]
- 95.Do KQ, Grima G, Benz B, Salt TE. Ann N Y Acad Sci. 2002;962:81. doi: 10.1111/j.1749-6632.2002.tb04058.x. [DOI] [PubMed] [Google Scholar]
- 96.Simon DI, Mullins ME, Jia L, Gaston B, Singel DJ, Stamler JS. Proc Natl Acad Sci USA. 1996;93:4736. doi: 10.1073/pnas.93.10.4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Suntsova TP, Beda NV, Nedospasov AA. Iubmb Life. 2002;54:281. doi: 10.1080/15216540215677. [DOI] [PubMed] [Google Scholar]
- 98.Stadtman ER, Levine RL. Amino Acids. 2003;25:207. doi: 10.1007/s00726-003-0011-2. [DOI] [PubMed] [Google Scholar]
- 99.Tapiero H, Mathe G, Couvreur P, Tew KD. Biomed Pharmacother. 2002;56:439. doi: 10.1016/s0753-3322(02)00284-6. [DOI] [PubMed] [Google Scholar]
- 100.Creager MA, Gallagher SJ, Girerd XJ, Coleman SM, Dzau VJ, Cooke JP. J Clin Invest. 1992;90:1248. doi: 10.1172/JCI115987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Moroz LL, Dahlgren RL, Boudko D, Sweedler JV, Lovell P. J Inorg Biochem. 2005;99:929. doi: 10.1016/j.jinorgbio.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 102.Morris SM., Jr J Nutr. 2004;134:2743S. doi: 10.1093/jn/134.10.2743S. [DOI] [PubMed] [Google Scholar]
- 103.Baydoun AR, Bogle RG, Pearson JD, Mann GE. Br J Pharmacol. 1994;112:487. doi: 10.1111/j.1476-5381.1994.tb13099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Deves R, Boyd CAR. Physiol Rev. 1998;78:487. doi: 10.1152/physrev.1998.78.2.487. [DOI] [PubMed] [Google Scholar]
- 105.Verrey F, Closs EI, Wagner CA, Palacin M, Endou H, Kanai Y. Pflugers Arch. 2004;447:532. doi: 10.1007/s00424-003-1086-z. [DOI] [PubMed] [Google Scholar]
- 106.Sloan JL, Mager S. J Biol Chem. 1999;274:23740. doi: 10.1074/jbc.274.34.23740. [DOI] [PubMed] [Google Scholar]
- 107.Escobales N, Rivera-Correa M, Altieri PI, Rodriguez JF. Amino Acids. 2000;19:451. doi: 10.1007/s007260070023. [DOI] [PubMed] [Google Scholar]
- 108.Nedvetsky PI, Sessa WC, Schmidt H. Proc Natl Acad Sci USA. 2002;99:16510. doi: 10.1073/pnas.262701999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wu GY, Morris SM. Biochemical J. 1998;336:1. doi: 10.1042/bj3360001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ratner S. Adv Enzymol Relat Areas Mol Biol. 1973;39:1. doi: 10.1002/9780470122846.ch1. [DOI] [PubMed] [Google Scholar]
- 111.Reutov VP. Biochemistry (Mosc) 2002;67:293. doi: 10.1023/a:1014832416073. [DOI] [PubMed] [Google Scholar]
- 112.Lincoln J, Hoyle CHV, Burnstock G. Nitric Oxide. Cambridge University Press; Cambridge: 1997. [Google Scholar]
- 113.Ford PC, Wink DA, Stanbury DM. FEBS Lett. 1993;326:1. doi: 10.1016/0014-5793(93)81748-o. [DOI] [PubMed] [Google Scholar]
- 114.Kharitonov VG, Sundquist AR, Sharma VS. J Biol Chem. 1994;269:5881. [PubMed] [Google Scholar]
- 115.Lewis RS, Deen WM. Chem Res Toxicol. 1994;7:568. doi: 10.1021/tx00040a013. [DOI] [PubMed] [Google Scholar]
- 116.Fukuto JM, Cho JY, Switzer CH. Nitric oxide. In: Ignarro LJ, editor. Biology and Pathobiology. Academic Press; San Diego: 2000. p. 23. [Google Scholar]
- 117.Moir JWB, Wood NJ. Cell Mol Life Sci. 2001;58:215. doi: 10.1007/PL00000849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Forde BG. Biochim Biophys Acta. 2000;1465:219. doi: 10.1016/s0005-2736(00)00140-1. [DOI] [PubMed] [Google Scholar]
- 119.Jensen FB. Comp Biochem Physiol, Part A: Mol Integr Physiol. 2003;135:9. doi: 10.1016/s1095-6433(02)00323-9. [DOI] [PubMed] [Google Scholar]
- 120.Wu GY, Meininger CJ. Annu Rev Nutr. 2002;22:61. doi: 10.1146/annurev.nutr.22.110901.145329. [DOI] [PubMed] [Google Scholar]
- 121.McKnight GM, Duncan CW, Leifert C, Golden MH. Br J Nutr. 1999;81:349. doi: 10.1017/s000711459900063x. [DOI] [PubMed] [Google Scholar]
- 122.Carossa S, Pera P, Doglio P, Lombardo S, Colagrande P, Brussino L, Rolla G, Bucca C. Eur J Clin Invest. 2001;31:876. doi: 10.1046/j.1365-2362.2001.00902.x. [DOI] [PubMed] [Google Scholar]
- 123.Palmerini CA, Palombari R, Perito S, Arienti G. Free Radic Res. 2003;37:29. doi: 10.1080/1071576021000028398. [DOI] [PubMed] [Google Scholar]
- 124.Sobko T, Reinders CI, Jansson EA, Norin E, Midtvedt T, Lundberg JO. Nitric Oxide. 2005;13:272. doi: 10.1016/j.niox.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 125.Goldstein S, Lind J, Merenyi G. Chem Rev. 2005;105:2457. doi: 10.1021/cr0307087. [DOI] [PubMed] [Google Scholar]
- 126.Beckman JS, Beckman TW, Chen J, Marshall PA, Freeman BA. Proc Natl Acad Sci USA. 1990;87:1620. doi: 10.1073/pnas.87.4.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Burrows CJ, Muller JG. Chem Rev. 1998;98:1109. doi: 10.1021/cr960421s. [DOI] [PubMed] [Google Scholar]
- 128.Reiter RJ. Prog Neurobiol. 1998;56:359. doi: 10.1016/s0301-0082(98)00052-5. [DOI] [PubMed] [Google Scholar]
- 129.Tamir S, de Rojas Walker T, Wishnok JS, Tannenbaum SR. Nitric Oxide. Pt B, Academic Press Inc.; San Diego: 1996. p. 230. [Google Scholar]
- 130.Maeda H, Akaike T. Biochemistry (Mosc) 1998;63:854. [PubMed] [Google Scholar]
- 131.Squadrito GL, Pryor WA. Free Radic Biol Med. 1998;25:392. doi: 10.1016/s0891-5849(98)00095-1. [DOI] [PubMed] [Google Scholar]
- 132.Bartosz G. Acta Biochim Pol. 1996;43:645. [PubMed] [Google Scholar]
- 133.Girotti AW. J Lipid Res. 1998;39:1529. [PubMed] [Google Scholar]
- 134.Groves JT. Curr Opin Chem Biol. 1999;3:226. doi: 10.1016/S1367-5931(99)80036-2. [DOI] [PubMed] [Google Scholar]
- 135.Schrammel A, Gorren ACF, Schmidt K, Pfeiffer S, Mayer B. Free Radic Biol Med. 2003;34:1078. doi: 10.1016/s0891-5849(03)00038-8. [DOI] [PubMed] [Google Scholar]
- 136.Goldstein S, Czapski G. J Am Chem Soc. 1996;118:3419. doi: 10.1021/ja00269a020. [DOI] [PubMed] [Google Scholar]
- 137.Keshive M, Singh S, Wishnok JS, Tannenbaum SR, Deen WM. Chem Res Toxicol. 1996;9:988. doi: 10.1021/tx960036y. [DOI] [PubMed] [Google Scholar]
- 138.Kirsch M, Fuchs A, de Groot H. J Biol Chem. 2003;278:11931. doi: 10.1074/jbc.M300237200. [DOI] [PubMed] [Google Scholar]
- 139.Sonnenschein K, de Groot H, Kirsch M. J Biol Chem. 2004;279:45433. doi: 10.1074/jbc.M405987200. [DOI] [PubMed] [Google Scholar]
- 140.Beda NV, Nedospasov AA. Russ J Bioorg Chem. 2006;32:1. doi: 10.1134/s1068162006010018. [DOI] [PubMed] [Google Scholar]
- 141.Frankenfeld CN, Rosenbaugh MR, Fogarty BA, Lunte SM. J Chromatogr A. 2006;1111:147. doi: 10.1016/j.chroma.2005.05.027. [DOI] [PubMed] [Google Scholar]
- 142.Zhang YH, Hogg N. Free Radic Biol Med. 2005;38:831. doi: 10.1016/j.freeradbiomed.2004.12.016. [DOI] [PubMed] [Google Scholar]
- 143.Stamler J, Simon D, Osborne J, Mullins M, Jaraki O, Michel T, Singel D, Loscalzo J. Proc Natl Acad Sci USA. 1992;89:444. doi: 10.1073/pnas.89.1.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Aukrust P, Svardal A, Muller F, Lunden B, Berge R, Froland S. Blood. 1995;86:1383. [PubMed] [Google Scholar]
- 145.Guo HY, Lee JD, Ueda T, Shan J, Wang JA. Jpn Heart J. 2003;44:865. doi: 10.1536/jhj.44.865. [DOI] [PubMed] [Google Scholar]
- 146.Aukrust P, Svardal AM, Muller F, Lunden B, Berge RK, Froland SS. Blood. 1995;86:1383. [PubMed] [Google Scholar]
- 147.Stamler JS, Lamas S, Fang FC. Cell. 2001;106:675. doi: 10.1016/s0092-8674(01)00495-0. [DOI] [PubMed] [Google Scholar]
- 148.Stamler JS. Cell. 1994;78:931. doi: 10.1016/0092-8674(94)90269-0. [DOI] [PubMed] [Google Scholar]
- 149.Padgett CM, Whorton AR. Cell Biochem Biophys. 1995;27:157. doi: 10.1007/BF02738108. [DOI] [PubMed] [Google Scholar]
- 150.Tanaka T, Nakamura H, Yodoi J, Bloom ET. Free Radic Biol Med. 2005;38:1231. doi: 10.1016/j.freeradbiomed.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 151.Tirmenstein MA, Nicholls-Grzemski FA, Schmittgen TD, Zakrajsek BA, Fariss MW. Antioxid Redox Signal. 2000;2:767. doi: 10.1089/ars.2000.2.4-767. [DOI] [PubMed] [Google Scholar]
- 152.Stamler JS, Simon DI, Osborne JA, Mullins ME, Jaraki O, Michel T, Singel DJ, Loscalzo J. Proc Natl Acad Sci USA. 1992;89:444. doi: 10.1073/pnas.89.1.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zhang YH, Hogg N. Am J Physiol Lung Cell Mol Physiol. 2004;287:L467. doi: 10.1152/ajplung.00350.2003. [DOI] [PubMed] [Google Scholar]
- 154.Crawford NM, Guo FQ. Trends Plant Sci. 2005;10:195. doi: 10.1016/j.tplants.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 155.Crawford JH, Chacko BK, Kevil CG, Patel RP. Antioxid Redox Signal. 2004;6:992. doi: 10.1089/ars.2004.6.992. [DOI] [PubMed] [Google Scholar]
- 156.Jourd’heuil D, Gray L, Grisham MB. Biochem Biophys Res Commun. 2000;273:22. doi: 10.1006/bbrc.2000.2892. [DOI] [PubMed] [Google Scholar]
- 157.Carver J, Doctor A, Zaman K, Gaston B. Nitric Oxide. Pt E, Elsevier Academic Press Inc.; San Diego: 2005. p. 95. [Google Scholar]
- 158.Satoh S, Kimura T, Toda M, Maekawa M, Ono S, Narita H, Miyazaki H, Murayama T, Nomura Y. J Neurochem. 1997;69:2197. doi: 10.1046/j.1471-4159.1997.69052197.x. [DOI] [PubMed] [Google Scholar]
- 159.Broniowska KA, Zhang YH, Hogg N. J Biol Chem. 2006;281:33835. doi: 10.1074/jbc.M603248200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Do KQ, Grima G, Benz B, Salt TE. Ann N Y Acad Sci. 2002;962:81. doi: 10.1111/j.1749-6632.2002.tb04058.x. [DOI] [PubMed] [Google Scholar]
- 161.Zhang YH, Hogg N. Proc Natl Acad Sci USA. 2004;101:7891. doi: 10.1073/pnas.0401167101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Greenacre SAB, Ischiropoulos H. Free Radic Res. 2001;34:541. doi: 10.1080/10715760100300471. [DOI] [PubMed] [Google Scholar]
- 163.Ischiropoulos H. Arch Biochem Biophys. 1998;356:1. doi: 10.1006/abbi.1998.0755. [DOI] [PubMed] [Google Scholar]
- 164.Ischiropoulos H, Zhu L, Chen J, Tsai M, Martin JC, Smith CD, Beckman JS. Arch Biochem Biophys. 1992;298:431. doi: 10.1016/0003-9861(92)90431-u. [DOI] [PubMed] [Google Scholar]
- 165.Beckman JS, Ischiropoulos H, Zhu L, Vanderwoerd M, Smith C, Chen J, Harrison J, Martin JC, Tsai M. Arch Biochem Biophys. 1992;298:438. doi: 10.1016/0003-9861(92)90432-v. [DOI] [PubMed] [Google Scholar]
- 166.Ischiropoulos H, Zhu L, Beckman JS. Arch Biochem Biophys. 1992;298:446. doi: 10.1016/0003-9861(92)90433-w. [DOI] [PubMed] [Google Scholar]
- 167.Yamakura F, Ikeda K. Nitric Oxide. 2006;14:152. doi: 10.1016/j.niox.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 168.Kato T, Kikugawa K. Food Chem Toxicol. 1992;30:617. doi: 10.1016/0278-6915(92)90196-r. [DOI] [PubMed] [Google Scholar]
- 169.Venitt S, Croftonsleigh C, Ooi SL, Bonnett R. Carcinogenesis. 1980;1:523. doi: 10.1093/carcin/1.6.523. [DOI] [PubMed] [Google Scholar]
- 170.Ohshima H, Friesen M, Brouet I, Bartsch H. Food Chem Toxicol. 1990;28:647. doi: 10.1016/0278-6915(90)90173-k. [DOI] [PubMed] [Google Scholar]
- 171.Pfeiffer S, Schmidt K, Mayer B. J Biol Chem. 2000;275:6346. doi: 10.1074/jbc.275.9.6346. [DOI] [PubMed] [Google Scholar]
- 172.Thomas DD, Espey MG, Vitek MP, Miranda KM, Wink DA. Proc Natl Acad Sci USA. 2002;99:12691. doi: 10.1073/pnas.202312699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Bian K, Gao ZH, Weisbrodt N, Murad F. Proc Natl Acad Sci USA. 2003;100:5712. doi: 10.1073/pnas.0931291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Eiserich JP, Hristova M, Cross CE, Jones AD, Freeman BA, Halliwell B, van der Vliet A. Nature. 1998;391:393. doi: 10.1038/34923. [DOI] [PubMed] [Google Scholar]
- 175.Radi R. Proc Natl Acad Sci USA. 2004;101:4003. doi: 10.1073/pnas.0307446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Brennan ML, Wu WJ, Fu XM, Shen ZZ, Song W, Frost H, Vadseth C, Narine L, Lenkiewicz E, Borchers MT, Lusis AJ, Lee JJ, Lee NA, Abu-Soud HM, Ischiropoulos H, Hazen SL. J Biol Chem. 2002;277:17415. doi: 10.1074/jbc.M112400200. [DOI] [PubMed] [Google Scholar]
- 177.Aslan M, Ryan TM, Townes TM, Coward L, Kirk MC, Barnes S, Alexander CB, Rosenfeld SS, Freeman BA. J Biol Chem. 2003;278:4194. doi: 10.1074/jbc.M208916200. [DOI] [PubMed] [Google Scholar]
- 178.Nicholls SJ, Shen ZZ, Fu XM, Levison BS, Hazen SL. Nitric Oxide. Pt E, Elsevier Academic Press Inc.; San Diego: 2005. p. 245. [Google Scholar]
- 179.Tabi T, Magyar K, Szoko E. Electrophoresis. 2005;26:1940. doi: 10.1002/elps.200410289. [DOI] [PubMed] [Google Scholar]
- 180.Tsikas D, Mitschke A, Suchy MT, Gutzki FM, Stichtenoth DO. J Chromatogr B. 2005;827:146. doi: 10.1016/j.jchromb.2005.03.039. [DOI] [PubMed] [Google Scholar]
- 181.Vallance P, Leone A, Calver A, Collier J, Moncada S. J Cardiovasc Pharmacol. 1992;20:S60. doi: 10.1097/00005344-199204002-00018. [DOI] [PubMed] [Google Scholar]
- 182.Rawal N, Rajpurohit R, Lischwe MA, Williams KR, Paik WK, Kim SD. Biochim Biophys Acta. 1995;1248:11. doi: 10.1016/0167-4838(94)00213-z. [DOI] [PubMed] [Google Scholar]
- 183.Miyazaki H, Matsuoka H, Cooke JP, Usui M, Ueda S, Okuda S, Imaizumi T. Circulation. 1999;99:1141. doi: 10.1161/01.cir.99.9.1141. [DOI] [PubMed] [Google Scholar]
- 184.Kielstein JT, Impraim B, Simmel S, Bode-Boger SM, Tsikas D, Frolich JC, Hoeper MM, Haller H, Fliser D. Circulation. 2004;109:172. doi: 10.1161/01.CIR.0000105764.22626.B1. [DOI] [PubMed] [Google Scholar]
- 185.Gokce N. J Nutrition. 2004;134:2807S. doi: 10.1093/jn/134.10.2807S. [DOI] [PubMed] [Google Scholar]
- 186.Stuhlinger MC, Abbasi F, Chu JW, Lamendola C, McLaughlin TL, Cooke JP, Reaven GM, Tsao PS. JAMA. 2002;287:1420. doi: 10.1001/jama.287.11.1420. [DOI] [PubMed] [Google Scholar]
- 187.Lin KY, Ito A, Asagami T, Tsao PS, Adimoolam S, Kimoto M, Tsuji H, Reaven GM, Cooke JP. Circulation. 2002;106:987. doi: 10.1161/01.cir.0000027109.14149.67. [DOI] [PubMed] [Google Scholar]
- 188.Maas R, Wenske S, Zabel M, Ventura R, Schwedhelm E, Steenpass A, Klemm H, Noldus J, Boger RH. Eur Urol. 2005;48:1004. doi: 10.1016/j.eururo.2005.07.017. [DOI] [PubMed] [Google Scholar]
- 189.Vallance P, Leone A, Calver A, Collier J, Moncada S. Lancet. 1992;339:572. doi: 10.1016/0140-6736(92)90865-z. [DOI] [PubMed] [Google Scholar]
- 190.Fliser D, Kronenberg F, Kielstein JT, Morath C, Bode-Boger SM, Haller H, Ritz E. J Am Soc Nephrol. 2005;16:2456. doi: 10.1681/ASN.2005020179. [DOI] [PubMed] [Google Scholar]
- 191.Abe T, Tohgi H, Murata T, Isobe C, Sato C. Neurosci Lett. 2001;312:177. doi: 10.1016/s0304-3940(01)02214-5. [DOI] [PubMed] [Google Scholar]
- 192.Kielstein JT, Zoccali C. Am J Kidney Dis. 2005;46:186. doi: 10.1053/j.ajkd.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 193.Scalera F, Martens-Lobenhoffer J, Tager M, Bukowska A, Lendeckel U, Bode-Boger SM. Biochem Biophys Res Commun. 2006;345:1075. doi: 10.1016/j.bbrc.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 194.Teerlink T. Vasc Med. 2005;10:S73. doi: 10.1191/1358863x05vm597oa. [DOI] [PubMed] [Google Scholar]
- 195.Bode-Boger SM, Boger RH, Kienke S, Junker W, Frolich JC. Biochem Biophys Res Commun. 1996;219:598. doi: 10.1006/bbrc.1996.0279. [DOI] [PubMed] [Google Scholar]
- 196.Lenzen H, Tsikas D, Boger RH. Eur J Clin Pharmacol. 2006;62:45. [Google Scholar]
- 197.Kurose I, Wolf R, Grisham MB, Granger DN. Am J Physiol Heart Circ Physiol. 1995;37:H2224. doi: 10.1152/ajpheart.1995.268.6.H2224. [DOI] [PubMed] [Google Scholar]
- 198.Kurose I, Wolf R, Grisham MB, Aw TY, Specian RD, Granger DN. Circ Res. 1995;76:30. doi: 10.1161/01.res.76.1.30. [DOI] [PubMed] [Google Scholar]
- 199.Macallister RJ, Whitley GSJ, Vallance P. Kidney Int. 1994;45:737. doi: 10.1038/ki.1994.98. [DOI] [PubMed] [Google Scholar]
- 200.Segarra G, Medina P, Vila JM, Chuan P, Domenech C, Torondel B, Lluch S. Am J Hypertens. 2001;14:1142. doi: 10.1016/s0895-7061(01)02208-7. [DOI] [PubMed] [Google Scholar]
- 201.Segarra G, Medina P, Ballester RM, Lluch P, Aldasoro M, Vila JM, Lluch S. Stroke. 1999;30:2206. doi: 10.1161/01.str.30.10.2206. [DOI] [PubMed] [Google Scholar]
- 202.Kielstein JT, Tsikas D, Fliser D. Eur J Clin Pharmacol. 2006;62:39. [Google Scholar]
- 203.Schwedhelm E. Vasc Med. 2005;10:S89. doi: 10.1177/1358836X0501000113. [DOI] [PubMed] [Google Scholar]
- 204.Causse E, Siri N, Arnal JF, Bayle C, Malatray P, Valdiguie P, Salvayre R, Couderc F. J Chromatogr B. 2000;741:77. doi: 10.1016/s0378-4347(00)00034-7. [DOI] [PubMed] [Google Scholar]
- 205.Trapp G, Sydow K, Dulay MT, Chou T, Cooke JP, Zare RN. J Sep Sci. 2004;27:1483. doi: 10.1002/jssc.200401918. [DOI] [PubMed] [Google Scholar]
- 206.Lucas KA, Pitari GM, Kazerounian S, Ruiz-Stewart I, Park J, Schulz S, Chepenik KP, Waldman SA. Pharmacol Rev. 2000;52:375. [PubMed] [Google Scholar]
- 207.Pyriochou A, Papapetropoulos A. Cell Signal. 2005;17:407. doi: 10.1016/j.cellsig.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 208.Wedel B, Humbert P, Harteneck C, Foerster J, Malkewitz J, Bohme E, Schultz G, Koesling D. Proc Natl Acad Sci USA. 1994;91:2592. doi: 10.1073/pnas.91.7.2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Wedel B, Harteneck C, Foerster J, Friebe A, Schultz G, Koesling D. J Biol Chem. 1995;270:24871. doi: 10.1074/jbc.270.42.24871. [DOI] [PubMed] [Google Scholar]
- 210.Zhao Y, Marletta MA. Biochemistry. 1997;36:15959. doi: 10.1021/bi971825x. [DOI] [PubMed] [Google Scholar]
- 211.Nioche P, Berka V, Vipond J, Minton N, Tsai AL, Raman CS. Science. 2004;306:1550. doi: 10.1126/science.1103596. [DOI] [PubMed] [Google Scholar]
- 212.Pellicena P, Karow DS, Boon EM, Marletta MA, Kuriyan J. Proc Natl Acad Sci USA. 2004;101:12854. doi: 10.1073/pnas.0405188101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Tomita T, Tsuyama S, Imai Y, Kitagawa T. J Biochem (Tokyo, Jpn) 1997;122:531. doi: 10.1093/oxfordjournals.jbchem.a021785. [DOI] [PubMed] [Google Scholar]
- 214.Tomita T, Ogura T, Tsuyama S, Imai Y, Kitagawa T. Biochemistry. 1997;36:10155. doi: 10.1021/bi9710131. [DOI] [PubMed] [Google Scholar]
- 215.Lewicki JA, Brandwein HJ, Mittal CK, Arnold WP, Murad F. J Cyclic Nucleotide Res. 1982;8:17. [PubMed] [Google Scholar]
- 216.Bellamy TC, Garthwaite J. Mol Cel Biochem. 2002;230:165. [PubMed] [Google Scholar]
- 217.Schmidt K, Desch W, Klatt P, Kukovetz WR, Mayer B. Naunyn Schmiedebergs Arch Pharmacol. 1997;355:457. doi: 10.1007/pl00004969. [DOI] [PubMed] [Google Scholar]
- 218.Stone JR, Marletta MA. Biochemistry. 1996;35:1093. doi: 10.1021/bi9519718. [DOI] [PubMed] [Google Scholar]
- 219.Makino R, Matsuda H, Obayashi E, Shiro Y, Iizuka T, Hori H. J Biol Chem. 1999;274:7714. doi: 10.1074/jbc.274.12.7714. [DOI] [PubMed] [Google Scholar]
- 220.Brandish PE, Buechler W, Marletta MA. Biochemistry. 1998;37:16898. doi: 10.1021/bi9814989. [DOI] [PubMed] [Google Scholar]
- 221.Kharitonov VG, Sharma VS, Magde D, Koesling D. Biochemistry. 1997;36:6814. doi: 10.1021/bi970201o. [DOI] [PubMed] [Google Scholar]
- 222.Bellamy TC, Garthwaite J. J Biol Chem. 2001;276:4287. doi: 10.1074/jbc.M006677200. [DOI] [PubMed] [Google Scholar]
- 223.Corbin JD, Francis SH. J Biol Chem. 1999;274:13729. doi: 10.1074/jbc.274.20.13729. [DOI] [PubMed] [Google Scholar]
- 224.Ellis G, Adatia I, Yazdanpanah M, Makela SK. Clin Biochem. 1998;31:195. doi: 10.1016/s0009-9120(98)00015-0. [DOI] [PubMed] [Google Scholar]
- 225.Yao DD, Vlessidis AG, Evmiridis NP. Microchim Acta. 2004;147:1. [Google Scholar]
- 226.Tsikas D. Free Radic Res. 2005;39:797. doi: 10.1080/10715760500053651. [DOI] [PubMed] [Google Scholar]
- 227.Jobgen WS, Jobgen SC, Li H, Meininger CJ, Wu G. J Chromatogr B. 2006 doi: 10.1016/j.jchromb.2006.07.018. [DOI] [Google Scholar]
- 228.Beale SC. Anal Chem. 1998;70:279R. [Google Scholar]
- 229.Righetti PG. Capillary Electrophoresis in Analytical Biotechnology. CRC Press; Boca Raton, FL: 1996. [Google Scholar]
- 230.Khaledi MG. High performance capillary electrophoresis. John Wiley & Sons; New York: 1998. [Google Scholar]
- 231.Dolnik V, Liu SR. J Sep Sci. 2005;28:1994. doi: 10.1002/jssc.200500243. [DOI] [PubMed] [Google Scholar]
- 232.Harris DS. Quantitative Chemical Analysis. W.H. Freeman and Company; 2002. [Google Scholar]
- 233.Hutterer KM, Jorgenson JW. Anal Chem. 1999;71:1293. doi: 10.1021/ac981221e. [DOI] [PubMed] [Google Scholar]
- 234.Timerbaev AR. Analyst. 2001;126:964. doi: 10.1039/b100594o. [DOI] [PubMed] [Google Scholar]
- 235.Timerbaev AR, Shpigun OA. Electrophoresis. 2000;21:4179. doi: 10.1002/1522-2683(200012)21:18<4179::AID-ELPS4179>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 236.Timerbaev AR. Electrophoresis. 2002;23:3884. doi: 10.1002/elps.200290008. [DOI] [PubMed] [Google Scholar]
- 237.Timerbaev AR. Electrophoresis. 2004;25:4008. doi: 10.1002/elps.200406111. [DOI] [PubMed] [Google Scholar]
- 238.Smith NW, Evans MB. J Pharm Biomed Anal. 1994;12:579. doi: 10.1016/0731-7085(93)e0012-c. [DOI] [PubMed] [Google Scholar]
- 239.Okamoto H, Timerbaev AR, Hirokawa T. J Sep Sci. 2005;28:522. doi: 10.1002/jssc.200400070. [DOI] [PubMed] [Google Scholar]
- 240.Timerbaev AR, Hirokawa T. Electrophoresis. 2006;27:323. doi: 10.1002/elps.200500320. [DOI] [PubMed] [Google Scholar]
- 241.Poinsot W, Lacroix M, Maury D, Chataigne G, Feurer B, Couderc F. Electrophoresis. 2006;27:176. doi: 10.1002/elps.200500512. [DOI] [PubMed] [Google Scholar]
- 242.Swinney K, Bornhop DJ. Electrophoresis. 2000;21:1239. doi: 10.1002/(SICI)1522-2683(20000401)21:7<1239::AID-ELPS1239>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 243.Swinney K, Bornhop D. Crit Rev Anal Chem. 2000;30:1. doi: 10.1021/ac000261r. [DOI] [PubMed] [Google Scholar]
- 244.Wildman BJ, Jackson PE, Jones WR, Alden PG. J Chromatogr. 1991;546:459. doi: 10.1016/s0021-9673(01)93044-4. [DOI] [PubMed] [Google Scholar]
- 245.Meulemans A, Delsenne F. J Chromatogr B. 1994;660:401. doi: 10.1016/0378-4347(94)00310-6. [DOI] [PubMed] [Google Scholar]
- 246.Wiklund NP, Leone AM, Gustafsson LE, Moncada S. Neuroscience. 1993;53:607. doi: 10.1016/0306-4522(93)90609-j. [DOI] [PubMed] [Google Scholar]
- 247.Boudko DY, Cooper BY, Harvey WR, Moroz LL. J Chromatogr B. 2002;774:97. doi: 10.1016/s1570-0232(02)00219-2. [DOI] [PubMed] [Google Scholar]
- 248.Boudko DY. Methods Mol Biol. 2004;279:9. doi: 10.1385/1-59259-807-2:009. [DOI] [PubMed] [Google Scholar]
- 249.Boudko DY, Moroz LL, Harvey WR, Linser PJ. Proc Natl Acad Sci USA. 2001;98:15354. doi: 10.1073/pnas.261253998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Kaniansky D, Masar M, Marak J, Bodor R. J Chromatogr A. 1999;834:133. doi: 10.1016/s0021-9673(98)00789-4. [DOI] [PubMed] [Google Scholar]
- 251.McReynolds JA, Gao L, Barber-Singh J, Shippy SA. J Sep Sci. 2005;28:128. doi: 10.1002/jssc.200401923. [DOI] [PubMed] [Google Scholar]
- 252.Morcos E, Wiklund NP. Acta Physiol Scand. 1999;167:54. doi: 10.1046/j.1365-201x.1999.600ah.x. [DOI] [PubMed] [Google Scholar]
- 253.Morcos E, Wiklund NP. Electrophoresis. 2001;22:2763. doi: 10.1002/1522-2683(200108)22:13<2763::AID-ELPS2763>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 254.Lee J, Ban E, Yi SY, Yoo YS. J Chromatogr A. 2003;1014:189. doi: 10.1016/s0021-9673(03)00943-9. [DOI] [PubMed] [Google Scholar]
- 255.Miyado T, Nagai H, Takeda S, Saito K, Fukushi K, Yoshida Y, Wakida SI, Niki E. J Chromatogr A. 2003;1014:197. doi: 10.1016/s0021-9673(03)00944-0. [DOI] [PubMed] [Google Scholar]
- 256.Gao L, Barber-Singh J, Kottegoda S, Wirtshafter D, Shippy SA. Electrophoresis. 2004;25:1264. doi: 10.1002/elps.200305840. [DOI] [PubMed] [Google Scholar]
- 257.Hirokawa T, Yoshioka M, Okamoto H, Timerbaev AR, Blaschke G. J Chromatogr B. 2004;811:165. doi: 10.1016/j.jchromb.2004.08.035. [DOI] [PubMed] [Google Scholar]
- 258.Miyado T, Tanaka Y, Nagai H, Takeda S, Saito K, Fukushi K, Yoshida Y, Wakida S, Niki E. J Chromatogr A. 2004;1051:185. [PubMed] [Google Scholar]
- 259.Vespalec R, Vlckova M, Horakova H. J Chromatogr A. 2004;1051:75. doi: 10.1016/j.chroma.2004.05.051. [DOI] [PubMed] [Google Scholar]
- 260.Faria NJR, Dobbie H, Slater JM, Shirley DG, Stocking CJ, Unwin RJ. Kidney Int. 2005;67:357. doi: 10.1111/j.1523-1755.2005.00089.x. [DOI] [PubMed] [Google Scholar]
- 261.Gaspar A, Juhasz P, Bagyi K. J Chromatogr A. 2005;1065:327. doi: 10.1016/j.chroma.2004.12.085. [DOI] [PubMed] [Google Scholar]
- 262.Meulemans A. J Chromatogr B. 1996;683:273. doi: 10.1016/0378-4347(96)00113-2. [DOI] [PubMed] [Google Scholar]
- 263.Meulemans A. J Chromatogr B. 2005;824:308. doi: 10.1016/j.jchromb.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 264.Miyado T, Tanaka Y, Nagai H, Takeda S, Saito K, Fukushi K, Yoshida Y, Wakida S, Niki E. J Chromatogr A. 2006;1109:174. doi: 10.1016/j.chroma.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 265.Macka M, Johns C, Doble P, Haddad PR. LC-GC North America. 2001;19:38. [Google Scholar]
- 266.Doble P, Haddad PR. J Chromatogr A. 1999;834:189. doi: 10.1016/s0021-9673(99)00431-8. [DOI] [PubMed] [Google Scholar]
- 267.Bao LY, Dasgupta PK. Anal Chem. 1992;64:991. [Google Scholar]
- 268.Bazzanella A, Lochmann H, Tomos AD, Bachmann K. J Chromatogr A. 1998;809:231. doi: 10.1016/s0021-9673(01)00627-6. [DOI] [PubMed] [Google Scholar]
- 269.Bazzanella A, Lochmann H, Mainka A, Bachmann K. Chromatographia. 1997;45:59. [Google Scholar]
- 270.Roder A, Bachmann K. J Chromatogr A. 1995;689:305. [Google Scholar]
- 271.Chen Z, Tang C, Yu JC. J High Resol Chromatogr. 1999;22:379. [Google Scholar]
- 272.Xu X, de Bruyn PCAM, de Koeijer JA, Logtenberg H. J Chromatogr A. 1999;830:439. [Google Scholar]
- 273.Dahlen J, Hagberg J, Karlsson S. Fresenius’ J Anal Chem. 2000;366:488. doi: 10.1007/s002160050098. [DOI] [PubMed] [Google Scholar]
- 274.Bord N, Cretier G, Rocca JL, Bailly C, Souchez JP. J Chromatogr A. 2005;1100:223. doi: 10.1016/j.chroma.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 275.Fung YS, Lau KM. Electrophoresis. 2003;24:3224. doi: 10.1002/elps.200305535. [DOI] [PubMed] [Google Scholar]
- 276.Johns C, Macka M, Haddad PR. Electrophoresis. 2004;25:3145. doi: 10.1002/elps.200405913. [DOI] [PubMed] [Google Scholar]
- 277.Kuhr WG, Yeung ES. Anal Chem. 1988;60:1832. doi: 10.1021/ac00168a038. [DOI] [PubMed] [Google Scholar]
- 278.Zhu RH, Kok WT. J Pharmac Biomed Anal. 1998;17:985. doi: 10.1016/s0731-7085(98)00065-x. [DOI] [PubMed] [Google Scholar]
- 279.Yang BC, Tan F, Guan YF. Trends Cardiovasc Med. 2006:77. [Google Scholar]
- 280.Prata C, Bonnafous P, Fraysse N, Treilhou M, Poinsot V, Couderc F. Electrophoresis. 2001;22:4129. doi: 10.1002/1522-2683(200111)22:19<4129::AID-ELPS4129>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 281.Lunn G, Hellwig LC. Handbook of Derivatization Reaction for HPLC. John Wiley & Sons; New York: 1998. [Google Scholar]
- 282.Albin M, Weinberger R, Sapp E, Moring S. Anal Chem. 1991;63:417. [Google Scholar]
- 283.Waterval JCM, Lingeman H, Bult A, Underberg WJM. Electrophoresis. 2000;21:4029. doi: 10.1002/1522-2683(200012)21:18<4029::AID-ELPS4029>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 284.Floyd PD, Moroz LL, Gillette R, Sweedler JV. Anal Chem. 1998;70:2243. doi: 10.1021/ac9713013. [DOI] [PubMed] [Google Scholar]
- 285.Moroz LL, Gillette R, Sweedler JV. J Experim Biol. 1999;202:333. doi: 10.1242/jeb.202.4.333. [DOI] [PubMed] [Google Scholar]
- 286.Zhang JY, Tian JN, Liu JQ, Gao H, Chen XG, Hu ZD. Microchim Acta. 2003;143:241. [Google Scholar]
- 287.Causse E, Terrier R, Champagne S, Nertz M, Valdiguie P, Salvayre R, Couderc F. J Chromatogr A. 1998;817:181. doi: 10.1016/s0021-9673(98)00363-x. [DOI] [PubMed] [Google Scholar]
- 288.Silva E, Quinones B, Freund N, Gonzalez LE, Hernandez L. Brain Res. 2001;923:45. doi: 10.1016/s0006-8993(01)03195-x. [DOI] [PubMed] [Google Scholar]
- 289.Silva E, Hernandez L, Quinonez B, Gonzalez LE, Colasante C. Neuroscience. 2004;124:395. doi: 10.1016/S0306-4522(03)00437-8. [DOI] [PubMed] [Google Scholar]
- 290.Nouadje G, Rubie H, Chatelut E, Canal P, Nertz M, Puig P, Couderc F. J Chromatogr A. 1995;717:293. doi: 10.1016/0021-9673(95)00747-3. [DOI] [PubMed] [Google Scholar]
- 291.Chang PL, Chiu TC, Chang HT. Electrophoresis. 2006;27:1922. doi: 10.1002/elps.200500496. [DOI] [PubMed] [Google Scholar]
- 292.Maeso N, Cifuentes A, Barbas C. J Chromatogr B. 2004;809:147. doi: 10.1016/j.jchromb.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 293.Zhao SL, Wang B, Yuan HY, Xiao D. J Chromatogr A. 2006;1123:138. doi: 10.1016/j.chroma.2006.05.038. [DOI] [PubMed] [Google Scholar]
- 294.Guijt RM, Evenhuis CJ, Macka M, Haddad PR. Electrophoresis. 2004;25:4032. doi: 10.1002/elps.200406156. [DOI] [PubMed] [Google Scholar]
- 295.Haber C, Jones WR, Soglia J, Surve MA, McGlynn M, Caplan A, Reineck JR, Krstanovic C. J Capillary Electrophor. 1996;3:1. [PubMed] [Google Scholar]
- 296.Huang XH, Pang TKJ, Gordon MJ, Zare RN. Anal Chem. 1987;59:2747. [Google Scholar]
- 297.Huang XH, Zare RN. Anal Chem. 1991;63:2193. doi: 10.1021/ac00002a020. [DOI] [PubMed] [Google Scholar]
- 298.Huang XH, Zare RN, Sloss S, Ewing AG. Anal Chem. 1991;63:189. doi: 10.1021/ac00002a020. [DOI] [PubMed] [Google Scholar]
- 299.Gordon MJ, Huang XH, Pentoney SL, Zare RN. Science. 1988;242:224. doi: 10.1126/science.242.4876.224. [DOI] [PubMed] [Google Scholar]
- 300.Gallagher PA, Oertel CM, Danielson ND. J Chromatogr A. 1998;817:31. [Google Scholar]
- 301.Gas B, Demjanenko M, Vacik J. J Chromatogr. 1980;192:253. [Google Scholar]
- 302.Pungor E, Pal F, Toth K. Magyar Kemiai Folyoirat. 1982;88:379. [Google Scholar]
- 303.Pungor E, Pal F, Toth K. Anal Chem. 1983;55:1728. [Google Scholar]
- 304.Zemann AJ, Schnell E, Volgger D, Bonn GK. Anal Chem. 1998;70:563. doi: 10.1021/ac9707592. [DOI] [PubMed] [Google Scholar]
- 305.Fracassi da Silva JA, do Lago CL. Anal Chem. 1998;70:4339. [Google Scholar]
- 306.Kaniansky D, Zelenska V, Masar M, Ivanyi F, Gazdikova S. J Chromatogr A. 1999;844:349. [Google Scholar]
- 307.Zemann AJ. TrAC Trends Anal Chem. 2001;20:346. [Google Scholar]
- 308.Zemann AJ. Electrophoresis. 2003;24:2125. doi: 10.1002/elps.200305476. [DOI] [PubMed] [Google Scholar]
- 309.Kuban P, Hauser PC. Electroanalysis. 2004;16:2009. [Google Scholar]
- 310.Sonlinova V, Kasicka V. J Sep Sci. 2006;29:1743. doi: 10.1002/jssc.200600167. [DOI] [PubMed] [Google Scholar]
- 311.Buchberger WW. J Chromatogr A. 2000;884:3. doi: 10.1016/s0021-9673(00)00283-1. [DOI] [PubMed] [Google Scholar]
- 312.Kaniansky D, Zelenska V, Baluchova D. Electrophoresis. 1996;17:1890. doi: 10.1002/elps.1150171214. [DOI] [PubMed] [Google Scholar]
- 313.Katzmayr MU, Klampfl CW, Buchberger W. J Chromatogr A. 1999;850:355. doi: 10.1016/s0021-9673(99)00168-5. [DOI] [PubMed] [Google Scholar]
- 314.Kaniansky D, Zelensky I, Hybenova A, Onuska FI. Anal Chem. 1994;66:4258. [Google Scholar]
- 315.Boudko DY, Harvey WR, Moroz LL. Soc Neurosci. 2001;27:75. [Google Scholar]
- 316.Wan QJ, Kuban P, Tanyanyiwa J, Rainelli A, Hauser PC. Anal Chim Acta. 2004;525:11. [Google Scholar]
- 317.Tuma P, Samcova E, Andelova K. J Chromatogr B. 2006;839:12. doi: 10.1016/j.jchromb.2005.12.020. [DOI] [PubMed] [Google Scholar]
- 318.Kaniansky D, Masar M, Bielcikova J, Ivanyi F, Eisenbeiss F, Stanislawski B, Grass B, Neyer A, Johnck M. Anal Chem. 2000;72:3596. doi: 10.1021/ac991236s. [DOI] [PubMed] [Google Scholar]
- 319.Coufal P, Zuska J, van de Goor T, Smith V, Gas B. Electrophoresis. 2003;24:671. doi: 10.1002/elps.200390079. [DOI] [PubMed] [Google Scholar]
- 320.Tanyanyiwa J, Schweizer K, Hauser PC. Electrophoresis. 2003;24:2119. doi: 10.1002/elps.200305422. [DOI] [PubMed] [Google Scholar]
- 321.Kvasnicka F, Voldrich M. J Chromatogr A. 2006;1103:145. doi: 10.1016/j.chroma.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 322.Kuban P, Abad-Villar EM, Hauser PC. J Chromatogr A. 2006;1107:159. doi: 10.1016/j.chroma.2005.12.060. [DOI] [PubMed] [Google Scholar]
- 323.Tanyanyiwa J, Leuthardt S, Hauser PC. Electrophoresis. 2002;23:3659. doi: 10.1002/1522-2683(200211)23:21<3659::AID-ELPS3659>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 324.Martin RS, Ratzlaff KL, Huynh BH, Lunte SM. Anal Chem. 2002;74:1136. doi: 10.1021/ac011087p. [DOI] [PubMed] [Google Scholar]
- 325.Ciszewski A, Milczarek G. Talanta. 2003;61:11. doi: 10.1016/S0039-9140(03)00355-2. [DOI] [PubMed] [Google Scholar]
- 326.Vitecek J, Petrlova J, Petrek J, Adam V, Potesil D, Havel L, Mikelova R, Trnkova L, Kizek R. Electrochim Acta. 2006;51:5087. [Google Scholar]
- 327.Zhang XJ. Front Biosci. 2004;9:3434. doi: 10.2741/1492. [DOI] [PubMed] [Google Scholar]
- 328.Bedioui F, Villeneuve N. Electroanalysis. 2003;15:5. [Google Scholar]
- 329.Casero E, Losada J, Pariente F, Lorenzo E. Talanta. 2003;61:61. doi: 10.1016/S0039-9140(03)00360-6. [DOI] [PubMed] [Google Scholar]
- 330.Holland LA, Leigh AM. Electrophoresis. 2002;23:3649. doi: 10.1002/1522-2683(200211)23:21<3649::AID-ELPS3649>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 331.Vandaveer WR, Pasas SA, Martin RS, Lunte SM. Electrophoresis. 2002;23:3667. doi: 10.1002/1522-2683(200211)23:21<3667::AID-ELPS3667>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 332.Vandaveer WR, Pasas-Farmer SA, Fischer DJ, Frankenfeld CN, Lunte SM. Electrophoresis. 2004;25:3528. doi: 10.1002/elps.200406115. [DOI] [PubMed] [Google Scholar]
- 333.Wang J. Electroanalysis. 2005;17:1133. [Google Scholar]
- 334.Olefirowicz TM, Ewing AG. Chimia. 1991;45:106. [Google Scholar]
- 335.Woolley AT, Lao KQ, Glazer AN, Mathies RA. Anal Chem. 1998;70:684. doi: 10.1021/ac971135z. [DOI] [PubMed] [Google Scholar]
- 336.Luo PF, Zhang FZ, Baldwin RP. Anal Chem. 1991;63:1702. [Google Scholar]
- 337.Gerhardt GC, Cassidy RM, Baranski AS. Anal Chem. 2000;72:908. doi: 10.1021/ac991129y. [DOI] [PubMed] [Google Scholar]
- 338.Wang J, Chen G, Pumera M. Electroanalysis. 2003;15:862. [Google Scholar]
- 339.Wang J, Chen G, Chatrathi MP, Musameh M. Anal Chem. 2004;76:298. doi: 10.1021/ac035130f. [DOI] [PubMed] [Google Scholar]
- 340.Garcia CD, Henry CS. Analyst. 2004;129:579. doi: 10.1039/b403529a. [DOI] [PubMed] [Google Scholar]
- 341.Wang J, Mannino S, Camera C, Chatrathi MP, Scampicchio M, Zima J. J Chromatogr A. 2005;1091:177. doi: 10.1016/j.chroma.2005.07.055. [DOI] [PubMed] [Google Scholar]
- 342.Xu JJ, Peng Y, Bao N, Xia XH, Chen HY. J Chromatogr A. 2005;1095:193. doi: 10.1016/j.chroma.2005.09.077. [DOI] [PubMed] [Google Scholar]
- 343.Hu S, Pang DW, Wang ZL, Cheng JK, Li ZW, Fan YZ, Hu HZ. Chem J Chinese Universities-Chinese. 1996;17:1207. [Google Scholar]
- 344.Zhang LY, Huang WH, Wang ZL, Cheng J. Anal Sci. 2002;18:1117. doi: 10.2116/analsci.18.1117. [DOI] [PubMed] [Google Scholar]
- 345.Chen JG, Woltman SJ, Weber SG. Advances in Chromatography. Vol. 36. Marcel Dekker; New York: 1996. 273 pp. [PubMed] [Google Scholar]
- 346.Sun XH, Yang XR, Wang EK. J Chromatogr A. 2003;1005:189. doi: 10.1016/s0021-9673(03)00927-0. [DOI] [PubMed] [Google Scholar]
- 347.Bao N, Xu JJ, Dou YH, Cai Y, Chen HY, Xia XH. J Chromatogr A. 2004;1041:245. doi: 10.1016/j.chroma.2004.04.036. [DOI] [PubMed] [Google Scholar]
- 348.Castilho TJ, Sotomayor MDT, Kubota LT. J Pharmac Biomed Anal. 2005;37:785. doi: 10.1016/j.jpba.2004.11.043. [DOI] [PubMed] [Google Scholar]
- 349.Powell PR, Paxon TL, Han KA, Ewing AG. Anal Chem. 2005;77:6902. doi: 10.1021/ac050963m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Chu QC, Guan YQ, Geng CH, Ye JN. Anal Lett. 2006;39:729. [Google Scholar]
- 351.Shiddiky MJA, Park H, Shim YB. Anal Chem. 2006;78:6809. doi: 10.1021/ac0606002. [DOI] [PubMed] [Google Scholar]
- 352.Weng QF, Xu GW, Yuan KL, Tang P. J Chromatogr B. 2006;835:55. doi: 10.1016/j.jchromb.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 353.Wise DD, Barkhimer TV, Brault PA, Kirchhoff JR, Messer WS, Hudson RA. J Chromatogr B. 2002;775:49. doi: 10.1016/s1570-0232(02)00269-6. [DOI] [PubMed] [Google Scholar]
- 354.Cahill PS, Wightman RM. Anal Chem. 1995;67:2599. doi: 10.1021/ac00111a017. [DOI] [PubMed] [Google Scholar]
- 355.Coltro WKT, da Silva JAF, da Silva HDT, Richter EM, Furlan R, Angnes L, do Lago CL, Mazo LH, Carrilho E. Electrophoresis. 2004;25:3832. doi: 10.1002/elps.200406091. [DOI] [PubMed] [Google Scholar]
- 356.Xia FQ, Jin WR, Yin XF, Fang ZL. J Chromatogr A. 2005;1063:227. doi: 10.1016/j.chroma.2004.12.022. [DOI] [PubMed] [Google Scholar]
- 357.Sevcikova P, Glatz Z. J Sep Sci. 2003;26:734. [Google Scholar]
- 358.Weng QF, Jin WR. J Chromatogr A. 2002;971:217. doi: 10.1016/s0021-9673(02)01002-6. [DOI] [PubMed] [Google Scholar]
- 359.Inoue T, Kirchhoff JR. Anal Chem. 2002;74:1349. doi: 10.1021/ac0108515. [DOI] [PubMed] [Google Scholar]
- 360.Wang A, Zhang L, Zhang S, Fang Y. J Pharmac Biomed Anal. 2000;23:429. doi: 10.1016/s0731-7085(00)00326-5. [DOI] [PubMed] [Google Scholar]
- 361.Owens GS, LaCourse WR. J Chromatogr B. 1997;695:15. doi: 10.1016/s0378-4347(97)00096-0. [DOI] [PubMed] [Google Scholar]
- 362.Huang XJ, Kok WT. J Chromatogr A. 1995;716:347. [Google Scholar]
- 363.Lin BL, Colon LA, Zare RN. J Chromatogr A. 1994;680:263. [Google Scholar]
- 364.Lunte SM, Oshea TJ. Electrophoresis. 1994;15:79. doi: 10.1002/elps.1150150112. [DOI] [PubMed] [Google Scholar]
- 365.Nyholm L. Analyst. 2005;130:599. doi: 10.1039/b415004j. [DOI] [PubMed] [Google Scholar]
- 366.Martin RS. Methods Mol Biol. 2006;339:85. doi: 10.1385/1-59745-076-6:85. [DOI] [PubMed] [Google Scholar]
- 367.Klampfl CW. Electrophoresis. 2006;27:3. doi: 10.1002/elps.200500523. [DOI] [PubMed] [Google Scholar]
- 368.Smyth WF. Electrophoresis. 2006;27:2051. doi: 10.1002/elps.200500524. [DOI] [PubMed] [Google Scholar]
- 369.Huck CW, Bakry R, Huber LA, Bonn GK. Electrophoresis. 2006;27:2063. doi: 10.1002/elps.200600046. [DOI] [PubMed] [Google Scholar]
- 370.Rubakhin SS, Page JS, Monroe BR, Sweedler JV. Electrophoresis. 2001;22:3752. doi: 10.1002/1522-2683(200109)22:17<3752::AID-ELPS3752>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 371.Page JS, Rubakhin SS, Sweedler JV. Anal Chem. 2002;74:497. doi: 10.1021/ac0156621. [DOI] [PubMed] [Google Scholar]
- 372.von Brocke A, Nicholson G, Bayer E. Electrophoresis. 2001;22:1251. doi: 10.1002/1522-2683(200105)22:7<1251::AID-ELPS1251>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 373.Smyth WF. Curr Pharm Anal. 2006;2:299. [Google Scholar]
- 374.Tsikas D. Curr Pharm Anal. 2005;1:15. [Google Scholar]
- 375.Xiao YX, Feng YP, Da SL, Yeung ES. Prog Chem. 2004;16:543. [Google Scholar]
- 376.Cannon DM, Winograd N, Ewing AG. Annu Rev Biophys Biomol Struct. 2000;29:239. doi: 10.1146/annurev.biophys.29.1.239. [DOI] [PubMed] [Google Scholar]
- 377.Lu X, Huang WH, Wang ZL, Cheng HK. Anal Chim Acta. 2004;510:127. [Google Scholar]
- 378.Woods LA, Roddy TP, Ewing AG. Electrophoresis. 2004;25:1181. doi: 10.1002/elps.200405842. [DOI] [PubMed] [Google Scholar]
- 379.Duffy CF, MacCraith B, Diamond D, O’Kennedy R, Arriaga EA. Lab on a Chip. 2006;6:1007. doi: 10.1039/b601896c. [DOI] [PubMed] [Google Scholar]
- 380.Gunasekera N, Olson KJ, Musier-Forsyth K, Arriaga EA. Anal Chem. 2004;76:655. doi: 10.1021/ac034916a. [DOI] [PubMed] [Google Scholar]
- 381.Woods LA, Powell PR, Paxon TL, Ewing AG. Electroanalysis. 2005;17:1192. doi: 10.1002/elan.200403240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Luzzi V, Lee CL, Allbritton NL. Anal Chem. 1997;69:4761. doi: 10.1021/ac970550o. [DOI] [PubMed] [Google Scholar]
- 383.Chen GY, Ewing AG. Crit Rev Neurobiol. 1997;11:59. doi: 10.1615/critrevneurobiol.v11.i1.40. [DOI] [PubMed] [Google Scholar]
- 384.Dahlgren RL, Page JS, Sweedler JV. Anal Chim Acta. 1999;400:13. [Google Scholar]
- 385.Ye XY, Kim WS, Rubakhin SS, Sweedler JV. Analyst. 2004;129:1200. doi: 10.1039/b409394a. [DOI] [PubMed] [Google Scholar]
- 386.Zhang X, Kim WS, Hatcher N, Potgieter K, Moroz LL, Gillette R, Sweedler JV. J Biol Chem. 2002;277:48472. doi: 10.1074/jbc.M209130200. [DOI] [PubMed] [Google Scholar]
- 387.Cruz L, Moroz LL, Gillette R, Sweedler JV. J Neurochem. 1997;69:110. doi: 10.1046/j.1471-4159.1997.69010110.x. [DOI] [PubMed] [Google Scholar]
- 388.Powell PR, Ewing AG. Anal Bioanal Chem. 2005;382:581. doi: 10.1007/s00216-005-3075-x. [DOI] [PubMed] [Google Scholar]
- 389.Parrot S, Bert L, Mouly-Badina L, Sauvinet V, Colussi-Mas J, Lambas-Senas L, Robert F, Bouilloux JP, Suaud-Chagny MF, Denoroy L, Renaud B. Cell Mol Neurobiol. 2003;23:793. doi: 10.1023/A:1025009221285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Poinsot V, Bayle C, Couderc F. Electrophoresis. 2003;24:4047. doi: 10.1002/elps.200305692. [DOI] [PubMed] [Google Scholar]
- 391.Kennedy RT, Watson CJ, Haskins WE, Powell DH, Strecker RE. Curr Opin Chem Biol. 2002;6:659. doi: 10.1016/s1367-5931(02)00373-3. [DOI] [PubMed] [Google Scholar]
- 392.Davies MI, Cooper JD, Desmond SS, Lunte CE, Lunte SM. Adv Drug Delivery Rev. 2000;45:169. doi: 10.1016/s0169-409x(00)00114-9. [DOI] [PubMed] [Google Scholar]
- 393.Sakamoto KI, Fujisawa H, Koizumi H, Tsuchida E, Ito H, Sadamitsu D, Maekawa T. J Neurotrauma. 1997;14:349. doi: 10.1089/neu.1997.14.349. [DOI] [PubMed] [Google Scholar]
- 394.Shou M, Smith AD, Shackman JG, Peris J, Kennedy RT. J Neurosci Methods. 2004;138:189. doi: 10.1016/j.jneumeth.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 395.Parrot S, Sauvinet V, Riban V, Depaulis A, Renaud B, Denoroy L. J Neurosci Methods. 2004;140:29. doi: 10.1016/j.jneumeth.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 396.Parrot S, Sauvinet V, Xavier JM, Chavagnac D, Mouly-Badina L, Garcia-Larrea L, Mertens P, Renaud B. Electrophoresis. 2004;25:1511. doi: 10.1002/elps.200305852. [DOI] [PubMed] [Google Scholar]
- 397.Gao L, Patterson EE, Shippy SA. Analyst. 2006;131:222. doi: 10.1039/b510156e. [DOI] [PubMed] [Google Scholar]
- 398.Guchardi R, Schwarz MA. Electrophoresis. 2005;26:3151. doi: 10.1002/elps.200410383. [DOI] [PubMed] [Google Scholar]
- 399.Grasemann H, Ratjen F. Pediatr Pulmonol. 1999;28:442. doi: 10.1002/(sici)1099-0496(199912)28:6<442::aid-ppul10>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 400.Govindaraju K, Cowley EA, Eidelman DH, Lloyd DK. Anal Chem. 1997;69:2793. doi: 10.1021/ac961249v. [DOI] [PubMed] [Google Scholar]
- 401.Vanderwinden JM. Acta Gastroenterol Belg. 1994;57:224. [PubMed] [Google Scholar]
- 402.Herulf M, Svenungsson B, Lagergren A, Ljung T, Morcos E, Wiklund NP, Lundberg JON, Weitzberg E. J Infect Dis. 1999;180:542. doi: 10.1086/314908. [DOI] [PubMed] [Google Scholar]
- 403.Stark ME, Szurszewski JH. Gastroenterology. 1992;103:1928. doi: 10.1016/0016-5085(92)91454-c. [DOI] [PubMed] [Google Scholar]
- 404.Iversen HH, Celsing F, Leone AM, Gustafsson LE, Wiklund NP. Br J Pharmacol. 1997;120:702. doi: 10.1038/sj.bjp.0700967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Monaghan JM, Cook K, Gara D, Crowther D. J Chromatogr A. 1997;770:143. doi: 10.1016/s0021-9673(96)01031-x. [DOI] [PubMed] [Google Scholar]
- 406.Leone AM, Francis PL, Rhodes P, Moncada S. Biochem Biophys Res Commun. 1994;200:951. doi: 10.1006/bbrc.1994.1542. [DOI] [PubMed] [Google Scholar]
- 407.Ferslew KE, Hagardorn AN, Robert TA. J Forensic Sci. 2001;46:615. [PubMed] [Google Scholar]
- 408.Morcos E, Wiklund NP. Methods Mol Biol. 2004;279:21. doi: 10.1385/1-59259-807-2:021. [DOI] [PubMed] [Google Scholar]
- 409.Nutku MS, Erim FB. J High Resolut Chromatogr. 1998;21:505. [Google Scholar]
- 410.Bayle C, Causse E, Couderc F. Electrophoresis. 2004;25:1457. doi: 10.1002/elps.200305874. [DOI] [PubMed] [Google Scholar]
- 411.Sbrana E, Paladini A, Bramanti E, Spinetti MC, Raspi G. Electrophoresis. 2004;25:1522. doi: 10.1002/elps.200305848. [DOI] [PubMed] [Google Scholar]
- 412.Carru C, Zinellu A, Sotgia S, Marongiu G, Farina MG, Usai MF, Pes GM, Tadolini B, Deiana L. J Chromatogr A. 2003;1017:233. doi: 10.1016/j.chroma.2003.08.020. [DOI] [PubMed] [Google Scholar]
- 413.Camera E, Picardo M. J Chromatogr B. 2002;781:181. doi: 10.1016/s1570-0232(02)00618-9. [DOI] [PubMed] [Google Scholar]
- 414.Vegvari A, Larsson AK, Hjerten S, Mannervik B. Chembiochem. 2002;3:1117. doi: 10.1002/1439-7633(20021104)3:11<1117::AID-CBIC1117>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 415.Misiti F, Meucci E, Zuppi C, Vincenzoni F, Giardina B, Castagnola M, Messana I. Biochem Biophys Res Commun. 2002;294:829. doi: 10.1016/S0006-291X(02)00552-1. [DOI] [PubMed] [Google Scholar]
- 416.Carru C, Zinellu A, Pes GM, Marongiu G, Tadolini B, Deiana L. Electrophoresis. 2002;23:1716. doi: 10.1002/1522-2683(200206)23:11<1716::AID-ELPS1716>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 417.Whiteman M, Siau JL, Halliwell B. J Biol Chem. 2003;278:8380. doi: 10.1074/jbc.M211086200. [DOI] [PubMed] [Google Scholar]
- 418.Massip C, Riollet P, Quemener E, Bayle C, Salvayre R, Couderc F, Causse E. J Chromatogr A. 2002;979:209. doi: 10.1016/s0021-9673(02)01502-9. [DOI] [PubMed] [Google Scholar]
- 419.Brundin L, Svenungsson E, Morcos E, Andersson M, Olsson T, Lundberg I, Wiklund NP. Ann Neurol. 1998;44:704. doi: 10.1002/ana.410440421. [DOI] [PubMed] [Google Scholar]
- 420.Brundin L, Morcos E, Olsson T, Wiklund NP, Andersson M. Acta Physiol Scand. 1999;167:13. [Google Scholar]
- 421.Brundin L, Morcos E, Olsson T, Wiklund NP, Andersson M. Eur J Neurol. 1999;6:585. doi: 10.1046/j.1468-1331.1999.650585.x. [DOI] [PubMed] [Google Scholar]
- 422.Brundin L, Andersson M, Wiklund NP, Olsson T. Nitric Oxide. 2000;4:271. [Google Scholar]
- 423.Svenungsson E, Andersson M, Brundin L, van Vollenhoven R, Khademi M, Tarkowski A, Greitz D, Dahlstrom M, Lundberg I, Klareskog L, Olsson T. Ann Rheum Dis. 2001;60:372. doi: 10.1136/ard.60.4.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 424.Sadamitsu D, Kuroda Y, Nagamitsu T, Tsuruta R, Inoue T, Ueda T, Nakashima K, Ito H, Maekawa T. Crit Care Med. 2001;29:77. doi: 10.1097/00003246-200101000-00018. [DOI] [PubMed] [Google Scholar]
- 425.Carlucci F, Tabucchi A, Biagioli B, Simeone F, Scolletta S, Rosi F, Marinello E. Biomed Pharmacother. 2002;56:483. doi: 10.1016/s0753-3322(02)00286-x. [DOI] [PubMed] [Google Scholar]
- 426.Panossian A, Hambartsumyan M, Panosyan L, Abrahamyan H, Mamikonyan G, Gabrielyan E, Amaryan G, Astvatsatryan V, Wikman G. Nitric Oxide. 2003;9:103. doi: 10.1016/j.niox.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 427.Pereira EC, Bertolami MC, Faludi AA, Sevanian A, Abdalla DSP. Free Radic Biol Med. 2004;37:1440. doi: 10.1016/j.freeradbiomed.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 428.Danilov AI, Andersson M, Bavand N, Wiklund NP, Olsson T, Brundin L. J Neuroimmunol. 2003;136:112. doi: 10.1016/s0165-5728(02)00464-2. [DOI] [PubMed] [Google Scholar]
- 429.Dasgupta PK, Bao LY. Anal Chem. 1993;65:1003. [Google Scholar]
- 430.Rhemrevboom MM. J Chromatogr A. 1994;680:675. [Google Scholar]
- 431.Janini GM, Chan KC, Muschik GM, Issaq HJ. J Chromatogr B. 1994;657:419. doi: 10.1016/0378-4347(94)00141-3. [DOI] [PubMed] [Google Scholar]
- 432.Ueda T, Maekawa T, Sadamitsu D, Oshita S, Ogino K, Nakamura K. Electrophoresis. 1995;16:1002. doi: 10.1002/elps.11501601167. [DOI] [PubMed] [Google Scholar]
- 433.Bjergegaard C, Moller P, Sorensen H. J Chromatogr A. 1995;717:409. [Google Scholar]
- 434.Trushina EV, Oda RP, Landers JP, McMurray CT. Electrophoresis. 1997;18:1890. doi: 10.1002/elps.1150181027. [DOI] [PubMed] [Google Scholar]
- 435.Friedberg MA, Hinsdale ME, Shihabi ZK. J Chromatogr A. 1997;781:491. doi: 10.1016/s0021-9673(97)00583-9. [DOI] [PubMed] [Google Scholar]
- 436.Bories PN, Scherman E, Dziedzic L. Clin Biochem. 1999;32:9. doi: 10.1016/s0009-9120(98)00090-3. [DOI] [PubMed] [Google Scholar]
- 437.Davies CA, Perrett D, Zhang Z, Nielsen BR, Blake DR, Winyard PG. Electrophoresis. 1999;20:2111. doi: 10.1002/(SICI)1522-2683(19990701)20:10<2111::AID-ELPS2111>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 438.Messana I, Rossetti DV, Misiti F, Vincenzoni F, Tellone E, Giardina B, Castagnola M. Electrophoresis. 2000;21:1606. doi: 10.1002/(SICI)1522-2683(20000501)21:8<1606::AID-ELPS1606>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 439.Govindaraju K, Toporsian M, Ward ME, Lloyd DK, Cowley EA, Eidelman DH. J Chromatogr B. 2001;762:147. doi: 10.1016/s0378-4347(01)00358-9. [DOI] [PubMed] [Google Scholar]
- 440.Badr O, Probert SD. Applied Energy. 1993;46:1. [Google Scholar]
- 441.Nicholson JP, Weston KJ, Fowler D. Atmos Environ. 2001;35:2009. [Google Scholar]
- 442.Thum T, Bauersachs J. J Chromatogr B. 2006 doi: 10.1016/j.jchromb.2006.07.023. [DOI] [Google Scholar]


