Abstract
Background
To elucidate the genes involved in the neoplastic transformation of B cells, global gene expression profiles were generated using Affymetrix U74Av2 microarrays, containing 12,488 genes, for four different groups of mouse B-cell lymphomas and six subtypes of pristane-induced mouse plasma cell tumors, three of which developed much earlier than the others.
Results
Unsupervised hierarchical cluster analysis exhibited two main sub-clusters of samples: a B-cell lymphoma cluster and a plasma cell tumor cluster with subclusters reflecting mechanism of induction. This report represents the first step in using global gene expression to investigate molecular signatures related to the role of cooperating oncogenes in a model of Myc-induced carcinogenesis. Within a single subgroup, e.g., ABPCs, plasma cell tumors that contained typical T(12;15) chromosomal translocations did not display gene expression patterns distinct from those with variant T(6;15) translocations, in which the breakpoint was in the Pvt-1 locus, 230 kb 3' of c-Myc, suggesting that c-Myc activation was the initiating factor in both. When integrated with previously published Affymetrix array data from human multiple myelomas, the IL-6-transgenic subset of mouse plasma cell tumors clustered more closely with MM1 subsets of human myelomas, slow-appearing plasma cell tumors clustered together with MM2, while plasma cell tumors accelerated by v-Abl clustered with the more aggressive MM3-MM4 myeloma subsets. Slow-appearing plasma cell tumors expressed Socs1 and Socs2 but v-Abl-accelerated plasma cell tumors expressed 4–5 times as much. Both v-Abl-accelerated and non-v-Abl-associated tumors exhibited phosphorylated STAT 1 and 3, but only v-Abl-accelerated plasma cell tumors lost viability and STAT 1 and 3 phosphorylation when cultured in the presence of the v-Abl kinase inhibitor, STI-571. These data suggest that the Jak/Stat pathway was critical in the transformation acceleration by v-Abl and that v-Abl activity remained essential throughout the life of the tumors, not just in their acceleration. A different pathway appears to predominate in the more slowly arising plasma cell tumors.
Conclusion
Gene expression profiling differentiates not only B-cell lymphomas from plasma cell tumors but also distinguishes slow from accelerated plasma cell tumors. These data and those obtained from the sensitivity of v-Abl-accelerated plasma cell tumors and their phosphorylated STAT proteins indicate that these similar tumors utilize different signaling pathways but share a common initiating genetic lesion, a c-Myc-activating chromosome translocation.
Background
Since the discovery that BALB/c mice predictably develop plasma cell tumors (PCTs) following the induction of intraperitoneal (ip) inflammatory granulomas by implanting plastic objects or injecting mineral oil or silicones, this model has been a valuable experimental system for studying plasma cell neoplasia [1]. The ip inflammatory tissue produces interleukin 6 (IL-6), insulin-like growth factor 1 (IGF1), and other cytokines that support the growth of TEPC-type PCTs that appear slowly, starting no earlier than 120 days after ip pristane injection. Infection with retroviruses containing oncogenes, e.g., Abelson Virus (v-Abl [2]), J3V1 virus (v-Raf plus v-Myc [3]), RIM virus (v-Ha-Ras plus c-Myc [4]), ABLMYC Virus (v-Abl plus c-Myc [5]), accelerates PCT formation, producing ABPC, J3PCs, RIMPCs and ABLMYCPCs, respectively, in as little as 12 days (see Figure 1A and Table 1). It has never been tested whether continued activity of these accelerating oncogenes was required subsequent to completion of transformation. iMycEμ (Myc/IgH-knock-in) BALB/c mice and IL-6-transgenic BALB/c mice also develop intraperitoneal PCTs iMycEμ PCs [6] and KiPCs [7], respectively, at a slightly accelerated rate following pristane injection. IL-6-transgenic BALB/c mice spontaneously develop PCTs in lymph nodes (IL6PCs [8]) but with latent periods no shorter than TEPCs. As with human multiple myeloma (MM), most of the tumors in the different groups of PCTs consist of neoplasms of plasma cells that are morphologically indistinguishable, although the cells in the accelerated tumors occasionally appear larger and more blastic. Other forms of B-cell lymphomas (BCL) also appear in the lymph nodes of iMyc mice, BCLEμ [9] and BCLCα [10].
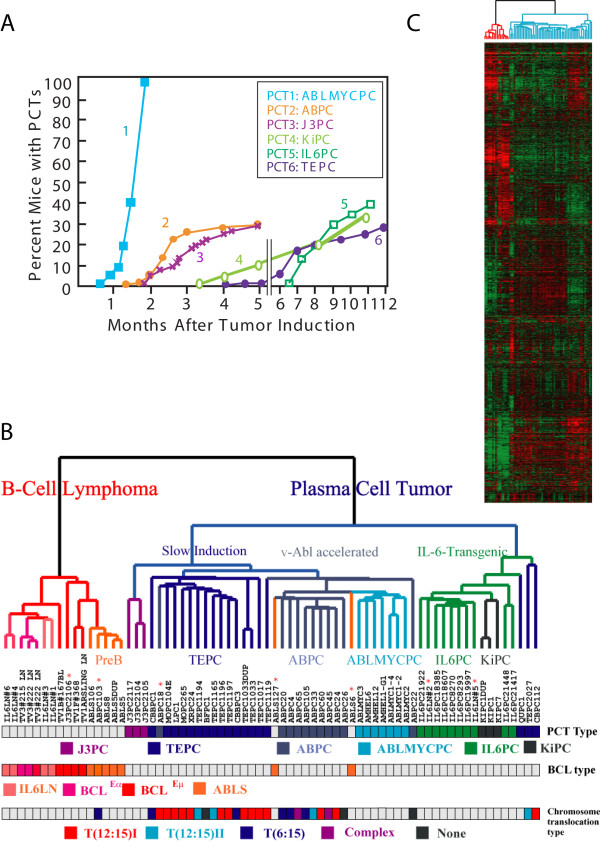
Figure 1.
Gene expression analysis of mouse plasmacytomas and B cell lymphomas. 70 RNA samples from murine B-cell malignancies, comprised of 6 different types of mouse plasma cell tumors and 4 different types of mouse B cell lymphomas, were used for global gene expression studies. A. Kinetics of appearance of 6 different subclasses of plasma cell tumors. Time course of appearance of PCTs after ip injection of pristane. IL6PCs arose spontaneously in lymph nodes of IL-6-transgenic mice, without pristane treatment, as the mice aged. See Table 1 for more details. B and C. Unsupervised hierarchical clustering. Using Affymetrix U74A v2 microarrays, 6424 genes were used for clustering of genes and samples after filtering out genes with more than 50% of A (absence) calls. Correlation-based (uncentered) average linkage clustering was performed on log base 2-transformed data previously centered to mean expression values of each gene. Gene expressions above this mean value are colored red; those expressed below are shown in green. The cluster dendrogram shows 2 major sample clusters: a PCT cluster and a B-cell lymphoma cluster. Samples are coded with 11 different colors based on previously assigned groups. Asterisks indicate samples that clustered in an unexpected manner (see text). In the dendrogram, the PCT samples are colored blue-green and the BCL samples are red-orange. In the first color bar, different viral and transgenic contributions to PCT development are distinguished by different colors. In the second color bar, the 4 types of BCLs are color-coded. In the third color bar, mouse PCTs previously characterized for the fine structure of their Myc-activating chromosomal translocations are identified in this dendrogram. PCT samples having the variant chromosomal translocation T(6;15) are shown in deep blue, type I T(12;15) are shown in red/orange, and type II T(12;15) are shown in light blue. PCT samples having complex chromosomal translocations [12] are shown in purple, and PCT samples having no identifiable chromosomal translocations are shown in pale black. Boxes without color indicate PCTs with unknown karyotypes.
Table 1.
Summary of mouse tumors studied by Affymetrix microarray profiling
| Tumors | Cited as | Exogenous gene effect | Tumor type | Site of origin | N |
| PCT-1 | ABLMYCPC | v-Abl + c-Myc | Virus-accelerated ip PCT | intraperitoneal | 7 |
| PCT-2 | ABPC | v-Abl | Virus-accelerated ip PCT | intraperitoneal | 12 |
| PCT-3 | J3PC | v-Raf + v-Myc | Virus-accelerated ip PCT | intraperitoneal | 4 |
| PCT-4 | KiPC | IL-6 transgene | Transgene-accelerated ip PCT | intraperitoneal | 3 |
| PCT-5 | IL6PC | IL-6 transgene | Spontaneous PCT | gut-associated lymphoid tissue | 8 |
| PCT-6 | TEPC, LPC, MOPC | none | Pristane oil-induced ip PCT | intraperitoneal | 18 |
| BCL-1 | IL6LN | IL-6 transgene | Spontaneous pre-malignant PCT | gut-associated lymphoid tissue | 6 |
| BCL-2 | Pre B v-Abl (a.k.a. ABLS) | v-Abl | Virus-accelerated Pre-B lymphoma | bone marrow + spleen | 6 |
| BCL-3 | BCLEμ | c-Myc in Igμ locus | Spontaneous BCL | lymph nodes | 3 |
| BCL-4 | BCLCα | c-Myc in Igα locus | Spontaneous BCL | lymph nodes | 3 |
Chromosomal translocations that deregulate expression of the proto-oncogene c-Myc constitute the earliest known tumorigenic step in PCT formation. More than 95% of PCTs, have chromosomal translocations, either the typical (85% of PCTs) T(12;15) or variants (15% of PCTs) T(6;15) or T(15;16) [11-13]. T(12;15) joins the immunoglobulin heavy chain locus and its strong enhancers to different regions: within the c-Myc locus (Class I) or upstream of it (Class II). T(6;15) joins the immunoglobulin kappa light chain locus with its enhancers to a site in the Pvt1 locus, between 200 and 300 kb 3' of the c-Myc locus [1,11]. Myc activation appears to be the hallmark of all typical BALB/c PCTs, presumably via Ig enhancer insertion near c-Myc. It has been assumed that the variant translocations also contribute to the transformation of plasma cells by upregulating c-Myc by some mechanism akin to but different from the 12;15 translocations. This notion has never been experimentally validated.
To investigate the mechanisms underlying Myc-induced B-cell neoplasia (particularly PCTs) in BALB/c mice, gene expression profiles for four groups of BCLs and for PCTs from six different induction protocols (Table 1) were generated. The analysis of the microarray gene expression data focused on four questions: 1) which genes differentiate BCLs from PCTs; 2) which genes and pathways are involved in PCT acceleration by the v-Abl oncogene; 3) are there genes that differ between PCTs induced by typical and variant c-Myc-activating chromosomal translocations; 4) how much similarity in gene expression profiles exists between mouse PCTs and human MMs? To answer the final question, a meta-analysis was performed by combining our mouse tumor expression data with data published for human multiple myelomas to determine similarities and differences between these plasma cell tumors in different species.
Results
Unsupervised clustering yielded two main clusters: B cell lymphoma cluster and plasma cell tumor cluster
To assess the relative similarities or dissimilarities in global gene expression patterns from six different subtypes of mouse PCT (PCT-1 – PCT-6, Table 1) and four different groups of mouse BCL (BCL-1 – BCL-4, Table 1), we first applied unsupervised hierarchical cluster analysis using correlation-based average linkage clustering. Characteristic expression signatures were searched for among the 6424 genes remaining after filtering out genes showing more than 50% absence (A) calls among the 70 RNA samples. This analysis revealed two principal groups, composed of BCLs and PCTs, as shown in Figures 1B and 1C.
With few exceptions, most of the PCTs, regardless of transgenic differences or viral agents accelerating appearance of PCTs, clustered together (blue-green dendrogram, Figures 1B and 1C). This cluster included both forms of PCTs that arose in IL-6-transgenic BALB/c mice: KiPCs, which arose intraperitoneally after ip pristane, and IL6PCs, which spontaneously arose in hyperplastic lymph nodes, independent of ip pristane. These results indicated that all PCTs share a common genetic expression signature. The IL-6-pre-malignant transgenic lymph node samples (IL6LN) did not cluster with the IL6PCs. Instead, these clustered with the BCLs (red-orange dendrogram, Figures 1B and 1C). ABLS are pre-B cell lymphomas induced by Abelson virus infection. Note that while the v-Abl-containing retroviruses also produced two subtypes of PCTs (ABPCs and ABLMYCPCs), the ABLSs clustered with BCLEμ and BCLCα, suggesting that cell type influenced gene expression more than v-Abl, in this situation.
The nature of c-Myc-activating chromosomal translocations, i.e., typical T(12;15) vs. variant T(6;15) [11,12], did not affect their inclusion in the PCT group, nor did it lead to subclusters within the ABPC or TEPC subgroups in this particular analysis (Figure 1B and 1C). Instead, tumor sample subclustering was influenced more by the agents used to accelerate PCT development, generating subclusters within the PCT cluster, each of which contained virtually only one PCT subclass, ABLMYCPC, ABPC, IL6PC, KiPC or TEPC. Note that ABPCs and ABLMYCPCs clustered with each other before clustering with other PCTs, indicating a greater degree of similarity between these two groups of PCTs, both of which were accelerated by v-Abl. The two forms of PCTs that arose in IL-6-transgenic BALB/c mice also clustered most closely with one another.
Constitutive expression of IL-6 in IL-6-transgenic mice triggers polyclonal plasmacytosis in lymphoid organs such as lymph nodes and spleen [14], and neoplastic transformation of these plasma cells occurs spontaneously over time [13]. Thus, RNA samples from 6 enlarged lymph nodes in young IL-6-transgenic BALB/c mice were provisionally designated IL6LN (rich in pre-malignant cells), while samples from 8 larger lymph nodes from older mice were designated IL6PC (now containing transformed plasma cells). As expected, hierarchical clustering grouped all 8 IL6PCs into the "Plasma Cell Tumor" cluster. However, two of the "IL6LN" samples, expected to be pre-malignant, owing to the younger ages of these transgenic mice, clustered with the IL6PC samples in the PCT group and are marked with asterisks in Figure 1B. Since the remaining four samples of IL6LN were tightly clustered with the bulk of the B-cell lymphomas in the BCL group, we took this finding as evidence that the two PCT-like IL6LN samples probably contained a significant percentage of transformed plasma cells.
Class comparison of the two main clusters revealed major, consistent gene expression differences between PCTs and BCLs. To gain biological insight into these differences, a two-sample t-test was applied to select the genes whose expression was significantly different between these two groups (926 genes, p < 1 × 10-5). Functional enrichment analysis [see Additional file 1] based on gene ontology (GO) terms showed that these expression differences were heavily enriched for B cell-specific genes that were generally downregulated in plasma cells [15-17]. Figure 2 presents heat maps of a subset of those genes from categories of gene functions assigned by Cancer Genome Anatomy Project (CGAP), illustrating that that PCTs and BCLs mobilize different signal transduction pathways. It is interesting to note that the majority of the gene differences consist of downregulation of genes in PCTs. It is likely that this reflects the wholesale focusing of the protein synthesis machinery of these antibody-secreting "factories" on the production of their characteristic protein.
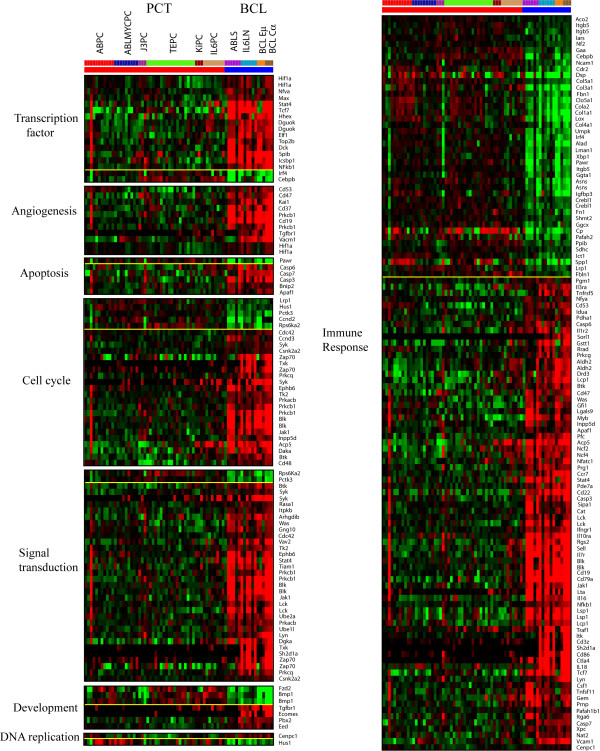
Figure 2.
Genes showing significant differences in expression between PCTs and BCLs. Class comparison between PCTs and BCLs showed 926 genes that showed significant differences (p < 1 × 10-5) in two sample t-test. The genes assigned to 8 different functional categories by CGAP are presented as a heat map of gene cluster. Additional details can be found in Additional file 1.
The genes with higher expression in PCTs include Cyclin D2 (Ccnd2), syndecan1 (Sdc1), Irf4, and Xbp1. Irf4 and syndecan1 (CD138) were previously reported to have increased expression in plasma cells [15,16,18-20]. Cyclin D2 was previously reported to be induced by c-Myc activation [21]. Several pro-collagens (Col1a1, Col1a2, Col3a1, and Col5a1) also showed higher expression in PCTs than in BCLs. As expected, transcription factors previously known to have higher expression in plasma cells, Xbp1, Cebpb, and Irf4 [15,16] also showed higher expression in PCT groups.
B-cell surface markers (e.g., CD79b, CD19, and CD22) were highly expressed in BCL samples, as expected. Expression of caspases (e.g., Casp3, Casp6 and Casp7) was higher in the BCL group, as was expression of the chemokines (Ccr7), cytokines (IL-16, IL18) and cytokine receptors (IL1r2, IL3ra, IL7r, and IL10ra). An additional relevant gene that was more highly expressed in the BCL group was Syk, a tyrosine kinase that is essential for B-cell antigen receptor triggering of cellular activation [22]. Genes involved in Jak-Stat signaling (Jak1, Stat4) and NFκB signaling (NFκB1) also showed higher expression in the BCL groups. None of these functions are thought to be active after B cells differentiate into plasma cells, which have lost nearly all their surface receptors and downregulated many other B-cell-specific genes. Although whole lymph nodes were used as samples in IL-6-transgenic PCTs and LN, the lower expression of B-cell surface markers seen in the expression profiles of the IL6PCs suggests that the main populations of cells in IL-6-transgenic mouse lymph nodes are plasma cells, which typically lose expression of most surface markers. Our results confirm the histological finding that percentage of B- and T lymphocytes decreased as the lymph nodes fill with plasma cells and PCTs [14].
Genes reflecting v-Abl-accelerated PCT induction
A direct comparison between two groups of PCTs: the rapid onset PCTs (ABPC and ABLMYC) and the slow onset PCTs, (TEPC, KiPC and IL6PC) was performed to get information on genes that might have contributed to the acceleration. A two-sample t-test between these two groups revealed 1195 genes with significant (p < 0.001) expression differences. Among these 1195 genes, 80 genes showed greater than 2-fold higher expression in the ABPC plus ABLMYC group, while 83 genes showed greater than 2-fold higher expression in the TEPC, KiPC and IL6PC group [see Additional file 2]. Several genes involved in cellular growth and proliferation (Cdkn2b, Tgfb1, Ptprc, Cd6, Gfi1, and Mybl2) showed higher expression in the ABPC plus ABLMYC group, as did many interferon-induced genes, presumably induced by the presence of retroviruses in this group. Several genes involved in cell death (Ada, Bid1, Gas6, Prkcζ,) were higher in TEPCs, KiPCs and IL6PCs. Table 2 lists a subset of these genes arranged by their categories of gene functions assigned by CGAP.
Table 2.
Genes showing significant differences in expression between rapid-forming PCTs (ABPC and ABLMYCPC) and slow-forming PCTs (TEPC, IL6PC and KiPC)
| Higher in slow-forming PCT (TEPC, IL6PC, KiPC | |||||||
| Gene function | Rapid PCT | Slow PCT | Fold difference (slow/rapid) | Description | Unigene ID | Gene symbol | p-value |
| Angiogenesis | |||||||
| 308.4 | 495.6 | 1.6 | Neogenin | Mm.42249 | Neo1 | 1.05E-05 | |
| Apoptosis | |||||||
| 2872.8 | 5249.1 | 1.8 | RIKEN cDNA A630035D09 gene | Mm.238213 | Bcl2l1 | 4.67E-05 | |
| Cell cycle | |||||||
| 583.6 | 2799.1 | 4.8 | Megakaryocyte-associated tyrosine kinase | Mm.2918 | Matk | < 1E-07 | |
| 270.1 | 912.6 | 3.4 | Protein kinase C, zeta | Mm.28561 | Prkcz | < 1E-07 | |
| 208.1 | 364.3 | 1.8 | Acid phosphatase, prostate | Mm.19941 | Acpp | 2.10E-06 | |
| 280.7 | 473.5 | 1.7 | Cyclin D1 | Mm.273049 | Ccnd1 | 7.12E-05 | |
| 1484.3 | 2441.1 | 1.6 | Casein kinase 1, epsilon | Mm.30199 | Csnk1e | 3.50E-06 | |
| Immune response | |||||||
| 204.1 | 535.4 | 2.6 | Prolactin receptor | Mm.10516 | Prlr | 5.4E-06 | |
| 124 | 267.7 | 2.2 | Transforming growth factor, beta receptor III | Mm.200775 | Tgfbr3 | 1.2E-06 | |
| 2872.8 | 5249.1 | 1.8 | RIKEN cDNA A630035D09 gene | Mm.238213 | Bcl2l1 | 4.67E-05 | |
| 707.2 | 1243.1 | 1.8 | AU RNA binding protein/enoyl-coenzyme A hydratase | Mm.252034 | Auh | 3.2E-06 | |
| 1000.1 | 1751.7 | 1.8 | Aldehyde dehydrogenase 2, mitochondrial | Mm.284446 | Aldh2 | 3.56E-05 | |
| 2990.5 | 5228.6 | 1.7 | Glucosidase, beta, acid | Mm.5031 | Gba | < 1E-07 | |
| 942.5 | 1629.7 | 1.7 | AU RNA binding protein/enoyl-coenzyme A hydratase | Mm.252034 | Auh | < 1E-07 | |
| 7349 | 12562.5 | 1.7 | CCAAT/enhancer binding protein (C/EBP), beta | Mm.347406 | Cebpb | 1E-07 | |
| 752.1 | 1285.1 | 1.7 | Profilin 1 | Mm.2647 | Pfn1 | 3.5E-06 | |
| 912.2 | 1538.3 | 1.7 | Aldehyde dehydrogenase 2, mitochondrial | Mm.284446 | Aldh2 | 6.2E-05 | |
| 2311.2 | 3862.5 | 1.7 | Epidermal growth factor receptor pathway substrate 15 | Mm.318250 | Eps15 | 1.24E-05 | |
| 166.2 | 274.5 | 1.7 | Microphthalmia-associated transcription factor | Mm.333284 | Mitf | 7.9E-06 | |
| 722.3 | 1097.6 | 1.5 | Amyloid beta (A4) precursor protein-binding, family B, member 1 | Mm.38469 | Apbb1 | 7E-06 | |
| 749.6 | 1135 | 1.5 | Glutathione synthetase | Mm.252316 | Gss | 7.61E-05 | |
| Transcription factor | |||||||
| 294 | 803.5 | 2.7 | AT motif binding factor 1 | Mm.196564 | Atbf1 | < 1e-07 | |
| 247.9 | 639.4 | 2.6 | E74-like factor 3 | Mm.3963 | Elf3 | 6.80E-06 | |
| 844.6 | 1883 | 2.2 | Pre B-cell leukemia transcription factor 1 | Mm.43358 | Pbx1 | < 1E-07 | |
| 765.9 | 1554.6 | 2.0 | POU domain, class 2, transcription factor 2 | Mm.208700 | Pou2f2 | 2.30E-06 | |
| 7349 | 12562.5 | 1.7 | CCAAT/enhancer binding protein (C/EBP), beta | Mm.347406 | Cebpb | 1.00E-07 | |
| 162.2 | 252.2 | 1.6 | RIKEN cDNA 2310011G17 gene | Mm.254233 | 2310011G17 Rik | 6.55E-05 | |
| Signal transduction | |||||||
| 583.6 | 2799.1 | 4.8 | Megakaryocyte-associated tyrosine kinase | Mm.2918 | Matk | < 1E-07 | |
| 270.1 | 912.6 | 3.4 | Protein kinase C, zeta | Mm.28561 | Prkcz | < 1E-07 | |
| 1484.3 | 2441.1 | 1.6 | Casein kinase 1, epsilon | Mm.30199 | Csnk1e | 3.50E-06 | |
| 456.5 | 729.4 | 1.6 | Homeodomain interacting protein kinase 3 | Mm.257925 | Hipk3 | 2.11E-05 | |
| Development | |||||||
| 844.6 | 1883 | 2.2 | Pre B-cell leukemia transcription factor 1 | Mm.43358 | Pbx1 | < 1E-07 | |
| Higher in rapid-forming PCT (ABPC and ABLMYCPC) | |||||||
| Gene function | Rapid PCT | Slow PCT | Fold difference (rapid/slow) | Description | Unigene ID | Gene symbol | p-value |
| Angiogenesis | |||||||
| 1868.3 | 902.8 | 2.1 | Plasminogen activator, urokinase | Mm.4183 | Plau | 8.29E-05 | |
| 9821.9 | 4360.2 | 2.3 | CD53 antigen | Mm.316861 | Cd53 | 3.40E-06 | |
| Apoptosis | |||||||
| 2588.3 | 1681.2 | 1.5 | BCL2-antagonist/killer 1 | Mm.2443 | Bak1 | 9.11E-05 | |
| Cell cycle | |||||||
| 2231.6 | 1126.6 | 2.0 | Proliferation-associated 2G4 | Mm.4742 | Pa2g4 | 2.76E-05 | |
| 4858.7 | 2552.3 | 1.9 | Adenylate kinase 2 | Mm.29460 | Ak2 | < 1E-07 | |
| 10477 | 6082.1 | 1.7 | CDC28 protein kinase regulatory subunit 2 | Mm.222228 | Cks2 | < 1E-07 | |
| 1371.5 | 869.3 | 1.6 | Casein kinase II, alpha 2, polypeptide | Mm.51136 | Csnk2a2 | 1.90E-06 | |
| Immune response | |||||||
| 3568.1 | 386 | 9.2 | CD6 antigen | Mm.290897 | Cd6 | 2.00E-07 | |
| 984.7 | 186.1 | 5.3 | Tubulin, alpha 3 | Mm.287784 | Tuba3 | < 1E-07 | |
| 2661.9 | 718.2 | 3.7 | BH3 interacting domain death agonist | Mm.235081 | Bid | < 1E-07 | |
| 7492.6 | 2554.5 | 2.9 | Acetyl-Coenzyme A dehydrogenase, medium chain | Mm.10530 | Acadm | < 1E-07 | |
| 3195.6 | 1346.2 | 2.4 | Inositol (myo)-1(or 4)-monophosphatase 1 | Mm.183042 | Impa1 | < 1E-07 | |
| 2455 | 1064.3 | 2.3 | RAS-related protein-1a | Mm.333868 | Rap1a | < 1E-07 | |
| 9821.9 | 4360.2 | 2.3 | CD53 antigen | Mm.316861 | Cd53 | 3.40E-06 | |
| 7479.7 | 3365.9 | 2.2 | Transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) | Mm.207996 | Tap1 | < 1E-07 | |
| 7347.8 | 3535.9 | 2.1 | Superoxide dismutase 1, soluble | Mm.276325 | Sod1 | < 1E-07 | |
| 365.9 | 179.9 | 2.0 | Growth factor independent 1 | Mm.2078 | Gfi1 | 1.30E-05 | |
| 2231.6 | 1126.6 | 2.0 | Proliferation-associated 2G4 | Mm.4742 | Pa2g4 | 2.76E-05 | |
| 1939.9 | 985.5 | 2.0 | RAS-related protein-1a | Mm.333868 | Rap1a | 4.00E-07 | |
| 450.3 | 261.9 | 1.7 | Ets variant gene 4 (E1A enhancer binding protein, E1AF) | Mm.5025 | Etv4 | 1.40E-06 | |
| 3031.7 | 1830.6 | 1.7 | Wiskott-Aldrich syndrome homolog (human) | Mm.4735 | Was | 5.13E-06 | |
| 3136.7 | 1944.6 | 1.6 | Chromosome segregation 1-like (S. cerevisiae) | Mm.22417 | Cse1l | 2.67E-05 | |
| 5738.7 | 3586.7 | 1.6 | Survival motor neuron 1 | Mm.2025 | Smn1 | 2.75E-05 | |
| 395.2 | 252.8 | 1.6 | Dihydrofolate reductase | Mm.23695 | Dhfr | 2.70E-06 | |
| 2017.5 | 1297.8 | 1.6 | FK506 binding protein 12-rapamycin associated protein 1 | Mm.21158 | Frap1 | 8.46E-05 | |
| 2588.3 | 1681.2 | 1.5 | BCL2-antagonist/killer 1 | Mm.2443 | Bak1 | 9.11E-05 | |
| 4410.1 | 2873.4 | 1.5 | Ribonucleotide reductase M1 | Mm.197486 | Rrm1 | 5.14E-05 | |
| 5971.6 | 3940.2 | 1.5 | Integrin beta 4 binding protein | Mm.271674 | Itgb4bp | 6.94E-05 | |
| 18860.1 | 12494.5 | 1.5 | Triosephosphate isomerase 1 | Mm.4222 | Tpi1 | 8.30E-06 | |
| Transcription factor | |||||||
| 6231.6 | 1885.8 | 3.3 | Lymphoblastomic leukemia | Mm.4925 | Lyl1 | < 1E-07 | |
| 3274.2 | 1544.3 | 2.1 | Chromodomain helicase DNA binding protein 1 | Mm.8137 | Chd1 | < 1E-07 | |
| 450.3 | 261.9 | 1.7 | Ets variant gene 4 (E1A enhancer binding protein, E1AF) | Mm.5025 | Etv4 | 1.40E-06 | |
| 3136.7 | 1944.6 | 1.6 | Chromosome segregation 1-like (S. cerevisiae) | Mm.22417 | Cse1l | 2.67E-05 | |
| Signal transduction | |||||||
| 4858.7 | 2552.3 | 1.9 | Adenylate kinase 2 | Mm.29460 | Ak2 | < 1E-07 | |
| 10477 | 6082.1 | 1.7 | CDC28 protein kinase regulatory subunit 2 | Mm.222228 | Cks2 | < 1E-07 | |
| 3031.7 | 1830.6 | 1.7 | Wiskott-Aldrich syndrome homolog (human) | Mm.4735 | Was | 5.13E-05 | |
| 1371.5 | 869.3 | 1.6 | Casein kinase II, alpha 2, polypeptide | Mm.51136 | Csnk2a2 | 1.90E-06 | |
| DNA replication | |||||||
| 3805.2 | 2420.4 | 1.6 | Small nuclear ribonucleoprotein polypeptide A' | Mm.821 | Snrpa1 | < 1e-07 | |
| 8247.5 | 4325.9 | 1.9 | Replication protein A1 | Mm.180734 | Rpa1 | 4.20E-06 | |
Class comparison between slow-forming PCTs (TEPC, IL6PC and KiPC) and rapid-forming PCTs (ABPC and ABLMYCPC) by two-sample t-test generated 1195 genes showing significant differences (p- value < 0.001). Among these 1195 genes, the genes showing more than 1.5 fold increase and p-value < 0.0001 were selected in this table. The genes in each category of gene function were assigned based on the annotation of CGAP.
Effects of v-Abl
The gene expression levels of Socs1 and Socs2 (Suppressors of Cell Signaling 1 and 2) showed very high levels in all Abelson virus-infected samples (ABPC, ABLMYCPC) compared to other PCTs [see Additional file 2]. Socs 1 & 2 are proteins that normally interrupt Jak/Stat signaling [23], but their growth-inhibiting effects are abrogated by the v-Abl protein [24,25]. These high levels of Socs expression are likely to reflect the continuous activation of Jak-STAT expected from v-Abl [26]. This hyperactive Jak/STAT pathway may be partly responsible for the rapid appearance of ABPCs and ABLMYCPCs compared to TEPCs, as shown in Figure 1A.
Sensitivity of v-Abl accelerated PCTs to treatment with STI-571
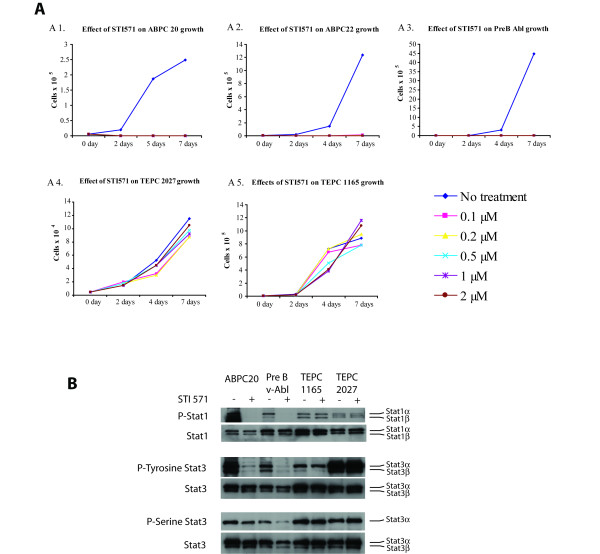
The main difference between ABPCs and TEPCs is the acceleration of plasma cell tumor formation by the v-Abl gene. The high levels of Socs1 &2 expression in ABPCs suggested that v-Abl was active in the fully mature PCTs. To confirm this finding and to determine whether fully transformed PCTs still retained a requirement for the v-Abl kinase that accelerated its appearance, cell survival analysis was performed after treatment of tissue culture lines of ABPC 20, ABPC 22, TEPC 1165, TEPC 2027 and pre B v-Abl lymphoma with 0.1 to 2.0 μM STI-571, an Abl kinase inhibitor [27]. Figure 3A shows a clear difference in the susceptibility of those cell lines to STI-571. ABPCs (ABPC20, ABPC22) and pre B cell lymphoma (pre B v-Abl) generated by Abelson virus infection showed higher susceptibility to STI-571 with complete inhibition of cell growth at 0.1 μM of STI-571 solution, while TEPC 1165 and TEPC 2027 showed no inhibition of cell growth even at 2 μM STI-571. Those data suggest that v-Abl is still required for cell survival and proliferation even in fully transformed ABPCs as was previously shown for v-Abl-induced pre B lymphomas [28].
Figure 3.
Greater sensitivity of ABPCs to STI-571 than TEPCs. A. Cell proliferation analysis of PCTs after treatment with STI-571. Five different cell lines including two ABPCs (ABPC20, ABPC22), two TEPCs (TEPC1165, TEPC 2027) and one pre B v-Abl lymphoma were treated with different concentrations of STI-571 at an initial concentration of 5 × 103 cells/well. Cell numbers were measured at 4 different times (0, 2, 4, and 7 days after treatment). B. Western blot analysis of PCTs for STAT1 phosphorylation and STAT3 phosphorylation. Samples for western blot were collected from untreated cells and cells after treatment with 5 μM STI-571 for 18 hours. Doublet bands are the α and β forms of the STAT proteins.
STI-571 inhibits Stat activation of v-Abl accelerated PCT and pre B lymphoma
The preceding analysis suggests that the v-Abl-induced PCTs utilize the Jak/STAT signaling pathways. As shown in Figure 3B, the PCT cell lines studied showed highly activated (phosphorylated) STAT1 and STAT3. However treatment with 5 mM STI-571 led to decreased phosphorylation of STAT1 and STAT3, but only in the cell lines induced with Abelson virus, ABPC 20 cell line and pre B v-Abl lymphoma. This dose of STI-571 had no effect on phosphorylation of STAT1 or STAT3 in TEPC 1165 and TEPC 2027. Presumably the latter PCTs also utilized the Jak/Stat signaling pathways, but they were not stimulated by Abl. What is more, 5 μM STI-571 completely inhibited the STAT1 phosphorylation in ABPC 20 and pre B v-Abl lymphoma and the STAT3 tyrosine phosphorylation in pre B v-Abl, and it virtually completely inhibited the tyrosine and serine phosphorylation of STAT3 in ABPC 20. Signals activating STAT3 can also come from other different pathways (e.g., those responsible for STAT phosphorylation in TEPC 1165 and TEPC 2027, such as Ras activation by c-Myc). In contrast, STAT3 activation of the pre B v-Abl line comes almost exclusively from v-Abl, as was shown in v-Abl-transformed pre B-cells [24]. STI-571's selective inhibition of Stat activation in ABPC 20 and pre B v-Abl lymphoma suggests that the v-Abl signaling pathways are still critical for viability, even in fully transformed tissue-cultured tumors.
Quantitative RT-PCR analysis
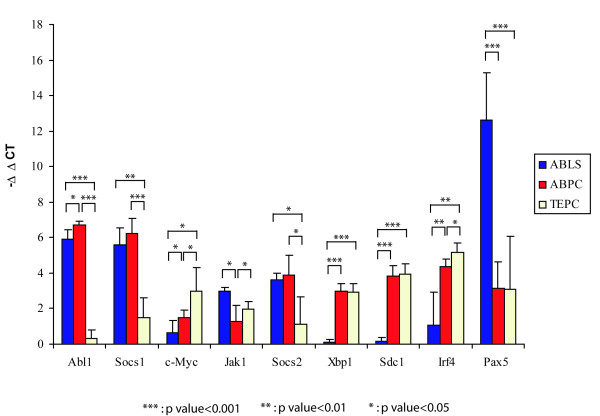
For validation of certain key findings in the microarray data, real time RT-PCR was performed for 9 selected genes that may play significant roles in PCTs (Figure 4). Thirteen different tumor RNA samples (6 ABPC, 4 TEPC and 3 ABLS) were analyzed, using RNA from the sample that showed the lowest expression as calibrator for each gene. Means for each of the three tumor types are shown in Figure 4, and individual results are presented in Additional file 3. These results paralleled the mRNA expression levels acquired via microarray analysis. c-Myc expression was generally highest in the four TEPC samples, while it showed lowest expression in the v-Abl-expressing ABLS (pre-B cell lymphomas). Socs1 and Socs2 expression was higher in ABPC and ABLS than in TEPC samples. The genes previously known to be plasma cell-specific transcription factors (Xbp1 and Irf4) and surface marker (Sdc1/CD138) showed higher expression in PCTs than in pre-B cell lymphomas (ABLS). Jak1 expression was higher in the pre-B lymphomas than in the PCTs.
Figure 4.
Quantitative real-time PCR analysis. qRT-PCR was performed on RNAs from 3 different groups of cultured cells to analyze relative mRNA content for 9 key genes: c-Myc, Socs1, Socs 2, Abl1, Jak1, Xbp1, Sdc1, Irf4, Pax5 as well as the "housekeeping gene", GAPDH, using primers and detection oligos designed in consultation with the manufacturer (ABI). The data was presented as mean ± standard deviation values of each group. qRT-PCR data for individual samples are presented as Additional file 3. All data were normalized to GAPDH. These values were then calibrated against the sample with the lowest expression and represented as -ΔΔCt values with standard errors.
Differences of gene expressions based on c-Myc gene chromosomal translocations
Cluster analysis suggested that the site of chromosomal translocation on chromosome 15 and the nature of its translocation partner chromosome did not affect the gene expression profile as much as additional oncogenes play in PCT induction. To test this concept more stringently, a two-sample t-test was used to search for differences in gene expression between typical T(12;15) and variant T(6;15) chromosomal translocations within a single subgroup of PCTs, ABPCs. Only 14 genes, most of which had low levels of expression, showed significant difference in expression, suggesting that the site of translocation had little effect on the mechanism of transformation of the PCTs [see Additional file 4]. Another t-test was performed to compare the effect of two different forms of T(12;15), class I or II, in TEPCs [see Additional file 4]. This showed 29 genes with significant differences in expression, also relatively low in expression, consistent with the previous comparison and our interpretation that the different forms of chromosomal translocation did not affect significant differences in mechanism of transformations or the type of cell being transformed. This analysis cannot rule out some differences in early neoplastic events, since we were only examining expression values from fully transformed tumors.
Meta-analysis of the mouse expression data co-mingled with expression pattern data for human plasma cell neoplasms
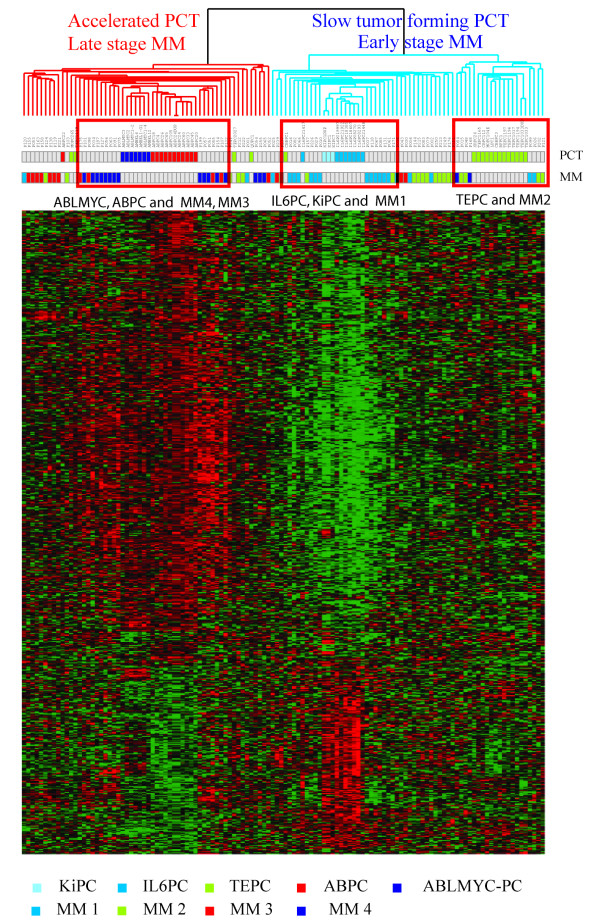
Having defined distinctive gene expression patterns among the PCT subtypes as well as between PCTs and BCLs, we next sought to examine how well our mouse models recapitulate human phenotypes as measured by relative similarities of gene expression patterns. Mouse plasmacytomas form in the intraperitoneal cavity, while human multiple myelomas are found in bone marrow, very different locations but not without potentially meaningful similarities: richness in adipocytes, plentiful reactive myeloid cells, abundant blood supply, among others. We used gene expression data from human B lymphocytes and human multiple myeloma isolates, and cross-compared them with the gene expression data from our models. Using genes that existed as orthologues in both human and mouse Affymetrix chips, unsupervised cluster analysis showed that BCLs (human and mouse) clustered separately from the PCTs and MMs [see Additional file 5]. Since the B-cell components in this series were small in number, we focused our analysis on the plasma cell components, human and murine. The human multiple myeloma samples had been previously assigned to MM1-MM4 subgroups, based on cluster analysis of gene expression patterns of bone-marrow plasma cells from patients with multiple myeloma [18,19]. When PCTs and MMs were re-analyzed without the B-cell components, there was co-stratification of mouse and human tumors in such a way that the more aggressive MM3 and MM4 co-clustered with the accelerated PCT-1 and -2 (ABLMYCPCs and ABPCs), while MM1 co-clustered with IL-6 transgenic PCTs (KiPCs and IL6PCs) (Figure 5). The slow-arising TEPCs (PCT-6) co-clustered with the less aggressive MM1-MM2 patients. This intriguing inter-species co-clustering of plasma cell neoplasms will be investigated in greater depth in another study that will incorporate more recent myeloma data [29] that present a new molecular classification of myelomas into seven subgroups.
Figure 5.
Meta-analysis of human multiple myeloma samples with mouse plasmacytomas. A total of 122 samples, comprised of 74 human samples and 48 mouse samples, were used for this meta-analysis. Human samples had previously classified into 4 different stages of multiple myeloma (MM1, MM2, MM3 and MM4, generally reflecting increasing severity or stage of disease [18]), and mouse samples were composed of 5 different PCT groups, ABPC and ABLMYCPC, "two accelerated subgroups [PCT-1 and -2] (Table 1)", and "TEPC, KiPC and IL6PC, the slower appearing subgroups [PCTs 4 – 6] (Table 1)". The stage of myeloma and group of plasma cell tumors to which each sample belongs is indicated by the color of the rows of boxes beneath sample names, with a color key beneath the heat map. The human and mouse data were normalized and standardized separately and then combined for analysis [40,41]. The genes for cluster analysis were selected by one-way ANOVA analysis (2266 genes, p-value < 0.001) of mouse PCTs, and 634 orthologs from these 2266 genes, present both in human and mouse array platforms were used for cluster analysis.
Discussion
Microarray expression profiling has been quite successful in differentiating subclasses within tumors with a similar histological/morphological diagnosis [18,30]. Tumors with common genetic lesions tend to cluster together, reflecting the fact that deregulation of one or more genes leads to downstream gene activation/inactivation cascades that are unique to that lesion. It has also made it possible to predict the prognosis of cancer patients and the likelihood of their responding to chemotherapy, based on gene expression patterns [18,31]. Major progress in experimental cancer research will require animal model systems and in vitro cell culture systems. Experimentally induced PCTs and related BCLs have long been, and continue to be, useful for the study of plasma cell neoplasms.
In this study, we generated gene expression profiles of mouse B-lymphocytic neoplasms, including four groups of PCTs that arose with different latent periods in chronic inflammatory tissue induced by ip pristane. We also studied two subgroups of PCTs that arose spontaneously in hyperplastic lymph nodes of IL-6-transgenic mice and B-cell lymphomas from iMyc gene-insertion mice. Statistical methods were employed to compare these expression profiles to search for specific gene expression signatures that might aid in understanding the molecular mechanisms at work in each subtype of B cell malignancy and the differences in tumor latent periods resulting from variation in the PCT induction schemes.
Classical unsupervised clustering analysis and statistical analysis of microarray data revealed a tight clustering of PCTs regardless of subtype. The expression patterns in this group showed major differences from those of the B-cell lymphoma group. As seen in earlier analyses [30], the stage of B cell differentiation strongly influenced the major gene expression differences. It is important to note that virtually all of the PCTs clustered together, including both intraperitoneal oil-induced PCTs as well as those that arose spontaneously in IL-6-transgenic lymph nodes, independent of ip pristane. These results indicated that all PCTs share a common genetic expression signature. It can also be seen that many genes present in the B-cell signature have been downregulated in the PCT signature, a well-known feature of the specialized plasma cell, which seems to reflect this cell's single-minded dedication to maximize synthesis of antibody Ig.
Many of the genes that emerged as part of the PCT signature after unsupervised clustering analysis had been recognized in previous studies of gene expression patterns as characteristic of this stage of B cell differentiation, before and after neoplastic transformation [17,19,32]. Irf4 and Syndecan1 are characteristically expressed in plasma cells, and cyclin D2 has been reported to have high expression in PCT, presumably secondary to deregulated c-Myc.
Subclusters within the PCT cluster were seen, chiefly reflecting the slow vs. rapid appearance of the PCTs and the nature of the accelerating oncogenes. Unlike human myelomas, mouse plasma cell tumors have a common initiating event, a Myc-activating chromosome translocation. We found that unsupervised clustering of these tumors did not reveal major differences in gene expression that could be associated with position of breakpoint and translocation on chromosome 15. That is, the nature of c-Myc-activating chromosomal translocations [i.e., typical T(12;15) vs. variant T(6;15)] did not affect their inclusion in the PCT group, nor did it lead to subclusters within the broad PCT grouping or the ABPC or TEPC subgroups, in this particular analysis. This supports our hypothesis that even T(6;15) translocations that occur within the Pvt-1 region initiate PCTs via induction of constitutive c-Myc expression, and that fine structural differences in the site of T(12;15) within or near the c-Myc locus itself do not materially affect the way Myc influences PCT induction. Of course, we know that additional genetic events are required for complete transformation of plasma cells, for Myc-activating chromosome translocations have been found in normal cells and organs that did not develop plasma cell tumors [33]. Future studies will focus on identifying these additional events that occur during tumor progression.
Our cluster analysis did, however, reflect differences in the cooperating genes used to accelerate tumorigenesis. The most striking gene expression changes associated with tumors accelerated by Abelson virus infection are the high levels of Socs1 expression. The Socs proteins are inhibitors of signaling pathways and are generally expressed at low levels in unstimulated cells. They become rapidly induced by cytokines, thereby inhibiting Jak-STAT signaling, forming a classic negative feedback loop [34]. It was previously shown that the v-Abl oncogene activates Jak-STAT signaling during transformation of pre-B cells in mice [24]. Although Socs-1 is highly expressed in v-Abl-transformed cells, it is unable to inhibit v-Abl-mediated Jak- STAT signaling. This is thought to be the result of one of the non-tyrosine kinase effects of v-Abl, such as phosphorylation of Socs-1 on non-tyrosine residues, leading to disruption of the interaction between Socs-1 and the elongin B/C complex and inhibition of Socs1-mediated proteasome targeting of activated Jaks [24]. STI-571/Imatinib (Gleevec, Novartis Pharma), at low concentrations, is a specific inhibitor of Abl kinase and shows effectiveness in the treatment of chronic myelogenous leukemia [27,35]. ABPC20, ABPC22 and pre B v-Abl are much more sensitive to killing by STI-571 than TEPC1165 or TEPC2027 (Figure 3A), indicating that the Abl kinase activity is still required for viability of PCTs induced with the assistance of Abelson Virus. Since nearly all ABPCs also show c-Myc-activating chromosome translocations, some cooperation between the signaling pathways of v-Abl and c-Myc must be responsible for the rapid transformation of ABPCs. Unexpectedly, however, this cooperation appears to be required even after full transformation is achieved and even after adaptation to growth in culture. It will be interesting to see if resistance to STI-571 develops in these cultures, as is often seen in human CML [31].
Stat4 is more highly expressed in the BCL group than in PCTs. However, the level of stat3 expression does not differ significantly between BCLs and PCTs, both groups showing relatively high expression levels. Jak1 also showed higher expression in the BCL group compared to PCTs, but Jak1 is relatively highly expressed, even in PCTs. The accelerating mechanisms engaged after v-Abl infection seems to utilize these pathways (Figure 3B), despite the concomitant induction of the counteracting Socs family of genes. These pathways are currently being studied in greater depth at the translational and post-translational levels within the PCT system, following up the leads afforded by our gene expression studies and the initial phosphorylation studies shown here, with the goals of understanding the mechanisms at work.
It has been illuminating to analyze our mouse expression data in conjunction with already published Affymetrix data from human multiple myeloma. Cluster analysis showed that human MM1 clustered most closely with PCT4 and PCT5, IL6PC and KiPC, the two groups of PCTs from IL-6-transgenic mice, while the more aggressive myeloma groups, MM3-MM4, clustered more tightly with PCT1 and PCT2, ABLMYCPC and ABPC, those with appearance accelerated by v-Abl activity. This similarity includes differences in expression of genes associated with proliferation. This was unexpected but significant, because plasma cell neoplasms are not generally associated with rapid proliferation. Instead, increased survival or escape from apoptosis is thought to be the chief mechanism responsible for the expansion of lymphocytes or plasma cells in lymph nodes or bone marrow, respectively. This similarity brings to mind the possibility that Imatinib, the activated Abl inhibitor, might be effective in treating aggressive myeloma patients.
This co-clustering suggests that different pathways can be utilized to achieve a similar outcome, namely transformation of plasma cells. Thus, the mouse PCT model, despite its biological differences from MM, offers an experimental model for studying the details of the etiology of plasma cell neoplasms with different degrees of aggressiveness, much as seen in human myelomas. This aspect of our study will be broadened to include new data on additional myeloma patients [29] in which expression data are used to define seven subgroups that differ in their molecular characteristics. This study will be the subject of a separate manuscript.
Conclusion
Lymphoid transformation and plasma cell tumor formation are complicated, multi-stage processes, so it is necessary to study these processes prospectively using research tools covering genome-wide changes in expression. The present study shows that gene expression profiling can differentiate B-cell lymphomas from plasma cell tumors and also distinguish slow from accelerated plasma cell tumors. These results and data obtained from the sensitivity of v-Abl-accelerated plasma cell tumors and their phosphorylated STAT proteins to the effects of STI-571 indicate that these otherwise similar tumors utilize different signaling pathways but share a common initiating genetic lesion, a c-Myc-activating chromosome translocation. This study of gene expression profiles of mouse B-cell lymphomas and several subclasses of plasma cell tumors provides data that offer clues for the understanding of B-cell neoplasia and plasma cell tumor formation and the interpretation of the prospective plasma cell tumor induction studies that are now under way.
Methods
Sample selection and RNA preparation
A total of 70 samples of RNA were prepared from transplanted mouse tissues. All solid PCT samples (except IL6PC) used for microarray hybridization had been transplanted at least once from the initial ip tumor tissue that arose following pristane injection. As summarized in Table 1, the four groups of BCLs and the six different PCT subtypes originated as follows. TEPCs (PCT-6) were obtained after ip injections of pristane in BALB/c mice [1]. ABPCs (PCT-2) and J3PCs (PCT-3) were obtained more rapidly (Figure 1A) by introducing Abelson virus [2] or J3V1 virus [3], retroviruses containing v-Abl, or v-Raf-1 and v-Myc genes, respectively, following the injection of pristane in BALB/c mice. ABLMYCPCs (PCT-1) originated even more rapidly in the pristane-conditioned peritoneum of BALB/c mice infected with ABLMYC virus [5], a retrovirus that expresses both v-Abl and c-Myc genes. IL6PCs (PCT-5) were obtained from hyperplastic lymph nodes that spontaneously developed in IL-6-transgenic BALB/c mice [8]. KiPCs (PCT-4) were generated by injecting pristane intraperitoneally into IL-6-transgenic BALB/c mice [7]. "Pre-malignant" IL6LN (BCL-1) were hyperplastic lymph nodes of IL-6-transgenic BALB/c mice [13]. ABLS (a.k.a. preB vAbl and BCL-2) were pre-B lymphomas that arose rapidly in lymphoid organs of Abelson virus-treated BALB/c mice [2]. BCLEμ (BCLs-3) are lymphoblastic B-cell lymphomas with a Burkitt-like morphology from lymph nodes of transgenic iMycEm mice [9]. BCLCα (BCL-4) were obtained from spleens of transgenic iMycCα mice [10]. All mice were maintained in our conventional colony on the NIH campus under Animal Study Protocol LG-028. Total RNA was prepared by grinding pieces of excised, snap frozen and otherwise unprocessed tissues in liquid nitrogen and extraction in TRIzol (Invitrogen, Carlsbad, CA) followed by further purification on RNAeasy columns (Qiagen, Inc., Valencia, CA).
Microarray hybridization
Affymetrix Murine Genome Set U74Av2 microarrays (Affymetrix Inc., West Sacramento, CA) were used for the hybridization of biotin-labeled cDNA probes synthesized from 5 μg of total RNA or 1 μg poly(A)+ RNA using a Superscript double-strand cDNA synthesis kit (Invitrogen, Carlsbad, CA), Bioarray High Efficiency RNA transcript labeling kit and Mg-catalyzed fragmentation kit (Enzo Biochemicals Inc., Farmingdale, NY) according to the manufacturers' instructions. Microarrays were stained with phycoerythrin-streptavidin (Molecular Probes, Carlsbad, CA), scanned with an Affymetrix GeneChip scanner and analyzed with Affymetrix Microarray Analysis Suite (MAS) version 5.0.
Statistical analysis of microarray data
BRB ArrayTools Version 3.0 [36] was used for the analysis of the MAS 5.0 data set. A log base 2 transformation was applied to the data set before arrays were normalized. Each array was normalized using median values of gene expression over the entire array (global normalization). A median array was selected as the reference array for normalization. Class comparison analysis was performed with the two-sample t-test of BRB ArrayTools with the estimation of false discovery rate. Cluster analysis was performed with Cluster and Treeview [37]. For the cluster analysis, the log base 2 transformed data were centered to the mean values of each gene's expression. Significantly enriched components defined in Gene Ontology (GO) were determined based on hypergeometric distribution analysis [38].
Meta-analysis of published data
The human multiple myeloma data and their classification into subclasses MM1 – MM4 was previously reported by Zhan et al. [18,19] using Affymetrix HuGeneFL Genome Array chips (Affymetrix, Inc., West Sacramento, CA). 3739 orthologues between the human FL microarray and the mouse U74Av2 microarray were identified based on curated Hu6800 orthology from Affymetrix [39]. Each data set was normalized separately and then combined together as reported earlier [40]. Before integrating the human data set with the mouse data set, the expression of each gene was standardized to a mean ± s.d. of 0 ± 1, independently in both data sets as described [41].
Cell culture, proliferation assay and inhibition by STI-571
This laboratory has adapted a number of PCTs, including TEPC 1165, TEPC 2027, ABPC22 and ABPC 20 studied here, to in vitro growth in RPMI 1640 plus 10% fetal bovine serum, 10 ng/ml IL-6, penicillin and streptomycin. Inhibition of proliferation by STI-571/Imatinib/Gleevec (Novartis, Basel, Switzerland) [27,35] was determined using several different concentrations of inhibitor (from 0.1 μM to 2 μM) on an initial cell suspension of 5 × 103 cells/well of a 24-well plate. Cell proliferation was determined by counting the cells at 2 days, 4 days and 7 days after treatment with STI-571.
Western blotting
Tissue cultured mouse PCT cells, with and without 5 μM STI-571, were harvested after 18 hours of treatment and were subjected to western blot analysis. Membranes were probed with the following antibodies: STAT1, STAT3, tyrosine 701-phosphorylated STAT1, tyrosine 705-phosphorylated STAT3 and serine 727-phosphorylated STAT3 (Cell Signaling, Beverly, MA).
Quantitative real-time PCR
Quantitative RT-PCR was performed using the ABI 7500 real time PCR system following the manufacturer's protocol. Mouse GAPDH primer and probes were used as the normalization control in quantitative analysis. The primers and probes used in this study were purchased from Applied Biosystems (Foster City, CA), and primer sequences are available upon request. The relative mRNA expression levels are expressed as -ΔΔCt, in which ΔCt is the difference between the threshold PCR cycle (Ct) value of each experimental mRNA and that of the corresponding internal control, GAPDH. The -ΔΔCt value is the difference between the ΔCt value of each tissue and that of the lowest-expressing sample, used as calibrator for that gene [42].
Authors' contributions
JFM and JDS conceived the project. JFM and JDO isolated the RNAs. ESP analyzed the data and wrote the manuscript. J-SL and HGW contributed additional statistical analyses. MP and SJ provided samples and interpretation. SG, HW, and FZ performed experiments, and JFM co-authored the manuscript and was responsible for project development. All authors read and approved the manuscript.
Supplementary Material
Supplementary Tables 1. Functional enrichment analysis of genes that showed significant differences in expression between plasma cell tumors and B-cell lymphomas. Supplementary Table 1A. Functional enrichment analysis according to GO Molecular Function category. Supplementary Table 1B. Functional enrichment analysis by GO Cellular Component category. Supplementary Table 1C. Functional enrichment analysis by GO Biological Process category. Supplementary Table 1D. Genes present in nine GO Molecular Function categories that were shown to be functionally enriched (see Supplementary Table 1A, above). Four tables showing lists of genes that showed significant (p < 0.005) differences in expression between plasma cell tumors and B-cell lymphomas characterized by their functional enrichment according to biological processes defined by the Gene Ontology Consortium. The enrichment of gene set was estimated by calculating the cumulated hypergeometric p values of the each biological process defined by the Gene Ontology Consortium http://www.geneontology.org. In Table 1D, 9 components from the Molecular Function GO category were selected to show the actual genes involved.
Supplementary Tables 2. Genes that showed significant (p < 0.001) differences in expression between rapid-forming plasma cell tumors (ABPC and ABLMYCPC) and slow-forming plasma cell tumors (TEPC, IL6PC and KiPC). Supplementary Table 2A. Genes that showed more than 2-fold higher expression in rapid-forming plasma cell tumors than in slow-forming plasma cell tumors. Supplementary Table 2B. Genes that showed more than 2-fold higher expression in slow-forming plasma cell tumors than in rapid-forming plasma cell tumors. Two tables showing lists of genes that showed significant (p < 0.001) differences in expression between rapid- and slow-forming plasma cell tumors.
Supplementary Figure 1. Quantitative RT-PCR of relative mRNA content in 13 mouse B-cell lymphomas and plasma cell tumors for 9 key genes. Results of quantitative RT-PCR validation of relative mRNA content in 13 mouse B-cell lymphomas and plasma cell tumors for 9 key genes: c-Myc, Socs1, Socs2, Abl1, Jak1, Xbp1, Sdc1, Irf4, Pax5 (BSAP) and the "housekeeping" gene, GAPDH.
Supplementary Tables 3. Genes with significant (p < 0.001) differences in gene expression associated with site of chromosomal translocation in two sample t-tests. Supplementary Table 3A. Genes that showed significant differences in expression between ABPCs with T(12;15) and ABPCs with T(6;15). Supplementary Table 3B. Genes that showed significant differences in expression between TEPCs with T(12;15 class I) and TEPC T(12;15 class II) chromosome translocations. Two tables showing lists of genes that showed significant (p < 0.001) differences in expression between similar subclasses of plasma cell tumors that differ in site of Myc-activating chromosomal translocation.
Supplementary Figure 2. Meta-analysis of global gene expression of 67 mouse samples (including 5 different subgroups of plasma cell tumors and B-cell lymphomas) combined with 123 human samples (including 5 different subgroups of multiple myelomas and tonsilar B cells). Heat map derived from a meta-analysis of a combination of expression data from human and mouse plasma cell dyscrasias.
Acknowledgments
Acknowledgements
This research was supported in part by the Intramural Research Program of the NIH and the National Cancer Institute, and we thank Gary Jones for excellent technical assistance. JDS and FZ were supported by National Institutes of Health grants CA55819 and CA97513, the Fund to Cure Myeloma and the Peninsula Community Foundation. We thank Novartis Pharma for the gift of SKI-571.
Contributor Information
Eun Sung Park, Email: espark@mdanderson.org.
John D Shaughnessy, Jr, Email: shaughnessyjohn@uams.edu.
Shalu Gupta, Email: guptashal@mail.nih.gov.
Hongyang Wang, Email: hw4f@virginia.edu.
Ju-Seog Lee, Email: jlee@mdanderson.org.
Hyun Goo Woo, Email: woohy@mail.nih.gov.
Fenghuang Zhan, Email: ZhanFenghuang@uams.edu.
James D Owens, Jr, Email: jow@helix.nih.gov.
Michael Potter, Email: potterm@mail.nih.gov.
Siegfried Janz, Email: siegfried-janz@uiowa.edu.
J Frederic Mushinski, Email: jm8p@nih.gov.
References
- Potter M. Neoplastic development in plasma cells. Immunological Reviews. 2003;194:177–195. doi: 10.1034/j.1600-065X.2003.00061.x. [DOI] [PubMed] [Google Scholar]
- Potter M, Sklar MD, Rowe WP. Rapid viral induction of plasmacytomas in pristane primed BALB/c mice. Science. 1973;182:592–594. doi: 10.1126/science.182.4112.592. [DOI] [PubMed] [Google Scholar]
- Troppmair J, Potter M, Wax JS, Rapp UR. An altered v-raf is required in addition to v-myc in J3V1 virus for acceleration of murine plasmacytomagenesis. Proc Natl Acad Sci USA. 1989;86:9941–9945. doi: 10.1073/pnas.86.24.9941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clynes R, Wax J, Stanton LW, Smith-Gill S, Potter M, Marcu KB. Rapid induction of IgM-secreting murine plasmacytomas by pristane and an immunoglobulin heavy-chain promoter/enhancer-driven c-myc/v-Ha-ras retrovirus. Proc Natl Acad Sci USA. 1988;85:6067–6071. doi: 10.1073/pnas.85.16.6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissinger EM, Mischak H, Largaespada DA, Kaehler DA, Mitchell T, Smith-Gill SJ, Risser R, Mushinski JF. Induction of plasmacytomas secreting antigen-specific monoclonal antibodies with a retrovirus expressing v-abl and c-myc. Proc Natl Acad Sci USA. 1991;88:8735–8739. doi: 10.1073/pnas.88.19.8735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SS, Shaffer AL, Kim JS, duBois W, Potter M, Potter M, Staudt LM, Janz S. Insertion of Myc into Igh accelerates peritoneal plasmacytomas in mice. Cancer Res. 2005;65:7644–7652. doi: 10.1158/0008-5472.CAN-05-1222. [DOI] [PubMed] [Google Scholar]
- Kovalchuk AL, Janz S. Switch-mediated CH deletions are a recurrent feature of Myc/CH translocations in peritoneal plasmacytomas in mice. Int J Cancer. 2002;101:423–426. doi: 10.1002/ijc.10638. [DOI] [PubMed] [Google Scholar]
- Kovalchuk AL, Kim JS, Park SS, Coleman AE, Ward JM, Morse HC, Kishimoto T, 3rd, Potter M, Janz S. IL-6 transgenic mouse model for extraosseous plasmacytoma. Proc Natl Acad Sci USA. 2002;99:1509–1514. doi: 10.1073/pnas.022643999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SS, Kim JS, Tessarollo L, Owens JD, Peng L, Han SS, Tae Chung S, Torrey TA, Cheung WC, Polakiewicz RD, McNeil N, Ried T, Mushinski JF, Morse HC, 3rd, Janz S. Insertion of c-Myc into Igh induces B-cell and plasma-cell neoplasms in mice. Cancer Res. 2005;65:1306–1315. doi: 10.1158/0008-5472.CAN-04-0268. [DOI] [PubMed] [Google Scholar]
- Cheung WC, Kim JS, Linden M, Peng L, Van Ness B, Polakiewicz RD, Janz S. Novel targeted deregulation of c-Myc cooperates with Bcl-X(L) to cause plasma cell neoplasms in mice. J Clin Invest. 2004;113:1763–1773. doi: 10.1172/JCI200420369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cory S. Activation of cellular oncogenes in hemopoietic cells by chromosome translocation. Adv Cancer Res. 1986;47:189–234. doi: 10.1016/s0065-230x(08)60200-6. Review. [DOI] [PubMed] [Google Scholar]
- Potter M, Wiener F. Plasmacytomagenesis in mice: model of neoplastic development upon chromosomal translocations. Carcinogenesis. 1992;13:1681–1697. doi: 10.1093/carcin/13.10.1681. [DOI] [PubMed] [Google Scholar]
- Kovalchuk AL, Kishimoto T, Janz S. Lymph nodes and Peyer's patches of IL-6 transgenic BALB/c mice harbor T(12,15) translocated plasma cells that contain illegitimate exchanges between the immunoglobulin heavy-chain mu locus and c-myc. Leukemia. 2000;14:1127–1135. doi: 10.1038/sj.leu.2401767. [DOI] [PubMed] [Google Scholar]
- Suematsu S, Matsuda T, Aozasa K, Akira S, Nakano N, Ohno S, Miyazaki J, Yamamura K, Hirano T, Kishimoto T. IgG1 plasmacytosis in interleukin 6 transgenic mice. Proc Natl Acad Sci USA. 1989;86:7547–7551. doi: 10.1073/pnas.86.19.7547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calame KL. Plasma cells: finding new light at the end of B cell development. Nature Immunology. 2001;2:1103–1108. doi: 10.1038/ni1201-1103. Review. [DOI] [PubMed] [Google Scholar]
- Lin K-I, Tunyaplin C, Calame K. Transcriptional regulatory cascades controlling plasma cell differentiation. Immunology Reviews. 2003;194:19–28. doi: 10.1034/j.1600-065X.2003.00040.x. Review. [DOI] [PubMed] [Google Scholar]
- Underhill GH, George D, Bremer EG, Kansas GS. Gene expression profiling reveals a highly specialized genetic program of plasma cells. Blood. 2003;101:4013–4021. doi: 10.1182/blood-2002-08-2673. [DOI] [PubMed] [Google Scholar]
- Zhan F, Hardin J, Kordsmeier B, Bumm K, Zheng M, Tian E, Sanderson R, Yang Y, Wilson C, Zangari M, Anaissie E, Morris C, Muwalla F, van Rhee F, Fassas A, Crowley J, Tricot G, Barlogie B, Shaughnessy J., Jr Global gene expression profiling of multiple myeloma, monoclonal gammopathy of undetermined significance, and normal bone marrow plasma cells. Blood. 2002;99:1745–1757. doi: 10.1182/blood.V99.5.1745. [DOI] [PubMed] [Google Scholar]
- Zhan F, Tian E, Bumm K, Smith R, Barlogie B, Shaughnessy J., Jr Gene expression profiling of human plasma cell differentiation and classification of multiple myeloma based on similarities to distinct stages of late-stage B-cell development. Blood. 2003;101:1128–1140. doi: 10.1182/blood-2002-06-1737. [DOI] [PubMed] [Google Scholar]
- Tarte K, Zhan F, De Vos J, Klein B, Shaughnessy J., Jr Gene expression profiling of plasma cells and plasmablasts: toward a better understanding of the late stages of B-cell differentiation. Blood. 2003;102:592–600. doi: 10.1182/blood-2002-10-3161. [DOI] [PubMed] [Google Scholar]
- Mai S, Hanley-Hyde J, Rainey GJ, Kuschak TI, Paul JT, Littlewood TD, Mischak H, Stevens LM, Henderson DW, Mushinski JF. Chromosomal and extrachromosomal instability of the cyclin D2 gene is induced by Myc overexpression. Neoplasia. 1999;1:241–52. doi: 10.1038/sj.neo.7900030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colucci F, Schweighoffer E, Tomasello E, Turner M, Ortaldo JR, Vivier E, Tybulewicz VL, Di Santo JP. Natural cytotoxicity uncoupled from the Syk and ZAP-70 intracellular kinases. Nat Immunol. 2002;3:288–294. doi: 10.1038/ni764. [DOI] [PubMed] [Google Scholar]
- Galm O, Yoshikawa H, Esteller M, Osieka R, Herman JG. SOCS-1, a negative regulator of cytokine signaling, is frequently silenced by methylation in multiple myeloma. Blood. 2003;101:2784–2788. doi: 10.1182/blood-2002-06-1735. [DOI] [PubMed] [Google Scholar]
- Danial NN, Rothman P. JAK-STAT signaling activated by Abl oncogenes. Oncogene. 2000;19:2523–2531. doi: 10.1038/sj.onc.1203484. Review. [DOI] [PubMed] [Google Scholar]
- Limnander A, Danial NN, Rothman PB. v-Abl signaling disrupts SOCS-1 function in transformed pre-B cells. Mol Cell. 2004;15:329–341. doi: 10.1016/j.molcel.2004.06.041. [DOI] [PubMed] [Google Scholar]
- Limnander A, Rothman PB. Abl oncogene bypasses normal regulation of Jak/STAT activation. Cell Cycle. 2004;3:1486–1488. doi: 10.4161/cc.3.12.1297. [DOI] [PubMed] [Google Scholar]
- Druker BJ, Tamura S, Buchdunger E, Ohno S, Segal GM, Fanning S, Zimmermann J, Lydon NB. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nat Med. 1996;2:561–566. doi: 10.1038/nm0596-561. [DOI] [PubMed] [Google Scholar]
- Engelman A, Rosenberg N. bcr/abl and src but not myc and ras replace v-abl in Lymphoid Transformation. Mol Cell Biol. 1990;10:4365–4369. doi: 10.1128/mcb.10.8.4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan F, Huang Y, Colla S, Stewart JP, Hanamura I, Gupta S, Epstein J, Yaccoby S, Sawyer J, Burington B, Anaissie E, Hollmig K, Pineda-Roman M, Tricot G, van Rhee F, Walker R, Zangari M, Crowley J, Barlogie B, Shaughnessy JD., Jr The molecular classification of multiple myeloma. Blood. 2006;108:2020–2028. doi: 10.1182/blood-2005-11-013458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RE, Staudt LM. Molecular diagnosis of lymphoid malignancies by gene expression profiling. Curr Opin Hematol. 2002;9:333–338. doi: 10.1097/00062752-200207000-00011. Review. [DOI] [PubMed] [Google Scholar]
- Villuendas R, Steegmann JL, Pollán M, Tracey L, Granda A, Fernández-Ruiz E, Casado LF, Martínez J, Martínez P, Lombardía L, Villalón L, Odriozola J, Piris MA. Analysis of genes involved in imatinib resistance in CML: a gene-expression profiling approach. Leukemia. 2006;20:1047–1054. doi: 10.1038/sj.leu.2404197. [DOI] [PubMed] [Google Scholar]
- Claudio JO, Masih-Khan E, Tang H, Gonçalves J, Voralia M, Li ZH, Nadeem V, Cukerman E, Francisco-Pabalan O, Liew CC, Woodgett JR, Stewart AK. A molecular compendium of genes expressed in multiple myeloma. Blood. 2002;100:2175–2186. doi: 10.1182/blood-2002-01-0008. [DOI] [PubMed] [Google Scholar]
- Janz S, Muller J, Shaughnessy J, Potter M. Detection of recombinations between c-myc and immunoglobulin switch alpha in murine plasma cell tumors and preneoplastic lesions by polymerase chain reaction. Proc Natl Acad Sci USA. 1993;90:7361–7365. doi: 10.1073/pnas.90.15.7361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottapel R, Ilangumaran S, Neale C, La Rose J, Ho JM, Nguyen MH, Barber D, Dubreuil P, de Sepulveda P. The tumor suppressor activity of SOCS-1. Oncogene. 2002;21:4351–4362. doi: 10.1038/sj.onc.1205537. [DOI] [PubMed] [Google Scholar]
- Druker BJ, Talpaz M, Resta DJ, Peng B, Buchdunger E, Ford JM, Lydon NB, Kantarjian H, Capdeville R, Ohno-Jones S, Sawyers CL. Efficacy and safety of a specific inhibitor of the BCR-ABL tyrosine kinase in chronic myeloid leukemia. N Engl J Med. 2001;344:1031–1037. doi: 10.1056/NEJM200104053441401. [DOI] [PubMed] [Google Scholar]
- BRB Array Tools http://linus.nci.nih.gov./BRB-ArrayTools.html
- Cluster and Treeview http://rana.lbl.gov/EisenSoftware.htm
- Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G. GO:Term Finder-open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Affymetrix http://www.affymetrix.com
- Lee JS, Chu IS, Mikaelyan A, Calvisi DF, Heo J, Reddy JK, Thorgeirsson SS. Application of comparative functional genomics to identify best-fit mouse models to study human cancer. Nat Genet. 2004;36:1306–1311. doi: 10.1038/ng1481. [DOI] [PubMed] [Google Scholar]
- Ellwood-Yen K, Graeber TG, Wongvipat J, Iruela-Arispe ML, Zhang J, Matusik R, Thomas GV, Sawyers CL. Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell. 2003;4:223–238. doi: 10.1016/S1535-6108(03)00197-1. [DOI] [PubMed] [Google Scholar]
- Livak KL, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Tables 1. Functional enrichment analysis of genes that showed significant differences in expression between plasma cell tumors and B-cell lymphomas. Supplementary Table 1A. Functional enrichment analysis according to GO Molecular Function category. Supplementary Table 1B. Functional enrichment analysis by GO Cellular Component category. Supplementary Table 1C. Functional enrichment analysis by GO Biological Process category. Supplementary Table 1D. Genes present in nine GO Molecular Function categories that were shown to be functionally enriched (see Supplementary Table 1A, above). Four tables showing lists of genes that showed significant (p < 0.005) differences in expression between plasma cell tumors and B-cell lymphomas characterized by their functional enrichment according to biological processes defined by the Gene Ontology Consortium. The enrichment of gene set was estimated by calculating the cumulated hypergeometric p values of the each biological process defined by the Gene Ontology Consortium http://www.geneontology.org. In Table 1D, 9 components from the Molecular Function GO category were selected to show the actual genes involved.
Supplementary Tables 2. Genes that showed significant (p < 0.001) differences in expression between rapid-forming plasma cell tumors (ABPC and ABLMYCPC) and slow-forming plasma cell tumors (TEPC, IL6PC and KiPC). Supplementary Table 2A. Genes that showed more than 2-fold higher expression in rapid-forming plasma cell tumors than in slow-forming plasma cell tumors. Supplementary Table 2B. Genes that showed more than 2-fold higher expression in slow-forming plasma cell tumors than in rapid-forming plasma cell tumors. Two tables showing lists of genes that showed significant (p < 0.001) differences in expression between rapid- and slow-forming plasma cell tumors.
Supplementary Figure 1. Quantitative RT-PCR of relative mRNA content in 13 mouse B-cell lymphomas and plasma cell tumors for 9 key genes. Results of quantitative RT-PCR validation of relative mRNA content in 13 mouse B-cell lymphomas and plasma cell tumors for 9 key genes: c-Myc, Socs1, Socs2, Abl1, Jak1, Xbp1, Sdc1, Irf4, Pax5 (BSAP) and the "housekeeping" gene, GAPDH.
Supplementary Tables 3. Genes with significant (p < 0.001) differences in gene expression associated with site of chromosomal translocation in two sample t-tests. Supplementary Table 3A. Genes that showed significant differences in expression between ABPCs with T(12;15) and ABPCs with T(6;15). Supplementary Table 3B. Genes that showed significant differences in expression between TEPCs with T(12;15 class I) and TEPC T(12;15 class II) chromosome translocations. Two tables showing lists of genes that showed significant (p < 0.001) differences in expression between similar subclasses of plasma cell tumors that differ in site of Myc-activating chromosomal translocation.
Supplementary Figure 2. Meta-analysis of global gene expression of 67 mouse samples (including 5 different subgroups of plasma cell tumors and B-cell lymphomas) combined with 123 human samples (including 5 different subgroups of multiple myelomas and tonsilar B cells). Heat map derived from a meta-analysis of a combination of expression data from human and mouse plasma cell dyscrasias.