Abstract
The basic helix–loop–helix (bHLH) transcription factor family regulates numerous developmental events in eukaryotic cells. In the model system, C. elegans, thirty-seven bHLH proteins have been identified via genome-wide sequence analysis and fourteen have been genetically characterized to date. These proteins influence cell fate specification of neural lineages and differentiation of myogenic lineages and have distinct roles in somatic gonadogenesis. We report here on the molecular characterization of HLH-17, a protein whose putative bHLH domain is homologous to the mammalian bHLH proteins BETA3 and bHLHB5. The gene hlh-17 is transcriptionally active at all developmental stages, with the highest steady state accumulation of hlh-17 mRNA during embryogenesis. An upstream hlh-17 sequence drives expression of GFP in the sheath cells of the cephalic sensilla. Finally, animals that are defective in HLH-17 via RNAi display egg-laying defects, while those carrying null mutations in hlh-17 do not develop beyond the L2 stage and are less attracted to potassium and sodium ions. We propose that hlh-17 affects the ability of C. elegans to respond to food cues, with possible downstream effects on insulin-signaling genes involved in the normal development and reproductive viability of the worm.
Keywords: Chemotaxis, Larval arrest, Neural differentiation, Quantitative RT-PCR, NeuroD, Insulin-dependent pathways
1. Introduction
The bHLH transcription factors contain a fifteen-aminoacid basic DNA-binding domain and a helix-loop-helix dimerization motif. This motif is comprised of two α-helices, separated by a variable loop, which present hydrophobic residues on their surface used for protein–protein interactions. Typically, individual bHLH proteins form either homodimers or heterodimers with other bHLH proteins and often switch between these dimerization states to differentially regulate gene expression. Consequently, bHLH proteins are historically classified by their DNA-binding properties, their ability to form homodimers and/or heterodimers with other bHLH proteins, and their functional and structural similarity (reviewed in Atchley and Fitch (1997) and Jones (2004)).
Members of the bHLH protein family are necessary for the timing of events in cellular differentiation and specification, cell growth and metabolism, and cell death. Animals as structurally diverse as humans and nematodes possess proteins with significant homology within specific bHLH domains and these homologous proteins often regulate similar processes in cellular development. One striking example of this evolutionary conservation of bHLH protein function is the neuronal differentiation factor, NeuroD. Homologs of NeuroD are found in most eukaryotic organisms and in mammals the proper expression and functioning of NeuroD is associated with normal pancreatic development and insulin expression. Mutations in NeuroD proteins manifest as type II diabetes mellitus in humans and in mice. NeuroD is also critical for post-natal brain development (Miyata et al., 1999) and is expressed in the olfactory bulb, the hippocampus, and the cerebellum (Lee et al., 1995). In the ear, NeuroD is believed to be essential for the maturation of primary neurons in the cochlea (reviewed by Fritzsch (2003)).
Recently, a negative regulator of NeuroD has been identified in hamster (BETA3) and in human and mouse (bHLHB5). None of these proteins bind to DNA in vitro, but they appear to inhibit NeuroD activity by forming competitive heterodimers with members of the bHLH E protein family (Miyata et al., 1999). The exact mechanism of inhibition in this scenario is uncertain; however, bHLHB5 is able to repress activity of the pax-6 promoter, a gene normally activated by NeuroD (Marsich et al., 2003), through a non-DNA binding mechanism. Pax genes encode proteins containing a paired domain and a homeodomain and are involved in the development of the eye, brain, kidney, and pancreas (Chalepakis et al., 1993; Mansouri et al., 1999).
In the soil nematode Caenorhabditis elegans, several bHLH proteins affecting neural development have been described previously, including the E/Da homolog HLH-2, which is expressed in neurons and neuronal precursors during embryonic development (Krause et al., 1997), the NeuroD homolog, CND-1, which is believed to specify the identity of ventral cord motor neurons (Hallam et al., 2000), and the PAX-6 homolog, VAB-3, which is required for head neuron specification (Chisholm and Horvitz, 1995). In this paper, we describe a novel C. elegans gene, hlh-17, which encodes a bHLH protein that is 54% homologous to Beta3/bHLHB5. Expression of hlh-17 mRNA has been detected in embryos and at all developmental stages in wild-type animals. Transcriptional fusions of the hlh-17 promoter to GFP show continuous expression in cells of the cephalic sensilla. Loss of HLH-17 activity through RNAi produces animals with egg-laying defects, while hlh-17 null animals show chemotaxis defects and arrest during early larval development. Taken together, our data suggest that HLH-17 is required for normal development and behavior.
2. Materials and methods
2.1. Nematode propagation
Wild-type C. elegans hermaphrodites were cultivated on solid nematode growth medium at 22 °C as previously described (Epstein and Shakes, 1995). Animal populations were synchronized by treatment with alkaline hypochlorite solution (Zhu et al., 1997), hatched on unseeded plates, and either frozen at −20°C or moved to seeded NGM plates. For the isolation of early embryonic RNA, L1-staged larvae were fed OP50 for 24 h at 25 °C. Plates were then inspected to ensure that animals contained early embryos but were not yet laying eggs. These animals were treated with alkaline hypochlorite and the recovered embryos were used for subsequent RNA isolation. For isolation of L2, L3, L4, and adult RNAs, animals were fed OP50 at 22 °C for 10, 20, 28, and 40 h, respectively, and, after visual confirmation of age, were then collected and frozen on dry ice and stored at −80 °C.
2.2. Nematode strains
All C. elegans strains used in this study were obtained from the C. elegans Genome Center, including the wild-type Bristol strain N2 and VC486 (ok487) IV/nT1[qIs51] (IV;V), an hlh-17 null mutant strain generated by the C. elegans Knockout Consortium. The consortium outcrossed the strain once and it was subsequently outcrossed four more times in our laboratory. An hlh-17::GFP construct was generated that contained 2.5-kbp upstream of the hlh-17 initiator codon by using the serial overlap extension PCR method (Hobert, 2002) and co-injected with 50–100 μg/mL of pRF-4, rol-6 marker into both syncytial gonad arms of wild-type L4 staged worms as previously described (Mello et al., 1991). Rolling F1 progeny were cloned onto separate plates and were screened for fluorescence.
2.3. Total RNA isolation and cDNA synthesis
Total RNA and cDNA were prepared as previously described (Williams et al., 2004). For PCR amplification of the cDNA, the cDNA synthesis reaction was diluted ten-fold in water and amplified in 30 cycles of PCR with gene-specific primers. In a 50 μL reaction, 1 μL of cDNA was amplified with 10 μM of each primer, 400 μM of each dNTP, 1.5 mM MgCl2, 20 mM Tris–HCl, pH 8.4, 50 mM KCl, and 2.5 units Taq DNA polymerase. cDNA was synthesized using the gene specific primer HLH17-UTR (5′-ATT TAT GGA AAC AGT TGA ATA ATT AAA T-3′) or the gene specific primer HLH17-exon2 rev (5′-GGC CAA AAG CAA CGT AGC AAT CTT GCT CAA C-3′). The product was purified from a 1.2% agarose 1X-TBE gel using gel extraction spin columns (Ambion, Inc.) and sequenced by a commercial laboratory.
2.4. Rapid amplification of cDNA ends
Total RNA was enriched for polyA(+) RNA using the RNeasy Total RNA Purification Kit (Qiagen, Inc.). Full-length, RNA ligase-mediated rapid amplification of the 5′ cDNA ends was performed using the GeneRacer Kit (Invitrogen Life Technologies). All steps prior to reverse transcription were carried out as described by the manufacturer. First strand and second strand syntheses were carried out as described for cDNA synthesis (Williams et al., 2004). The initial amplification by PCR was further amplified by a nested reaction using the 5′ RACE specific primer provided by the manufacturer and the 3′ gene specific primers “HLH17-UTR” (5′-ATT TAT GGA AAC AGT TGA ATA ATT AAA T-3′) or “HLH17-exon2 rev” (5′-GGC CAA AAG CAA CGT AGC AAT CTT GCT CAA C-3′). The products from nested PCR were purified from 1.2% agarose 1X-TBE gels and sequenced using the two 3′ primers used for cDNA amplification or the primer “hlh17-f-rtPCR” (5′-ATG GGG TCC CTG GGG ACT CTC CTC GCG-3′).
2.5. Quantitative real-time PCR
cDNA was prepared as described above and was diluted (1:20 v /v) in water. PCR was performed on the Mx3000P Instrument (Stratagene) in the RCMI Cellular and Molecular Biology Core Facility with the QPCR Master Kit (Stratagene). Each reaction contained 1 μL of diluted cDNA, a total volume of 6 μL of gene specific primers (hlh17-qPCR-f:5′-TGATACTGCTTTCGCTGAGCCT-3′; hlh17-qPCR-r:5′-TCTTGCTCAACTTCCTCACGGA-3′) and 1 μL of molecular beacon (hlh17-qPCR-probe: 5′-AACCGCCTTAAACGAGGCACTTGACGATCTGCGAGGGCGGTT-3′). Actual ratios of forward and reverse primers were optimized to yield maximum product. Reaction cycle consisted of a melting step of 95 °C for 10 min, followed by 35 cycles of 95 °C for 30 s, 52 °C for 1 min, and 72 °C for 1 min. All reactions were performed in triplicate and data were normalized to levels of hlh-1 cDNA (primer sequences-hlh1-f-rtPCR: 5′-TCC AAC TGC ACC TAC CAC TT-3′; hlh1-r-rtPCR:5′-TCT GTG GCA TTT GGT GTG CTC T-3′; and hlh1-probe: 5′-ACC GTC CGC CAA CTC GTC AGA CGT GAA GCC AAT CAT TAG GAC GGT-3′).
2.6. Double-stranded RNA interference
Double-stranded RNA was generated in vitro as previously described and was injected into the syncytial gonad of L4/early adult stage animals (Fire et al., 1998). Animals were allowed to recover for 5 h and were then transferred to plates seeded with lawns of E. coli strain HT115(DE3) producing hlh-17 dsRNA. Injected animals were fed the dsRNA (Timmons et al., 2001) while laying eggs for 12–24 h. Progeny were cultivated on separate plates, were also fed bacteria producing the dsRNA, and were scored for obvious changes in phenotype and behavior. To generate the feeding construct, genomic DNA sequence of hlh-17 was amplified by PCR, cloned into the vector L4440 and transformed into E. coli strain HT115(DE3). Double-stranded RNA synthesis was induced with isopropyl-β-d-thiogalactoside (Timmons et al., 2001). The injected dsRNA corresponded to 102 nucleotides in exon 3 of hlh-17, while the feeding construct contained exons one and two and the intervening intron. Neither of these sequencings is significantly homologous to other genes in C. elegans.
2.7. Chemotaxis assays
N2 and VC486 embryos, obtained by the alkaline hypochlorite method as in Section 2.1, were allowed to grow to the L1 stage over a 16-h period at 20 °C. These L1 animals were assayed for their response to the following chemicals: 5 M sodium chloride, 10% 2-nonanone, 5 M potassium acetate, and 50 μM copper acetate. Petri dishes 10 cm in diameter were filled with 8 mL of agar medium [2% agar, 10 mM HEPES, and 0.25% Tween 20] (Uchida et al., 2003). A straight line was drawn across the center of the plate, with a dot marked 1 cm from the end of the line. At least 12 h before the assay, a 1 μL drop of the chemical was applied to one of the marked dots and labeled A for attractant. Immediately preceding the assay, a 1 μL drop of ethanol was applied to the Petri dish on the remaining dot and labeled C for counter-attractant. A 1 μL drop of 1mM sodium azide was applied to both A and C. At time= 0, animals were placed in the middle of the two dots. The worms were allowed to roam freely for an hour at 20 °C, then the number of immobilized worms within a 0.5 cm radius of A and C were counted. The chemotaxis index (CI) was calculated as CI= [(total # of worms at A)−(total # of worms at C)]/total # of worms assayed (Bargmann et al., 1993).
3. Results
3.1. Molecular analysis of hlh-17 expression
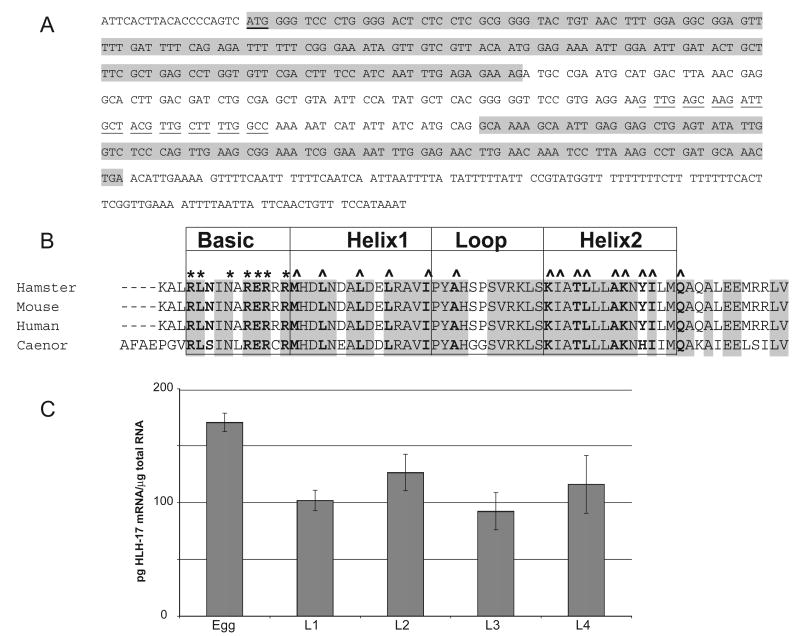
As indicated in Wormbase (www.wormbase.org), the gene sequence hlh-17 is predicted to encode a bHLH domain protein (GenBank accession no. Z82267). Once we confirmed hlh-17 activity by RT-PCR (data not shown), we used 5′-RACE and quantitative RT-PCR (qRT-PCR) to identify the transcription start site and any 5′ untranslated sequences and to determine the temporal expression pattern of hlh-17. Sequencing a mixed population cDNA library produced by 5′-RACE indicates that the hlh-17 transcript begins 19 nucleotides upstream of the transcription initiator codon and contains 3 exons (Fig. 1A). Analysis and ClustalW alignment (Chenna et al., 2003) of the predicted amino acid sequence indicates that hlh-17 is homologous to the mammalian proteins Beta3/bHLHB5. While the mammalian proteins are 100% identical to one another within the bHLH domain, the amino acid sequence of the bHLH domain from C. elegans is 74% identical to the mammalian sequences (Fig. 1B). The entire protein sequence is significantly shorter than the mammalian protein and is 54% homologous to the full-length mammalian proteins. For qRT-PCR, total RNA was isolated from synchronized cultures of C. elegans. Developmental stages were confirmed, prior to RNA isolation, by visual inspection of the developing gonad. cDNA was synthesized from total RNA and was amplified in real-time reactions. As indicated in Fig 1C, the highest level of steady-state mRNA accumulation was seen in embryos. During larval development and in adults (data not shown), hlh-17 mRNA levels did not significantly change.
Fig. 1.
Molecular analysis of hlh-17. (A) cDNA sequence as indicated by 5′-RACE. The hlh-17 transcript starts 19-bp upstream of the predicted initiator codon, which is indicated by bold, underlined characters. The figure depicts the three exons in the cDNA, where exons 1 and 3 are shaded and exon 2 is not. Predicted codons are separated by a space. The sequence complementary to the primer used to sequence the RACE product is underlined, while other primer sequences are indicated in Materials and methods. This figure also depicts the 3′ untranslated region, which is not shaded. Sequences of the 3′ end of the full length cDNAs were obtained by using a 15-nucleotide primer that started immediately after the initiator codon. (B) Alignment of predicted amino acid residues for the bHLH region of HLH-17 and the mammalian homologs. The residues with predicted structural function are indicated in bold and residues that are identical in all four proteins are indicated by shading. Those residues predicted to influence DNA binding are marked by an asterisk (*), while those predicted to influence dimerization are indicated by a caret (^). HLH-17 is 74% identical to the mammalian proteins within the bHLH domain and the 12 residues immediately C-terminal to helix 2. (C) Quantitative RT-PCR analysis of changes in gene expression during C. elegans development.
3.2. Loss of hlh-17 activity via RNA interference
To determine the effects of silencing hlh-17 expression, wild-type, early L4 stage hermaphrodites were injected with, fed, or injected with and fed hlh-17 dsRNA. All progeny, regardless of the initial method of dsRNA delivery, were also fed hlh-17 dsRNA throughout their life cycle and observed for morphological and behavioral defects. Silencing of hlh-17 resulted in the egg-laying defective (egl) phenotype that is most easily scored as the “bag-of-worms” phenotype (Chen and Caswell-Chen, 2003). Under normal conditions, wild-type hermaphrodites lay embryos during gastrulation (Fig. 2A,B) and typically retain an average of between 15 and 20 developing embryos at one time. Many of the progeny of hlh-17(RNAi) hermaphrodites retained their embryos to much later stages of development (see Fig. 2C,D) and consistently accumulated above-average numbers of embryos (data not shown). Feeding or injecting dsRNA resulted in slightly fewer egl progeny than combining the two methods together (see Fig. 2E). Typically, C. elegans adults retain embryos under the following conditions: if the vulval and uterine egg-laying muscles do not function, when the vulva has not properly developed during the fourth larval molt, and if the animals cannot eat, such as when food becomes scarce or the pharynx cannot pump. During our assays, each animal was assured an adequate food supply as each animal was cultivated on its own NGM plate containing a full lawn of E. coli. Likewise, both the organization of the vulva and pharyngeal pumping of hlh-17(RNAi) animals appeared wild-type under a stereomicroscope. Because the egl phenotype may be quantitated by measuring changes in the total number of progeny, we assayed hlh-17(RNAi) animals for changes in viable progeny. Those hlh-17(RNAi) animals that displayed the egl phenotype produced fewer progeny than did the progeny of control animals (see Fig. 2E.). We also observed that many progeny, but not all, would hatch while still inside the hermaphrodite and would then crawl out of the vulva. Furthermore, hlh-17(RNAi) animals would initiate egg-laying at approximately the same time as control animals. Over the next 30 h, they would gradually retain embryos for longer and longer periods, until bagging occurred. Together, these observations support earlier observations that the morphology of the vulva is not affected by hlh-17 activity. Finally, the progeny of hlh-17(RNAi) animals had normal lifespans and less than 5% of them displayed the egl phenotype.
Fig. 2.
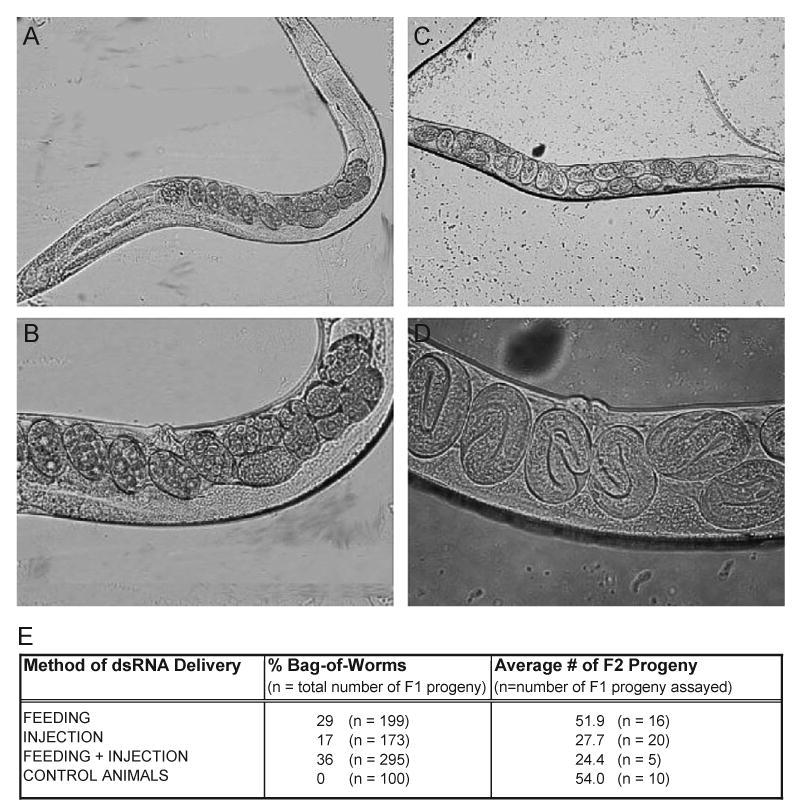
Silencing hlh-17 expression via RNA interference. In all panels, anterior is to the left and dorsal is up. (A and B) An adult wild-type worm contains an average of between 15 and 20 fertilized oocytes undergoing embryogenesis, which are laid prior to gastrulation. (C and D) Adult hlh-17 (RNAi) animals retain an average of 40 embryos beyond gastrulation onto hatching. Embryos retained here are in three-fold stage. In all cases, animals were well fed throughout the course of the experiment.
3.3. The hlh-17 promoter is active in neuronal tissues of the head
To visualize the spatial expression pattern of hlh-17, a 2.5-kbp promoter region upstream of the hlh-17 initiator ATG was used to drive the expression of GFP in wild-type animals (see Fig. 3). GFP expression was detected in four cells near the nerve ring (arrowheads in Fig 3B) as well as in two dendrites that extend toward the mouth (arrows in Fig 3A,B) displaying a pattern similar to that seen in C. elegans when amphid neurons are stained. Interestingly, the GFP shows a web-like pattern that seems to extend away from each of the cells (see arrowhead in Fig 3C). Weak GFP expression can also be seen in cells of developing embryos (Fig. 3D). We have recently identified these cells as sheath cells of the cephalic sensilla (shown in Wormatlas, www.wormatlas.org). Expression in these cells is detected at all developmental stages and in both males and hermaphrodites.
Fig. 3.
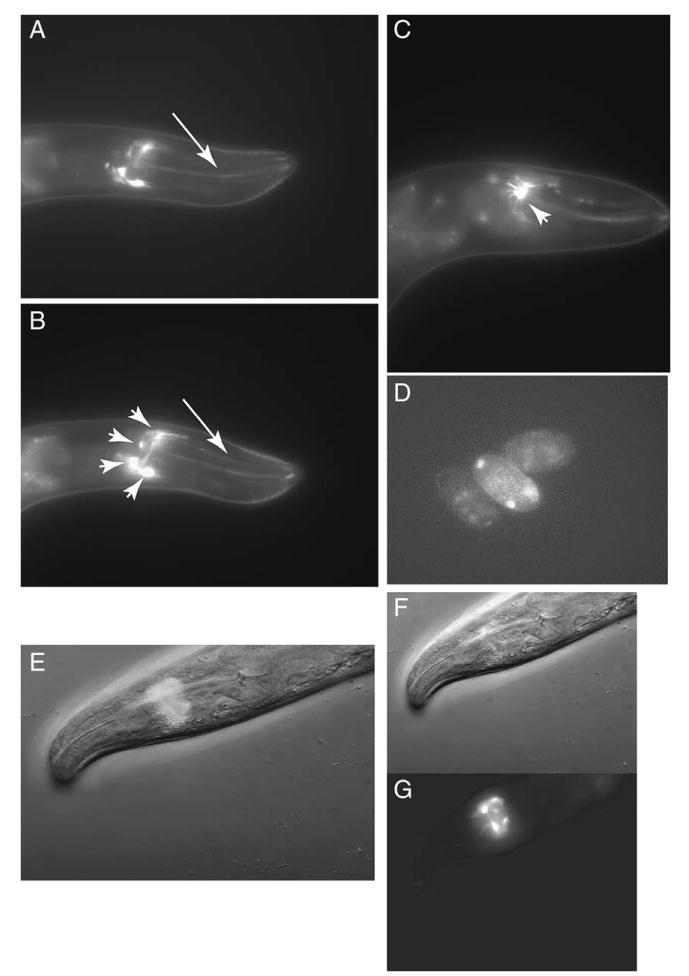
Promoter activity of hlh-17. (A, B, and C) An hlh-17 promoter fusion to GFP shows expression in head cells, with fluorescence extending from four cells (arrowheads in B and C) in the nerve ring to two dendrites (arrows in A and B) that extend towards the mouth. A through C are images of the same animal in different focal planes. (D) Some GFP signaling is detected in embryonic cells. (E) GFP/brightfield merge of F and G showing the location of GFP signaling within the nerve ring.
3.4. The hlh-17 gene product, HLH-17, is required for normal development
The normal life cycle of C. elegans consists of four stages punctuated by molts, referred to as larval stages L1 through L4. After hatching in the absence of food, the L1 stage animals do not develop further until fed. Once food is supplied, L1 animals will continue to develop normally, undergoing the first molt to the L2 stage within 15 h at 20 °C. To further characterize HLH-17 activity, we requested that the C. elegans Gene Knockout Consortium (http://celeganskoconsortium.omrf.org) generate a strain carrying a null allele of hlh-17. We received this strain (VC486), outcrossed it four times to the wild-type background, and performed a preliminary analysis of its behavior. The homozygous deletion of hlh-17 is lethal; however, VC486 is balanced by a GFP-marked chromosomal translocation. Animals that are heterozygous for the hlh-17 null allele and carrying this balancer are viable and age normally. Greater than 95% of the animals carrying homozygous null alleles for hlh-17 do not develop beyond L1 stage, even in the presence of food. Those that manage to age beyond L1 stage take much more time to do so, often not proceeding to L2 until after 30 h of feeding at 20 °C. Generally, the hlh-17 homozygous mutants persist as late L1s or early L2s for 60–96 h after feeding. At the time of death, the animals have many vacuoles in their bodies and seem to die due to some general metabolic defect since no single tissue is more affected than others. Therefore, hlh-17 encodes a vital gene product required for the progression of C. elegans beyond early larval development.
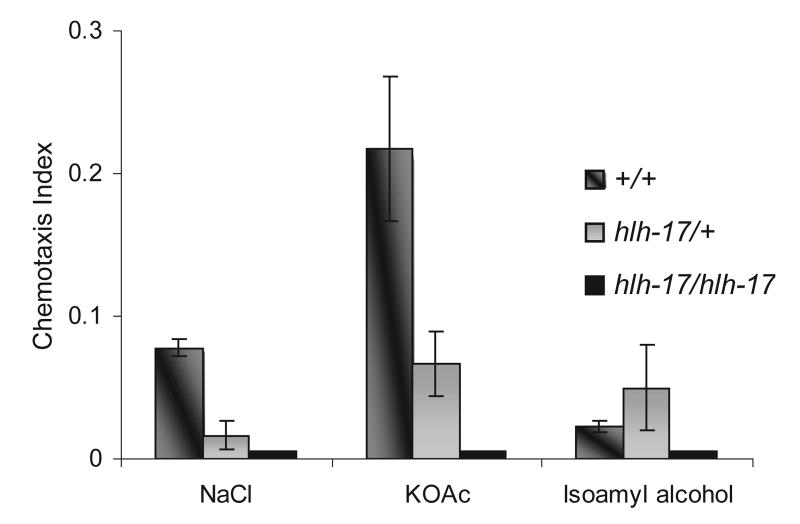
Interestingly, C. elegans' ability to molt through its early larval stages and to lay eggs may both involve its ability to sense its food source. RNAi-mediated silencing of hlh-17, which is expressed throughout the life cycle, produced worms with a phenotype similar to that seen in starved adults; additionally, the hlh-17 null mutant arrests at the L1 stage like hatched embryos that have not been fed. These food-modulated behaviors and the expression of hlh-17 in neuronal support cells in the head make it feasible that the sensory ability of the hlh-17 mutant may be affected. We used chemotaxis assays to measure the effect of hlh-17 on chemotactic responses of wild-type and hlh-17 mutant L1 animals. We reasoned that the response to food cues would be effectively regulated by chemosensory responses rather than odorsensory ones and so we assayed water-soluble chemicals that are known attractants and that are detected by at least one chemosensory neuron in C. elegans. As indicated in Fig. 4, animals that are defective in HLH-17 activity are not as strongly attracted to potassium or sodium ions as wild-type animals. Because hlh-17 mutant L1s become increasingly lethargic the longer they remain arrested at L1, chemotaxis assays were performed within 20 h after hatching. Nevertheless, the defect in chemotaxis is not due to locomotion defects since hlh-17 animals are responsive to both anterior and posterior touches and are able to move freely on plates. Interestingly, wild-type animals are less responsive to chemo-attractants during the L1 stage than they are at L4 stage (In our studies, the chemotaxis index in L4 stage wild-type animals was closer to 0.8 for all three chemicals assayed in Fig. 4—data not shown.). Assays with sodium acetate produced similar results to those with sodium chloride. Neither wild-type nor hlh-17 mutant animals at L1 stage showed an attraction for lysine, biotin, or cAMP (data not shown).
Fig. 4.
Loss of hlh-17 activity results in L1 animals with abnormal chemotactic responses. Animals were assayed for their response to sodium, potassium, and isoamyl alcohol. hlh-17 mutant heterozygotes and homozygotes were significantly less attracted to potassium and sodium than wild-type animals. All animals displayed the same response to isoamyl alcohol. Homozygous mutants were unresponsive to all three chemicals.
4. Discussion
4.1. Potential roles for hlh-17
Sequence analysis showed that the bHLH protein HLH-17 is significantly identical within the bHLH domain to BETA3/bHLHB5, negative regulators of insulin expression mostly found in mammalian brain, pancreas, kidney and lung (Peyton et al., 1996; Xu et al., 2002). It is generally believed that the Beta3/bHLHB5 proteins act to repress transcription of NeuroD responsive promoters via a non-DNA-binding mechanism. Since proteins with a conserved bHLH domain often perform similar functions in development, HLH-17 may function in C. elegans in a role analogous to that of BETA3/bHLHB5 in mammals.
The conserved insulin signaling pathways in C. elegans are often triggered by environmental cues, especially cues that signal the availability of food. These cues are often processed by ciliated sensory neurons in the head, which release insulin-like signals that are transduced to target cells, ultimately regulating metabolism, reproduction, and lifespan (Nelson and Padgett, 2003). Alcedo and Kenyon (2003) have provided direct evidence for the link between sensory neurons and longevity and have suggested that the ASI gustatory neurons in the head of C. elegans promote longevity by influencing insulin/IGF-1 signaling. Much of the research on insulin signaling pathways in C. elegans have supported this link between life span regulation and metabolism. Mutations in the insulin receptor gene, daf-2, or in the phosphatidylinositol 3-kinase catalytic subunit gene, age-1, for example, affect larval arrest, dauer diapause, adult behavior, reproduction and longevity (Tissenbaum and Ruvkun, 1998; Gems et al., 1998). Mutations in daf-2 have been grouped into two classes. Animals carrying class one mutations in daf-2 form constitutive dauer larvae, a stress-resistant, developmentally arrested larval stage that is normally induced by the unavailability of food, high temperature or crowding. Animals belonging to this class of daf-2 mutants also have increased lifespan and show increased tolerance to thermal stress. Animals that carry class two mutations often arrest as embryos or as L1 or L2 larvae. Those that age to adulthood often have smaller brood sizes and either produce progeny at later points in their life cycle or retain their developing embryos longer than wild-type animals (Gems et al., 1998).
Interestingly, the data presented here suggest that HLH-17 may indirectly influence insulin signaling. Like class two daf-2 mutants, animals carrying null mutations in hlh-17 arrest as L1 larvae and occasionally escape to L2 stage. These animals also show significant defects in gustatory responses to sodium and potassium ions and in the olfactory response to isoamyl alcohol when compared to wild-type animals at the L1 stage. Because greater than 90% of the hlh-17 null mutant animals arrested as early larvae, it was not possible to compare the lifespan of these animals to those with active HLH-17. RNAi allowed us to investigate the developmental role of HLH-17. Despite a functional vulva, as indicated by the early ability to lay eggs and by the later ability of hatched L1 larvae to crawl out of the vulva, animals that were hlh-17(RNAi) displayed the egl, bag-of-worms phenotype. Animals that bagged retained above-average numbers of embryos that developed beyond gastrulation, often as late as hatching. Like the class two daf-2 mutants, hlh-17(RNAi) animals had smaller brood sizes.
The sensory organs of C. elegans are arranged in groups known as sensilla. Each consists of at least one ciliated nerve ending, one non-neuronal socket cell, and one non-neuronal sheath cell. Socket cells act to join the sensillum to the hypodermis, while the sheath cell envelops the endings of neurons (see www.wormatlas.org; White et al., 1986). Generally, there are two general types of sensillum. Those that have channels exposed to the animals' external environment are considered to be chemosensory. The second class have no such channel and are considered to be mechanosensory in function (White et al., 1986). Most of our understanding of sensory perception in C. elegans originate from studies on the chemosensory sensilla known as amphids. Amphid neurons are required for normal attraction to or repulsion from water-soluble and volatile chemicals, including the pheromone responsible for signaling entry into dauer stage through use of the TGF-β and cyclic GMP pathways (Ren et al., 1996; Birnby et al., 2000). The ciliated amphid neurons and their supporting sheath cells are also required for normal lifespan (Apfeld and Kenyon, 1999).
Given the phenotypes of hlh-17 null and RNAi animals, we were slightly surprised to find that the hlh-17 promoter is not active in the amphids but, rather, in the cephalic sensilla. The functions of the cephalic sensilla are largely unknown, but the cephalic neurons are one of three classes of dopaminergic neurons believed to function redundantly to sense mechanosensory stimulus from bacteria. Under normal growth conditions, wild-type, well-fed animals that are removed from food and re-introduced to an environment lacking food will significantly slow their movements (Sawin et al., 2000). Likewise, wild-type animals that have been briefly starved will significantly slow their movements when re-introduced to their food source (Duerr et al., 1999). The mechanosensory response elicited in part by the cephalic neurons are believed to mediate the motor circuits to control these behavioral changes (Sawin et al., 2000). In the mature nervous system, sheath cells envelop the sensory endings and couple them to the hypodermal openings formed by socket cells (Ward et al., 1975; Perkins et al., 1986). The cephalic sheath cells may also provide substrates for early axon guidance during the building of the nerve ring (Wadsworth et al., 1996), a bundle of approximately 100 axons that act as the principle circumferential tract in the C. elegans nervous system (Antebi et al., 1997). These four cells have flat sheet-like processes that partly envelop the neuropile of the nerve ring and have narrow extensions that extend radially to interpose themselves at muscle–hypodermis boundaries. The expression of hlh-17 in the cephalic sheath cells suggests either that the cephalic neurons are able to influence insulin-signaling pathways or that loss of hlh-17 activity negatively affects the guidance of axons from other sensilla.
We propose that HLH-17 functions to modulate the behavioral and developmental response to food cues. We further propose that downstream targets of HLH-17 are genes involved in the insulin-dependent metabolic pathway and, as a result, HLH-17 indirectly affects reproduction and aging.
4.2. Further characterization of hlh-17
Our data suggest a link between hlh-17 activity and insulin-signaling pathways and that changes in HLH-17 activity may also influence dauer formation. To further substantiate these possibilities, we used the C. elegans SAGE (Serial Analysis of Gene Expression) database to identify dauer genes that were expressed in patterns similar to hlh-17 (see Jones et al. (2001) and http://elegans.bcgsc.ca/home/sage.html). Due to the low levels of hlh-17 mRNA, limited to four cells in the heads of larvae and adults, hlh-17 activity is not detected in most developmentally staged animals and is only weakly detected in two-week-old dauer larvae. Sequence-specific SAGE tags for hlh-17 are detected in FACS-sorted pan-neuronal cells of the C. elegans embryo, but not in FACS-sorted ciliated neurons. SAGE data also show that many genes involved in dauer decisions are expressed with hlh-17 in FACS-sorted pan-neuronal cells (including, in decreasing order by the number of sequence specific tags, daf-21, dao-5, daf-19, daf-1, daf-5, daf-4, daf-12, and daf-16). It is possible that HLH-17 activity is required for some of these genes to properly function in the cephalic sensilla. It would be interesting to see if HLH-17 influences dauer formation or if hlh-17 expression is altered in daf mutants.
If, as our data imply, HLH-17 functions in a manner similar to Beta3/bHLHB5 in mammalian cells, it would act as a negative regulator of transcription by forming competitive heterodimers with other bHLH proteins. While expression of cnd-1 and hlh-2, which encode the C. elegans homologs of NeuroD and E proteins, respectively, are not expressed in pan-neuronal cells, we reasoned that potential dimerization partners of hlh-17 (1) would be another bHLH protein, (2) would be present in at least some of the same FACS sorted cell types as HLH-17 during SAGE analysis, and (3) might also be present in old dauer larvae during SAGE analysis. By these criteria, we have used the SAGE database to identify all bHLH genes (search parameter included gene designations hlh-1 through hlh-29) that were expressed with similar patterns either in FACS-sorted cells or in dauer larvae as hlh-17. We identified three HLH genes (hlh-20, hlh-19, and hlh-13) that were similarly expressed in both FACS-sorted cells and dauer larvae, while three (hlh-11, hlh-29, and hlh-6) were similarly expressed in dauer larvae. Interestingly, hlh-20 encodes a bHLH protein that is homologous to the mammalian sterol regulatory element binding protein (SREBP). Overexpression of the SREBP isoform C in mammals is associated with increased insulin resistance (Horton et al., 2002). In C. elegans, hlh-20 is required for normal lipid metabolism and for normal larval and embryonic development (www.wormbase.org). Our current studies are focused on determining if these or other HLH proteins will heterodimerize with hlh-17 in vitro.
Acknowledgments
We thank Michael Krause (National Institutes of Health, NIDDK), Andy Fire (Stanford University School of Medicine, Department of Pathology and Genetics), and Kenneth Samuel (Morgan State University, Department of Biology) for helpful discussions and technical assistance and Johnson lab members for critical review of the manuscript. We also thank Dr. Zeynep Altun and WormAtlas (www.wormatlas.org) for help with identification of cells expressing hlh-17::GFP constructs. The qRT-PCR was performed in the Cellular and Molecular Biology Core of the RCMI Biomedical Research Center (supported by NIH RCMI grant #G12 RR17581-02). The C. elegans strains used in this work were provided by Caenorhabditis Genetics Center. Portions of the work described here served to partially fulfill the requirements for the Master of Science degree by T.M. This work was funded by NSF grant #MCB0212336 and by NIH RCMI grant #5G12RR017581 to C.J.
Abbreviations
- bHLH
basic helix-loop-helix
- cDNA
complementary DNA
- kbp
kilobase pair
- RT-PCR
reverse-transcription polymerase chain reaction
- bps
base pairs
References
- Alcedo J, Kenyon C. Regulation of C. elegans longevity by specific gustatory and olfactory neurons. Neuron. 2003;41:45–55. doi: 10.1016/s0896-6273(03)00816-x. [DOI] [PubMed] [Google Scholar]
- Antebi A, Norris CR, Hedgecock EM. Cell and growth cone migrations. In: Riddle DL, Blumenthal T, Meyer BJ, Preiss JR, editors. C elegans II. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1997. pp. 583–609. [PubMed] [Google Scholar]
- Apfeld J, Kenyon C. Regulation of lifespan by sensory perception in Caenorhabditis elegans. Nature. 1999;402:804–809. doi: 10.1038/45544. [DOI] [PubMed] [Google Scholar]
- Atchley WR, Fitch WM. A natural classification of the basic helix–loop–helix class of transcription factors. Proc Natl Acad Sci U S A. 1997;94:5172–5176. doi: 10.1073/pnas.94.10.5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann CI, Hartwieg E, Horvitz HR. Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell. 1993;74:515–527. doi: 10.1016/0092-8674(93)80053-h. [DOI] [PubMed] [Google Scholar]
- Birnby DA, Link EM, Vowels JJ, Tian H, Colacurcio PL, Thomas JH. A transmembrane guanylyl cyclase (DAF-11) and Hsp90 (DAF-21) regulate a common set of chemosensory behaviors in Caenorhabditis elegans. Genetics. 2000;155:85–104. doi: 10.1093/genetics/155.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalepakis G, Stoykova A, Wijnholds J, Tremblay P, Gruss P. Pax: gene regulators in the developing nervous system. J Neurobiol. 1993;24:367–1384. doi: 10.1002/neu.480241009. [DOI] [PubMed] [Google Scholar]
- Chen JJ, Caswell-Chen EP. Why Caenorhabditis elegans adults sacrifice their bodies to progeny. Nematology. 2003;5:641–645. [Google Scholar]
- Chenna R, et al. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm AD, Horvitz HR. Patterning of the Caenorhabditis elegans head region by the Pax-6 family member vab-3. Nature. 1995;377:52–55. doi: 10.1038/377052a0. [DOI] [PubMed] [Google Scholar]
- Duerr JS, et al. The cat-1 gene of Caenorhabditis elegans encodes a vesicular monoamine transporter required for specific monoamine-dependent behaviors. J Neurosci. 1999;19:72–84. doi: 10.1523/JNEUROSCI.19-01-00072.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein HF, Shakes DC. Caenorhabditis elegans: Modern Biological Analysis of an Organism. Academic Press; San Diego: 1995. pp. 3–29. [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double stranded RNA in C. elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Fritzsch B. Development of inner ear afferent connections: forming primary neurons and connecting them to the developing sensory epithelia. Brain Res Bull. 2003;60:423–433. doi: 10.1016/s0361-9230(03)00048-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gems D, et al. Two pleiotropic classes of daf-2 mutation affect larval arrest, adult behavior, reproduction and longevity in Caenorhabditis elegans. Genetics. 1998;150:129–155. doi: 10.1093/genetics/150.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallam SJ, Singer E, Waring DA, Jin YS. The C. elegans NeuroD homolog cnd-1 functions in multiple aspects of motor neuron fate specification. Development. 2000;127:4239–4252. doi: 10.1242/dev.127.19.4239. [DOI] [PubMed] [Google Scholar]
- Hobert O. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques. 2002;32:728–730. doi: 10.2144/02324bm01. [DOI] [PubMed] [Google Scholar]
- Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S. An overview of the basic helix loop helix proteins. Genome Res. 2004;5:226–231. doi: 10.1186/gb-2004-5-6-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SJ, et al. Changes in gene expression associated with developmental arrest and longevity in Caenorhabditis elegans. Genome Res. 2001;11:1346–1352. doi: 10.1101/gr.184401. [DOI] [PubMed] [Google Scholar]
- Krause M, et al. A C. elegans E/Daughterless bHLH protein marks neuronal but not striated muscle development. Development. 1997;124:2179–2189. doi: 10.1242/dev.124.11.2179. [DOI] [PubMed] [Google Scholar]
- Lee JE, Hollenberg SM, Snider L, Turner DL, Lipnick N, Weintraub H. Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix–loop–helix protein. Science. 1995;268:836–844. doi: 10.1126/science.7754368. [DOI] [PubMed] [Google Scholar]
- Mansouri A, Goudreau G, Gruss P. Pax genes and their role in organogenesis. Cancer Res. 1999;59:1707s–1709s. [PubMed] [Google Scholar]
- Marsich E, Vetere A, Di Piazza M, Tell G, Paoletti S. The pax6 gene is activated by the basic helix–loop–helix transcription factor Neurod/Beta2. Biochem J. 2003;376:707–715. doi: 10.1042/BJ20031021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T, Maeda T, Lee JE. NeuroD is required for differentiation of the granule cells in the cerebellum and hippocampus. Genes Dev. 1999;13:1647–1652. doi: 10.1101/gad.13.13.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DW, Padgett RW. Insulin worms its way into the spotlight. Genes Dev. 2003;17:813–818. doi: 10.1101/gad.1090203. [DOI] [PubMed] [Google Scholar]
- Perkins LA, Hedgecock EM, Thomson JN, Culotti JG. Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev Biol. 1986;117:456–487. doi: 10.1016/0012-1606(86)90314-3. [DOI] [PubMed] [Google Scholar]
- Peyton M, Stellrecht CM, Naya FJ, Huang HP, Samora PJ, Tsai MJ. BETA3, a novel helix–loop–helix protein, can act as a negative regulator of BETA2 and MyoD-responsive genes. Mol Cell Biol. 1996;16:626–633. doi: 10.1128/mcb.16.2.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren P, Lim CS, Johnsen R, Albert PS, Pilgrim D, Riddle DL. Control of C. elegans larval development by neuronal expression of a TGF-beta homolog. Science. 1996;274:1389–1391. doi: 10.1126/science.274.5291.1389. [DOI] [PubMed] [Google Scholar]
- Sawin ER, Ranganathan R, Horvitz HR. C. elegans locomotory rate is modulated by the environment through a dopaminergic pathway and by experience through a serotonergic pathway. Neuron. 2000;26:619–631. doi: 10.1016/s0896-6273(00)81199-x. [DOI] [PubMed] [Google Scholar]
- Tissenbaum HA, Ruvkun G. An insulin-like signaling pathway affects both longevity and reproduction in Caenorhabditis elegans. Genetics. 1998;148:703–717. doi: 10.1093/genetics/148.2.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Uchida O, Nakano H, Koga M, Ohshima Y. The C. elegans che-1 gene encodes a zinc-finger transcription factor required for specification of the ASE chemosensory neurons. Development. 2003;130:1215–1224. doi: 10.1242/dev.00341. [DOI] [PubMed] [Google Scholar]
- Wadsworth WG, Bhatt H, Hedgecock EM. Neuroglia and pioneer neurons express UNC-6 to provide global and local netrin cues for guiding migrations in C. elegans. Neuron. 1996;16:35–46. doi: 10.1016/s0896-6273(00)80021-5. [DOI] [PubMed] [Google Scholar]
- Ward S, Thomson N, White JG, Brenner S. Electron microscopical reconstruction of the anterior sensory anatomy of the nematode Caenorhabditis elegans. J Comp Neurol. 1975;160:313–337. doi: 10.1002/cne.901600305. [DOI] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- Williams TR, Lee TM, Johnson CM. Glaucoma studies in the eyeless worm: stress responsiveness and temporal expression of the Caenorhabditis elegans myocilin-like gene, cof-2. Cell Mol Biol. 2004;50(6):723–731. [PubMed] [Google Scholar]
- Xu ZP, Dutra A, Stellrecht CM, Wu C, Piatigorsky J, Saunders GF. Functional and structural characterization of the human gene BHLHB5, encoding a basic helix–loop–helix transcription factor. Genomics. 2002;80:311–318. doi: 10.1006/geno.2002.6833. [DOI] [PubMed] [Google Scholar]
- Zhu J, et al. End-1 encodes an apparent GATA factor that specifies the endoderm precursor in Caenorhabditis elegans embryos. Genes Dev. 1997;11:2883–2896. doi: 10.1101/gad.11.21.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]