Abstract
We argue that if there is a category of slightly deleterious mutations, then there should be a category of slightly advantageous back-mutations. We show that when there are both slightly deleterious and advantageous back-mutations, there is likely to be an increase in the rate of evolution after a population size expansion. This increase in the rate of evolution is short-lived. However, we show how its signature can be captured by comparing the rate of evolution in species that have undergone population size expansion versus contraction. We test our model by comparing the pattern of evolution in pairs of island and mainland species in which the colonization event was either island-to-mainland (population size expansion) or mainland-to-island (contraction). We show that the predicted pattern of evolution is observed.
It is now well established that there is a category of mutations that are slightly deleterious; these are deleterious mutations whose fate is determined by a combination of random genetic drift and selection. Four lines of evidence suggest that these mutations exist. First, in many species, nonsynonymous mutations segregate at lower allelic frequencies on average than silent mutations; this has been observed in humans (1), Drosophila (2, 3), and various bacteria (4, 5). There is also evidence that some mutations in noncoding DNA are slightly deleterious; in Drosophila, mutations in introns and intergenic DNA tend to segregate at lower frequencies than synonymous mutations (6), and, in humans, mutations in conserved noncoding sequences (7) or sites (8) segregate at lower frequencies than mutations outside these sequences or sites. Second, species that are expected to have small effective population sizes have higher ratios of nonsynonymous to synonymous substitutions (9–15). A similar effect is apparent in the sequences upstream and downstream of protein-coding sequences (16), and within conserved nongenic sequences in mammals (17); in both cases, these sequences appear to be much less well conserved, relative to intron sequences, in hominids, which have small effective population sizes, than in rodents, which probably have large effective population sizes. Third, regions of the genome with small effective population sizes tend to show a higher ratio of nonsynonymous to synonymous substitutions; this is particularly apparent on the sex-specific chromosomes (18–20). Finally, in some species the ratio of nonsynonymous to synonymous polymorphism is greater than the ratio of nonsynonymous to synonymous substitution. This is particularly evident in the mitochondrial genome (21–23). Such a pattern is consistent with the presence of slightly deleterious nonsynonymous mutations, because these contribute to polymorphism but rarely become fixed.
There is, therefore, considerable evidence that slightly deleterious mutations exist. However, models of molecular evolution in which all mutations are deleterious predict a decline in fitness, which is unrealistic, and one might reasonably expect there to be slightly advantageous mutations if there are slightly deleterious mutations for two reasons. First, models of molecular evolution that contain advantageous mutations generally predict there to be a class of mutations that are slightly advantageous (24–26). Second, simple back-mutation is expected to generate slightly advantageous mutations. For example, let us imagine that a site is fixed for C, and that a new T mutation occurs that is slightly deleterious with a disadvantage of −s. Let us imagine that this T mutation spreads through the population and becomes fixed. If a new C mutation then occurs at this site, it will be slightly advantageous with an advantage of +s, unless the relative fitnesses of the C and T alleles have changed. Such a change in fitness could occur because of a change in the environment or the fixation of mutations at other sites which have epistatic interactions with the alleles at a site of interest. However, it seems likely that fitnesses will remain approximately constant, at least over moderate time scales. Models of evolution in which there are slightly deleterious mutations and slightly advantageous mutations are familiar in the study of synonymous codon evolution (27, 28), but they have rarely been considered in studies of protein evolution (although see refs. 24–26 and 29–31).
Although we expect slightly advantageous mutations to exist, currently there is only limited evidence for their existence in real populations; analyses of McDonald–Kreitman (32) type data within the context of the selection models proposed by Sawyer and Hartl (33) and Sawyer et al. (34) suggest that there are weakly selected advantageous mutations (34–37). However, these data are also always consistent with strongly advantageous mutations.
Here, we undertake to test for the presence of slightly advantageous mutations by exploiting a property of models of molecular evolution that contain both slightly deleterious and slightly advantageous mutations. Under the “classic” nearly neutral model of molecular evolution (38), all mutations are assumed to be neutral, slightly deleterious, or strongly deleterious, leading to the prediction that the rate of substitution will be higher for species and genomic regions with small effective population sizes. This prediction arises from the idea that purifying selection is less efficient against slightly deleterious mutations in small populations than in large ones, allowing some proportion of mildly deleterious mutations to drift to fixation in small populations. However, as Gillespie (24) first noted, this simple pattern is not always predicted if the distribution of fitness effects allows some proportion of mutations to be advantageous. If there is an expansion in effective population size, there will be a temporary increase in the rate of substitution because of the fixation of advantageous mutations that were previously effectively neutral. As a result, the rate of substitution is higher in the population that has the larger effective population size for some time after the expansion. Gillespie (24) did not formally demonstrate this behavior, but Takano-Shimizu (39) has shown theoretically that this increase in the rate of substitution occurs in a model in which there are slightly deleterious and slightly advantageous back-mutations.
There are potentially many different models we could consider in which there are both slightly deleterious and advantageous mutations (24). Here, we choose the simplest model, which has the clearest biological basis, a model in which each slightly deleterious mutation has a corresponding slightly advantageous back-mutation: i.e., if an A mutation at a site fixed for G is deleterious, the G mutation at the same site fixed for A is advantageous with the same absolute strength of selection. This model was previously considered by Takano-Shimizu (39). We confirm his result that there is an instantaneous increase in the rate of substitution after an increase in effective population size and show that this increase can be substantial but that it is relatively short-lived. However this pattern is seen only if some mutations are allowed to be slightly advantageous; if all mutations are assumed to be deleterious the rate of substitution is always predicted to correlate negatively with effective population size, regardless of whether an expansion in effective population size has occurred or not. We use a comparative framework to examine cases where there has been an expansion in population size and those where there has been a contraction, by choosing pairs of island and mainland species and classifying them according to the direction of colonization, island-to-mainland or mainland-to-island. We demonstrate statistically significant support for models of molecular evolution that allow adaptive evolution through back-mutation.
Theory
Slightly Deleterious and Advantageous Mutations.
Here. we investigate a simple two-allele model in which the population size is assumed to have been constant for a long time, so the system is at statistical equilibrium with respect to mutation, selection, and genetic drift before there is an instantaneous change to a new effective population size. We assume that the organism is haploid, although the model would behave identically if the organism was diploid and there was genic selection. Let us imagine that each site can be occupied by one of two alleles, A1, which has an advantage of +s over A2, and A2, which has a disadvantage of −s compared with A1. Let us assume that the mutation rate from A1 to A2 is equal to that between A2 and A1 and that the mutation rate is sufficiently low that Neu ≪ 1, where Ne is the effective population size, and u is the nucleotide mutation rate. Because Neu ≪ 1, any one site will typically be fixed for either the A1 or A2 allele. This model is identical to the classic models of synonymous codon evolution in which codons may be either preferred or unpreferred (27, 28).
Let us begin by considering the system before the population size expansion. The change in the proportion of sites fixed for the A1 allele f can be modeled by the differential equation
where Q(S) = S/(1 − e−S), is approximately N times the probability that a new mutation with selection strength S = 2Ne s will be fixed (40), and N is the census population size. It is not difficult to show that the proportion of sites fixed for the A1 allele at equilibrium is
(27, 28). The rate of substitution at a site in this system is
which simplifies at equilibrium to
Now, let us consider the dynamics of the system after the change in population size. Let us assume that the effective population size becomes α times larger than it was before the change in the population size t generations in the past. Solving Eq. 1, and noting that we are assuming that the population was at equilibrium before the increase/decrease in population size, we have
where φ(αS0) = Q(αS0) + Q(−αS0), τ = ut, and S0 is the value of S before the change in the population size. Substituting the proportion of sites fixed for the A1 allele after the change in population size given by Eq. 5 into Eq. 3, we derive an expression for the rate of substitution after a population size change
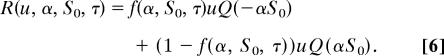 |
We have so far derived expressions for the rates of substitution at equilibrium and after a population size change, for the case when all mutations are subject to the same strength of selection. However, in reality, there is likely to be a distribution of fitness effects. We assume here that the absolute strength of selection follows a Γ-distribution. This distribution has been widely used to model the distribution of fitness effects because it is simple, flexible, and seems to fit the available data fairly well (41) (although see ref. 42. The Γ-distribution is governed by a shape parameter, β and the mean strength of selection, S̄:
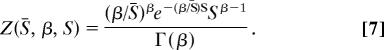 |
Under this distribution Eqs. 4 and 6 become, respectively,
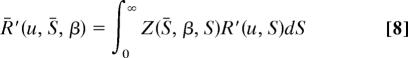 |
and
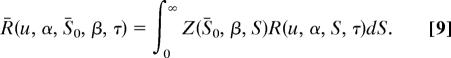 |
Rate of Evolution.
We investigated our model over a range of Γ-distribution shape parameters. For each value of the shape parameter, we found the mean strength of selection, which at equilibrium, would yield a rate of evolution relative to the neutral value of 0.05, the value of the nonsynonymous to synonymous substitution rate ratio (ω) that we observe in our data analysis (see below). However, the results are almost completely independent of the mean strength of selection; the mean can vary by several orders of magnitude and have essentially no effect on the results either qualitatively or quantitatively; our analysis is therefore applicable to cases in which ω is much higher or lower than we have investigated.
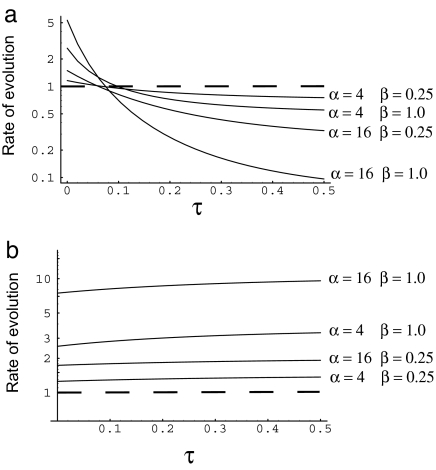
In Table 1, we give the rate of evolution immediately after a change in effective population size relative to the rate before the change. Several points are apparent; first, the rate of evolution increases if the population size decreases, as we would expect; selection becomes less effective and more mutations therefore become effectively neutral. This is also the pattern we would expect if there were only slightly deleterious mutations (see below). If there is an increase in population size, but it is less <2-fold, then the rate of evolution decreases, as Takano-Shimizu (39) previously demonstrated. This is because an increase in population size has two effects; the fixation probability of deleterious mutations decreases, whereas the fixation probability of advantageous mutations increases. For small increases, the change in fixation probability is greater for the deleterious mutations, and, hence, the rate of substitution slows. If the increase in population size is >2-fold, then there is an increase in the rate of evolution. The magnitude of this increase depends on the shape parameter of the Γ-distribution; the smaller the shape parameter, the smaller the increase. However, the increase in the rate of substitution can be very substantial; if there has been an 8-fold increase in effective population size, then there would be a 1.6-fold increase in rate if the shape parameter was 0.25 and a 2.7-fold increase if the shape parameter was 1.
Table 1.
The instantaneous change in the rate of evolution after a population size change
| α | β = 0.25 | β = 0.5 | β = 0.75 | β = 1.0 |
|---|---|---|---|---|
| 0.25 | 1.3 | 1.6 | 2.0 | 2.5 |
| 0.5 | 1.1 | 1.2 | 1.3 | 1.5 |
| 1 | 1.0 | 1.0 | 1.0 | 1.0 |
| 1.5 | 0.99 | 0.97 | 0.96 | 0.95 |
| 2 | 1.0 | 1.0 | 1.0 | 1.0 |
| 4 | 1.2 | 1.3 | 1.4 | 1.5 |
| 8 | 1.6 | 2.1 | 2.4 | 2.7 |
| 16 | 2.6 | 3.8 | 4.7 | 5.3 |
| 32 | 4.8 | 7.4 | 9.3 | 11 |
The increase in the substitution rate is due to the fixation of slightly advantageous back-mutations, but the rate of evolution slows as these get fixed (Fig. 1a). After this, the rate of substitution declines to below the value it was before the increase in population size. Surprisingly the length of time this increase persists is largely independent of both the magnitude of the increase in the population size and of the shape of the distribution of fitness effects; for all combinations of parameters the increase lasts ≈0.1/u generations.
Fig. 1.
The rate of evolution after a population size increase (a) or decrease (b), relative to the rate before the change in population size.
If the population size decreases, there is a jump in the rate of evolution, after which the rate continues to increase slowly until a new equilibrium state is reached (Fig. 1b). This slow increase is due to the gradual fixation of slightly deleterious mutations, which can substitute back to the slightly advantageous allele.
Slightly Deleterious Mutations.
We have so far considered a model in which both slightly deleterious and advantageous mutations can occur at a site. A model in which there are only neutral, slightly deleterious, and strongly deleterious mutations, which we might call the “classic” nearly neutral model, behaves quite differently. Let us assume there are no advantageous mutations, so that all mutations are deleterious, although some may be sufficiently weakly selected so they behave as either effectively neutral or slightly deleterious. If we assume, as above, that the distribution of fitness effects is a Γ-distribution, then it can be shown that the rate of evolution is
(43–45) where k is a constant and β is the shape parameter of the Γ-distribution. Hence, the rate of evolution after a change in population size, relative to that before is simply α−β, where α is the increase in population size. In this case, if the population increases in size, the rate of evolution decreases and vice versa. To give some numerical examples, imagine that the shape parameter of the Γ-distribution is 0.5. If the population doubles in size, the rate of evolution will decrease by 30%. Conversely, if the population size decreases by half, the rate of evolution will increase by 40%.
Testing the Model.
The model above suggests a number of tests that we might use to differentiate a model in which some mutations are slightly advantageous from one in which there are only slightly deleterious mutations. First, the rate of evolution is expected to increase after an increase in population size. Second, the rate of evolution should decline along a lineage that has expanded. And third, the rate of evolution should increase through time if there has been a decrease in population size. Unfortunately, none of these predictions is easy to test; the increase in the rate of evolution after a population size expansion is short-lived, and there are statistical difficulties with trying to test correlations between the rate of evolution and time because the estimate of the rate depends on time.
However, there is a test we can conduct that appears to be robust; we predict that the ratio (X) of the divergence along a lineage with large Ne, such as a mainland lineage, over the divergence along a lineage with the small Ne, such as an island lineage, will depend on whether we are considering an expansion along the lineage with large Ne (island-to-mainland colonization) or contraction along the lineage with small Ne (mainland-to-island colonization). We predict under our model that Xim the value of X for island-to-mainland colonizations, will be >Xmi, because the former involves a period of adaptive evolution because of slightly advantageous back-mutations.
To demonstrate that Xim > Xmi, we first need to derive expressions for the amount of DNA sequence divergence that has occurred since the two lineages being considered diverged. We assume that one of these lineages remains at a constant population size, whereas the other expands or contracts; we assume that this change in population size occurred very soon after speciation, because this seems reasonable for the data we are considering. The divergence along the equilibrium and expanded/contracted lineages are, respectively:
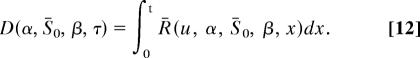 |
We can now write expressions for Xim and Xmi:
where Ni and Nm are the effective population sizes of the island and mainland species, and s̄ is the mean strength of selection.
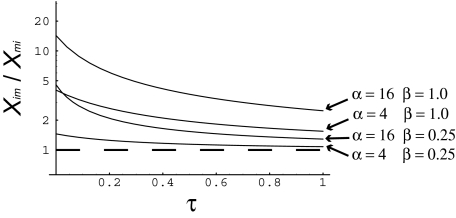
Fig. 2 shows the ratio of Xim over Xmi plotted against time, for two distributions of fitness effects and two levels of difference in effective population size between the mainland and island populations. It is evident that Xim is always >Xmi, and the difference is often substantial. This suggests that, if there are substitutions of advantageous back-mutations occurring, after an expansion in population size, we should be able to detect them as a difference between Xim and Xmi. The magnitude of Xim/Xmi depends on two factors. Xim is substantially >Xmi if the mainland and island are substantially different in population size and if the shape parameter of the distribution of fitness effects is large. In contrast, under the classic nearly neutral model, in which there are only neutral, slightly deleterious mutations and strongly deleterious mutations, then Xim = Xmi. In the section below, we test whether Xim > Xmi in real populations.
Fig. 2.
The ratio of Xim/Xmi for two DFEs and two values of α, the ratio of the mainland and island population sizes.
Results
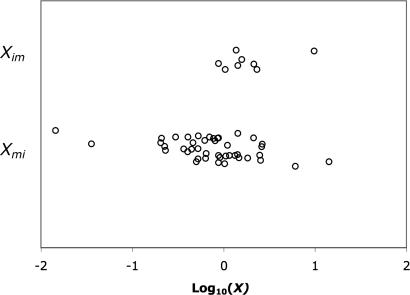
We predict that the rate of evolution in mainland species relative to the rate in the island species should be greater when the colonization has been from island-to-mainland, and hence has involved a population size expansion, than when the colonization was mainland-to-island and has involved a population size decrease. To test this, we compiled eight cases of island-to-mainland colonization (Table 2) and compared these cases to 43 examples of mainland-to-island colonization compiled by Woolfit and Bromham (15). We consider the ratio of the nonsynonymous and synonymous substitution rates, ω, rather than the rate of nonsynonymous substitution, to control for differences in the mutation rate that might exist between island and mainland species. The values of ω for the island-to-mainland examples are given in Table 2, and the values of Xim are plotted with the values Xmi in Fig. 3. It is evident that on average Xim is >Xmi. The difference between Xim and Xmi is large, the geometric mean of Xim is more than twice that of Xmi (1.9 versus 0.73), and the difference is significant (one-tailed Mann–Whitney test P = 0.006).
Table 2.
Details of the island-to-mainland data sets
| Data set | Group | Island | Mainland | Gene | ω (island) | ω (mainland) |
|---|---|---|---|---|---|---|
| Chameleons I | Lizard | Madagascar | Africa | ND2 | 0.053 | 0.11 |
| Chameleons II | Lizard | Madagascar | Africa | ND4 | 0.040 | 0.057 |
| Chameleons III | Lizard | Madagascar | Africa | ND4 | 0.033 | 0.052 |
| Anolis I | Lizard | Jamaica | North and South America | ND2 | 0.071 | 0.074 |
| Anolis II | Lizard | Cuba | North America | ND2 | 0.053 | 0.073 |
| Monarcha | Bird | Solomon Islands | Australia | ND2 | 0.043 | 0.10 |
| Coraria | Plant | Pacific island | South America | rbcL and matK | 0.21 | 2.0 |
| Eleutherodactylus | Frog | Cuba | Central America | c-myc | 0.16 | 0.14 |
Fig. 3.
Log10(X) plotted for cases of island-to-mainland colonization (Xim, upper set of points) and mainland-to-island colonization (Xmi, lower set of points). Scatter on the y axis within each group of points was introduced to separate the data.
There are two potential problems with the chameleon comparisons. First, Madagascar is not a small island, and, as a consequence, there might not have been a significant population size expansion when Africa was colonized. Second, the evidence suggesting that chameleons colonized Africa from Madagascar is not very strong. This biogeographical history is supported by a phylogeny including morphological and molecular data (46), but for all of the colonizations to be in the opposite direction would only require one further colonization event. We therefore repeated the analysis, removing the chameleon data sets; Xim is still significantly >Xmi (P = 0.026).
Additionally, our data set is very biased toward certain groups of organisms, particularly vertebrates, and within the vertebrates, reptiles. However, the difference is still significant if we restrict our comparison to vertebrates (P = 0.011) and nearly significant even if we exclude the chameleons (P = 0.065). The differences are also nearly significant if we restrict the comparison to just reptiles (clades P = 0.080) but we have relatively little data: 5 cases of island-to-mainland colonization and 10 cases in the opposite direction.
Discussion
We have argued, as others have before (24, 29, 31, 38), that, if there are slightly deleterious mutations, then there are likely to be slightly advantageous mutations. We have investigated a model in which slightly deleterious mutation has a corresponding slightly advantageous back-mutation. We show that these slightly advantageous mutations manifest themselves as an increase in the rate of evolution after a population size increase. We show that this increase in the rate of substitution can be detected by comparing the rates of sequence divergence in closely related pairs of island and mainland species. We predict under our model that the ratio of the rate of evolution in the mainland species over the rate in the island species should be greater for cases in which the direction of colonization was island-to-mainland and hence involved a population size expansion, compared with a mainland-to-island colonization leading to a contraction of the island population. We tested this prediction using DNA sequence data and observed the predicted pattern.
Although we do observe the result we expected in our data analysis, it is also evident that our model fails to fit the data perfectly. Although Xim is predicted to be >Xmi, we also expect both Xim and Xmi to be <1 once enough time has elapsed [supporting information (SI) Fig. 4]. However, Xim is significantly >1 (Wilcoxon signed rank test gives P = 0.023), and Xim is <1 for only a single data set. The majority of Xmi values are <1, as Woolfit and Bromham (15) showed, but the skew is not as strong as one might expect; Xmi < 1 for only 27 of 43 comparisons (excluding their Anole data set, which was, in fact, an island-to-mainland colonization event) (P = 0.063). The relatively high values of both Xim and Xmi might be due to other adaptive substitutions that are not simply back-mutations. If the rate of adaptive substitution is limited by the rate of mutation, we expect bigger populations to have higher rates of adaptive substitution for two reasons. First, the mutation rate to any particular advantageous mutation is higher in larger populations because there are more alleles to mutate; and second, selection is effective on a greater proportion of mutations in large populations. Both of these effects will tend to increase both Xim and Xmi.
There are potentially several other explanations for the difference between Xim and Xmi. First, we might expect rates of adaptive substitution to increase, or selective constraint to decrease, in a novel habitat. This would tend to increase Xim and decrease Xmi because, respectively, the mainland and island species are experiencing new environments and therefore an increase in the rate of evolution. However, this seems an unlikely explanation for the following reason. Consider mainland-to-island colonizations; Xmi will tend to be decreased by both slightly deleterious mutations and novelty-of-habitat mutations. However, the geometric mean value of Xmi is not much <1 (0.73 with 95% bootstrap confidence intervals of 0.50 and 1.0), which means the number of novelty-of-habitat mutations cannot be very large if slightly deleterious mutations tend to have a large effect on the substitution rate. We expect slightly deleterious mutations to have a substantial effect on the rate because the average shape parameter for the Γ-distribution estimated from mtDNA is 0.93 (31); this means, for example, that a 2-fold reduction in effective population size will generate a 2-fold difference in the substitution rate (Eq. 10). Hence, there cannot be many novelty-of-habitat mutations under this hypothesis, which means that Xim and Xmi cannot be very different. We demonstrate the limitations of this hypothesis more quantitatively in SI Text.
It is also possible that the difference between Xim and Xmi is due to the fixation of slightly deleterious mutations during the bottleneck in population size that is expected to have occurred during colonization; this will tend to elevate Xim and decrease Xmi. This also seems an unlikely explanation, because we expect the duration of the bottleneck to have been short relative to the time scale over which we have estimated the substitution rates.
We have considered a simple model of evolution in which each slightly deleterious mutation has a corresponding slightly advantageous back-mutation. Back-mutations would seem to be the most obvious source of slightly advantageous mutations. However, it is possible that a slightly deleterious mutation at one site can be compensated for by an advantageous mutation at another site, as appears to be the case in the evolution of RNA molecules (47) and possibly proteins (48). It seems likely that these compensatory mutations will be less common than back-mutations. Furthermore, these compensatory mutations are likely to be weakly selected unless one advantageous mutation can compensate for the effects of many deleterious mutations, so our central conclusion, that there is a category of slightly advantageous mutations, remains supported.
There are a number of potential problems and limitations with our data analysis. First, our analysis is dominated by data from mitochondrial DNA. Second, we have assumed that synonymous mutations are neutral. Although selection on synonymous codon use is well documented in bacteria (49) and the nuclear DNA of many eukaryotes (50), there is no evidence of selection on synonymous codon use in mitochondrial DNA, which make up the bulk of our data. If there is selection on synonymous codon use, we might expect an increase in the rate of synonymous substitution during population size expansion for the same reasons that we expect an increase in the rate of protein evolution. How ω behaves will depend on the distribution of fitness effects of both nonsynonymous and synonymous mutations. Second, our data analysis is not the balanced design we would like; ideally we would compare cases of island-to-mainland colonization and mainland-to-island colonization for species from the same genus involving the same islands and mainland. It is therefore possible that our results are due to various biases in our data set; for example, it might be that the difference between the mainland and island species' population sizes is actually smaller for the island-to-mainland colonization cases, than for the mainland-to-island cases. Fourth, Bazin et al. (51) have recently shown there is no correlation between census population size and mitochondrial DNA diversity (although see ref. 52 for a counterexample). This suggests that an increase in census population may not lead to an increase in effective population size for mitochondrial DNA.
Although there is little evidence of widespread adaptive evolution in hominids (53, 54), it has been estimated that maybe 50% of amino acid (55, 56) and 20% of all (6) substitutions are a consequence of adaptive evolution in Drosophila. The level of adaptive evolution might be even higher in bacteria (4) and viruses (57, 58). Therefore, a question of great interest is how much of the adaptive evolution we detect is due to back-mutation and how much is in response to changes in the environment. The estimates of adaptive evolution cited above were obtained by comparing levels of polymorphism and substitution using the McDonald–Kreitman approach (32). It can be shown that this method does not detect slightly advantageous back-mutations when the population size is stationary (J.C. and A.E.-W., unpublished data). However, it may be that some of the adaptive evolution that we detect is a consequence of back-mutations when there has been population size expansion. This remains to be investigated.
Materials and Methods
We test for the presence of advantageous back-mutations by comparing Xim and Xmi. We took our values of Xmi from the study of Woolfit and Bromham (15), who compared the nonsynonymous over synonymous rate ratio, ω, values from island and mainland species. The direction of colonization was mainland-to-island in all but one of their 44 cases (M. Woolfit, personal communication). Unfortunately, island-to-mainland colonization is relatively rare, and we were able to compile only eight (largely) independent cases. The comparisons are not entirely independent because the outgroup from Chameleons I, Rhampholeon spectrum, is one of the mainland species in Chameleons II. This level of nonindependence is not likely to be a major problem. Details of the data sets are given in Table 2 and SI Table 3. The evidence for the direction of colonization is as follows: Chameleons (46), Anole lizards (59), Monarcha birds (60), Coraria plants (61), and Eleutherodactylus frogs (62). In each case, the direction of colonization is inferred from a phylogeny by using parsimony.
In each data set, we have one or more island species, one or more mainland species, and at least one outgroup species. For those data sets in which we had more than two island and two mainland species available, we selected two island and two mainland species such that the basal branch to the island and mainland clades was relatively short; we did this to maximize Xim and hence maximize the chances of detecting adaptive evolution via back mutation. However, we have basal branch information for only three of our comparisons (for the Monarcha, the basal branch is so short that Xim is undefined), and the results are qualitatively similar to those for the complete clades, so we present only the complete clade analysis.
For each data set we used PAML (63) to estimate ω across the tree. We allowed the outgroup, the island and the mainland clades/lineages, to have their own ω value. The ω values were estimated by using the F4 × 3 model, as used by Woolfit and Bromham (15). We also note that the majority of data analyzed by Bromham and Woolfit came from mitochondrial DNA, just as our data does. We analyzed ω values rather than dN to control for any differences in the mutation rate between lineages.
Supplementary Material
Acknowledgments
We thank John Welch, who suggested the comparative analysis, and John Welch, Brian Charlesworth, Deborah Charlesworth, Nina Stoletzki, and three anonymous referees for helpful comments on the manuscript. This work was supported by the National Evolutionary Synthesis Center and the Biotechnology and Biological Sciences Research Council.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0705456104/DC1.
References
- 1.Cargill M, Altshuler D, Ireland J, Sklar P, Ardlie K, Patil N, Lane CR, Lim EP, Kalyanaraman N, Nemesh J, et al. Nat Genet. 1999;22:231–238. doi: 10.1038/10290. [DOI] [PubMed] [Google Scholar]
- 2.Akashi H. Genetics. 1999;151:221–238. doi: 10.1093/genetics/151.1.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fay J, Wycoff GJ, Wu C-I. Nature. 2002;415:1024–1026. doi: 10.1038/4151024a. [DOI] [PubMed] [Google Scholar]
- 4.Charlesworth J, Eyre-Walker A. Mol Biol Evol. 2006;23:1348–1356. doi: 10.1093/molbev/msk025. [DOI] [PubMed] [Google Scholar]
- 5.Hughes AL. Genetics. 2005;169:533–538. doi: 10.1534/genetics.104.036939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andolfatto P. Nature. 2005;437:1149–1152. doi: 10.1038/nature04107. [DOI] [PubMed] [Google Scholar]
- 7.Drake JA, Bird C, Nemesh J, Thomas DJ, Newton-Cheh C, Reymond A, Excoffier L, Attar H, Antonarakis SE, Dermitzakis ET, Hirschhorn JN. Nat Genet. 2006;38:223–227. doi: 10.1038/ng1710. [DOI] [PubMed] [Google Scholar]
- 8.Asthana S, Noble WS, Kryukov G, Grant CE, Sunyaev S, Stamatoyannopoulos JA. Proc Natl Acad Sci USA. 2007;104:12410–12415. doi: 10.1073/pnas.0705140104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bromham L, Leys R. Mol Biol Evol. 2005;22:1393–1402. doi: 10.1093/molbev/msi133. [DOI] [PubMed] [Google Scholar]
- 10.Johnson KP, Seger J. Mol Biol Evol. 2001;18:874–881. doi: 10.1093/oxfordjournals.molbev.a003869. [DOI] [PubMed] [Google Scholar]
- 11.Li W-H, Tanimura M, Sharp PM. J Mol Evol. 1987;25:330–342. doi: 10.1007/BF02603118. [DOI] [PubMed] [Google Scholar]
- 12.Ohta T. Proc Natl Acad Sci USA. 1993;90:4548–4551. doi: 10.1073/pnas.90.10.4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohta T. J Mol Evol. 1995;40:56–63. doi: 10.1007/BF00166595. [DOI] [PubMed] [Google Scholar]
- 14.Woolfit M, Bromham L. Mol Biol Evol. 2003;20:1545–1555. doi: 10.1093/molbev/msg167. [DOI] [PubMed] [Google Scholar]
- 15.Woolfit M, Bromham L. Proc Biol Sci. 2005;272:2277–2282. doi: 10.1098/rspb.2005.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keightley PD, Lercher MJ, Eyre-Walker A. PLoS Biol. 2005;3:e42. doi: 10.1371/journal.pbio.0030042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keightley PD, Kryukov GV, Sunyaev S, Halligan DL, Gaffney DJ. Genome Res. 2005;15:1373–1378. doi: 10.1101/gr.3942005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berlin S, Ellegren H. J Mol Evol. 2006;62:66–72. doi: 10.1007/s00239-005-0067-6. [DOI] [PubMed] [Google Scholar]
- 19.Bachtrog D. Genome Res. 2005;15:1393–1401. doi: 10.1101/gr.3543605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bartolome C, Charlesworth B. Genetics. 2006;174:2033–2044. doi: 10.1534/genetics.106.064113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gerber AS, Loggins R, Kumar S, Dowling T. Ann Rev Genet. 2001;35:539–566. doi: 10.1146/annurev.genet.35.102401.091106. [DOI] [PubMed] [Google Scholar]
- 22.Nachman MW. Genetica. 1998;102:61–69. [PubMed] [Google Scholar]
- 23.Rand DM, Kann LM. Genetica. 1998;102/103:393–407. [PubMed] [Google Scholar]
- 24.Gillespie JH. Genetics. 1994;138:943–952. doi: 10.1093/genetics/138.3.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohta T, Tachida H. Genetics. 1990;126:219–229. doi: 10.1093/genetics/126.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tachida H. Genetics. 1991;128:183–192. doi: 10.1093/genetics/128.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bulmer M. Genetics. 1991;129:897–907. doi: 10.1093/genetics/129.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li W-H. J Mol Evol. 1987;24:337–345. doi: 10.1007/BF02134132. [DOI] [PubMed] [Google Scholar]
- 29.DePristo MA, Weinreich DM, Hartl DL. Nat Rev Genet. 2005;6:678–687. doi: 10.1038/nrg1672. [DOI] [PubMed] [Google Scholar]
- 30.Loewe L, Charlesworth B, Bartolome C, Noel V. Genetics. 2006;172:1079–1092. doi: 10.1534/genetics.105.047217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piganeau G, Eyre-Walker A. Proc Natl Acad Sci USA. 2003;100:10335–10340. doi: 10.1073/pnas.1833064100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McDonald JH, Kreitman M. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- 33.Sawyer SA, Hartl DL. Genetics. 1992;132:1161–1176. doi: 10.1093/genetics/132.4.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sawyer S, Kulathinal RJ, Bustamante CD, Hartl DL. J Mol Evol. 2003;57:S154–S164. doi: 10.1007/s00239-003-0022-3. [DOI] [PubMed] [Google Scholar]
- 35.Bustamante CD, Fledel-Alon A, Williamson SH, Nielsen R, Todd Hubisz M, Glanowski S, Tanenbaum DM, White TJ, Sninsky JJ, Hernandez RD, et al. Nature. 2005;437:1153–1157. doi: 10.1038/nature04240. [DOI] [PubMed] [Google Scholar]
- 36.Bustamante CD, Nielsen R, Sawyer SA, Olsen KM, Purugganan MD, Hartl DL. Nature. 2002;416:531–534. doi: 10.1038/416531a. [DOI] [PubMed] [Google Scholar]
- 37.Sawyer SA, Parsch J, Zhang Z, Hartl DL. Proc Natl Acad Sci USA. 2007;104:6504–6510. doi: 10.1073/pnas.0701572104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohta T. Ann Rev Ecol Syst. 1992;23:263–286. [Google Scholar]
- 39.Takano-Shimizu T. Genetics. 1999;153:1285–1296. doi: 10.1093/genetics/153.3.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kimura M. The Neutral Theory of Molecular Evolution. Cambridge, UK: Cambridge Univ Press; 1983. [Google Scholar]
- 41.Eyre-Walker A, Woolfit M, Phelps T. Genetics. 2006;173:891–900. doi: 10.1534/genetics.106.057570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Loewe L, Charlesworth B. Biol Lett. 2006;2:426–430. doi: 10.1098/rsbl.2006.0481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chao L, Carr DE. Evolution (Lawrence, Kans) 1993;47:688–690. doi: 10.1111/j.1558-5646.1993.tb02124.x. [DOI] [PubMed] [Google Scholar]
- 44.Kimura M. Proc Natl Acad Sci USA. 1979;76:3440–3444. doi: 10.1073/pnas.76.7.3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ohta T. In: Molecular Evolution and Polymorphism. Kimura M, editor. Mishima, Japan: Natl Inst Genetics; 1977. pp. 148–167. [Google Scholar]
- 46.Raxworthy CJ, Forstner MR, Nussbaum RA. Nature. 2002;415:784–787. doi: 10.1038/415784a. [DOI] [PubMed] [Google Scholar]
- 47.Chen Y, Carlini DB, Baines JF, Parsch J, Braverman JM, Tanda S, Stephan W. Genes Genet Syst. 1999;74:271–286. doi: 10.1266/ggs.74.271. [DOI] [PubMed] [Google Scholar]
- 48.Choi SS, Li W, Lahn BT. Nat Genet. 2005;37:1367–1371. doi: 10.1038/ng1685. [DOI] [PubMed] [Google Scholar]
- 49.Sharp PM, Burgess CJ, Lloyd AT, Mitchell KJ. Hatfield DL, Lee BJ, Pirtle RM. Transfer RNA in Protein Synthesis. Boca Raton, FL: CRC; 1992. [Google Scholar]
- 50.Duret L. Curr Opin Genet Dev. 2002;12:640–649. doi: 10.1016/s0959-437x(02)00353-2. [DOI] [PubMed] [Google Scholar]
- 51.Bazin E, Glemin S, Galtier N. Science. 2006;312:570–572. doi: 10.1126/science.1122033. [DOI] [PubMed] [Google Scholar]
- 52.Mulligan CJ, Kitchen A, Miyamoto MM. Science. 2006;314:1390a. doi: 10.1126/science.1132585. [DOI] [PubMed] [Google Scholar]
- 53.Chimpanzee Sequencing and Analysis Consortium. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 54.Zhang L, Li W-H. Mol Biol Evol. 2005;22:2504–2507. doi: 10.1093/molbev/msi240. [DOI] [PubMed] [Google Scholar]
- 55.Bierne N, Eyre-Walker A. Mol Biol Evol. 2004;21:1350–1360. doi: 10.1093/molbev/msh134. [DOI] [PubMed] [Google Scholar]
- 56.Smith NGC, Eyre-Walker A. Nature. 2002;415:1022–1024. doi: 10.1038/4151022a. [DOI] [PubMed] [Google Scholar]
- 57.Nielsen R, Yang Z. Mol Biol Evol. 2003;20:1231–1239. doi: 10.1093/molbev/msg147. [DOI] [PubMed] [Google Scholar]
- 58.Williamson SH. Mol Biol Evol. 2003;20:1318–1325. doi: 10.1093/molbev/msg144. [DOI] [PubMed] [Google Scholar]
- 59.Nicholson KE, Glor RE, Kolbe JJ, Larson A, Hedges SB, Losos JB. J Biogeogr. 2005;32:929–938. [Google Scholar]
- 60.Filardi CE, Moyle RG. Nature. 2005;438:216–219. doi: 10.1038/nature04057. [DOI] [PubMed] [Google Scholar]
- 61.Yokoyama J, Suzuki M, Iwatsuki K, Hasebe M. Mol Phyl Evol. 2000;14:11–19. doi: 10.1006/mpev.1999.0672. [DOI] [PubMed] [Google Scholar]
- 62.Crawford AJ, Smith EN. Mol Phyl Evol. 2005;35:536–555. doi: 10.1016/j.ympev.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 63.Yang Z. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.