Abstract
Plant roots harbor a large diversity of microorganisms that have an essential role in ecosystem functioning. To better understand the level of intimacy of root-inhabiting microbes such as arbuscular mycorrhizal fungi and bacteria, we provided 13CO2 to plants at atmospheric concentration during a 5-h pulse. We expected microbes dependent on a carbon flux from their host plant to become rapidly labeled. We showed that a wide variety of microbes occurred in roots, mostly previously unknown. Strikingly, the greatest part of this unsuspected diversity corresponded to active primary consumers. We found 17 bacterial phylotypes co-occurring within roots of a single plant, including five potentially new phylotypes. Fourteen phylotypes were heavily labeled with the 13C. Eight were phylogenetically close to Burkholderiales, which encompass known symbionts; the others were potentially new bacterial root symbionts. By analyzing unlabeled and 13C-enriched RNAs, we demonstrated differential activity in C consumption among these root-inhabiting microbes. Arbuscular mycorrhizal fungal RNAs were heavily labeled, confirming the high carbon flux from the plant to the fungal compartment, but some of the fungi present appeared to be much more active than others. The results presented here reveal the possibility of uncharacterized root symbioses.
Keywords: ribosomal RNA, stable isotope probing, symbiosis, arbuscular mycorrhiza, endophytes
Plants are the dominant primary producers in most terrestrial ecosystems. In the soil, they are escorted by a myriad of microorganisms living freely or in intimate interaction with their roots (1, 2). These microorganisms can be pathogenic, parasitic, saprotrophic, or mutualistic. Among the root symbionts, arbuscular mycorrhizal (AM) fungi are well known and have been observed colonizing the roots of most plant species in many ecosystems (3). Recent studies show that high diversity is the norm even where plant diversity is low (4–6). These AM fungi are biotrophs, unable to grow in the absence of a living plant, and often display a broad host range although there is growing evidence for differences in host preference (5–8). They have been demonstrated to improve plant mineral nutrition (3) and stress resistance (3). AM fungi are important for the global carbon cycle because up to 20% of photoassimilates can be translocated to them (9). We also know that the diversity of AM fungi can determine plant community structure and ecosystem productivity (10). The plant–bacteria symbioses are variably documented. The best studied symbiosis is the rhizobium–legume interaction, but a single plant root can harbor a large variety of fungi (1) and bacteria (2), as well as several different archaea (2). So far we have no information about the functions of most of these root-living microbes. The strategy chosen herein, stable isotope probing (SIP)–RNA analysis, enabled us to highlight an unsuspected diversity of microbes living in roots. We identified microbes that are active and direct utilizers of photosynthetic carbon from the plant by demonstrating a differential carbon flow to them.
Results and Discussion
Several methods have been developed recently to analyze the functional diversity of microorganisms in situ without preliminary cultivation or isolation. Here we use SIP–RNA (11) based on the fractionation of heavily labeled [13C]RNAs from a mixture after providing CO2 enriched in the stable isotope 13C. The enriched RNAs can be assigned to microbial taxon by analyzing the small subunit ribosomal RNA (SSU rRNA). RNA becomes labeled more rapidly and heavily than DNA because ribosomes are more abundant and turn over faster than DNA, and transcription is not semiconservative. Hence, SIP–RNA allows analysis of shorter term responses than SIP–DNA (i.e., primary consumers are targeted before the label can reach secondary consumers).
We used a pulse of 13CO2 to label turfs lifted from an upland grassland in the United Kingdom (experiment A) and a regenerating peatland in France (experiment B). The vegetation included grasses (predominantly Agrostis capillaris, Festuca rubra, Poa pratensis) and white clover (Trifolium repens) for the upland grassland and Agrostis stolonifera, Eriophorum angustifolium, and Hydrocotyle vulgaris for the peatland. The purpose of experiment A was to identify and compare the AM fungal communities associated with the co-occurring species, A. capillaris and T. repens, and to characterize their behavior with regard to the carbon flux from the plant. Experiment B surveyed a wider range of microorganisms (bacteria as well as AM fungi) associated with the roots of A. stolonifera. To focus on the primary consumers of current photosynthates, the time of exposure to 13C assimilates must be reduced to a minimum. In a previous field experiment that provided a 13CO2 pulse at atmospheric concentration to plants, the maximum enrichment of microbial RNA was reached 3 h after the end of a 6-h pulse (12). Furthermore, microorganisms have been demonstrated to rapidly metabolize 13C-labeled primary metabolites released directly in soil (13). Bearing these results in mind, we reduced the pulse to 5 h with harvesting directly after completion.
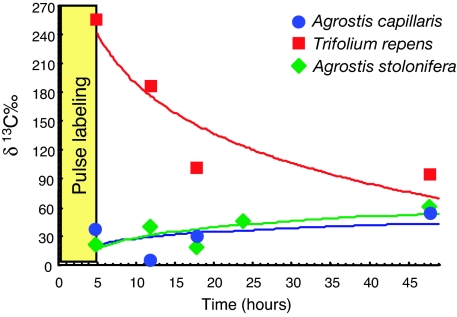
The overall isotopic signature (δ 13C) demonstrated that the roots of all three plant species were enriched in 13C immediately after the end of the pulse labeling (Fig. 1). Carbon translocation was especially rapid to the roots of T. repens, consistent with the high AM mycelial carbon respiration that was recently demonstrated in this species (14). It is likely that demand by rhizobia in root nodules also contributed to the early accumulation of 13C in clover roots. By contrast, 13C built up in Agrostis roots over several days, although our results demonstrated that microbes still exploited a 13C-rich pool of recent photosynthates.
Fig. 1.
Isotopic signatures (δ 13C‰) of roots after 13CO2 pulse labeling. Each dot represents a mean of four measures (i.e., not true replicates). Results of δ 13C‰ were obtained for T. repens (squares) and A. capillaris (circles) in experiment A and from A. stolonifera (diamonds) in experiment B. Fitted regression curves are shown. Natural values of δ 13C‰ (control before labeling) were −35 for T. repens roots, −28 for A. capillaris, and −30 for A. stolonifera.
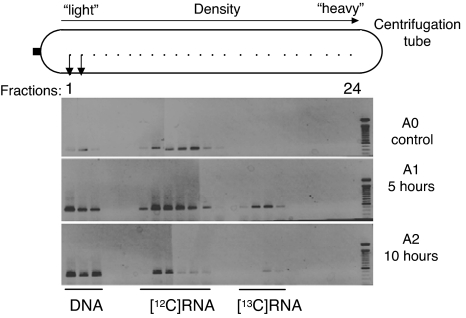
To identify the AM fungal and bacterial communities colonizing the roots, we used primers specific for the amplification of the SSU rRNA of AM fungi (4) and specific for bacteria (modified from ref. 15). Isopycnic ultracentrifugation of the RNA extracts by using cesium trifluoroacetate (CsTFA) was followed by fractionation of the gradient and RNA precipitation. Direct PCR on each fraction showed that only low-density fractions 1, 2, and 3 gave a positive signal indicating the presence of DNA (data not shown). This DNA was used to assess the overall diversity of AM fungi in the roots for experiment A. From these direct PCR amplifications, it can be concluded that fractions 4–24 did not contain detectable DNA, so any amplification in the RT-PCR reflected RNA only. In the unlabeled control, RT-PCR products were seen in fractions 7–13 (Fig. 2). After the 13CO2 pulse, additional bands appeared in the higher density fractions (fractions 15–18 for A1 and 17–18 for A2) (Fig. 2), which we interpret as being amplified from the 13C-labeled (heavy) RNA, whereas unlabeled RNA was found in fractions 7–12 (Fig. 2). We argue that light RNA did not contaminate the heavy fractions because no positive clones were recovered from fractions 17 and 18 of the unlabeled control. The gap between the light and heavy RNAs (e.g., A1 fraction 14) (Fig. 2) indicates that the labeling was intense. This finding is supported by estimated buoyant densities of 1.78–1.80 g/ml for the unlabeled and 1.82–1.85 g/ml for the labeled RNA, comparable with values reported for pure [12C]rRNA and pure [13C]rRNA, respectively (16). In addition, there was no delay: heavy RNA was found immediately after the end of the 5-h pulse labeling. Similar results were obtained in experiment B. The main conclusion from these observations is that the AM fungi were preferentially using assimilates provided by plants (labeled molecules), rather than previously fixed carbon (unlabeled). Traces of heavy RNA were still detected 43 h after the end of the 13CO2 pulse labeling (experiment A) (data not shown). For the bacteria, we found similar results, with positive RT-PCRs and heavily labeled RNA in fraction 17 (heavy RNAs). Compared with the results for the AM fungi, the only difference is a positive signal that is uninterrupted from fractions 5–17 (experiment B) (data not shown). Thus, depending on the bacteria and their ecological status, different proportions of 13C-labeled photosynthates were incorporated. From an ecological point of view, bacterial symbionts (i.e., root-inhabiting bacteria, whether mutualistic or parasitic) can be expected to be heavily labeled in the same way as the AM fungi, whereas saprotrophic bacteria should receive little direct photosynthate.
Fig. 2.
RT-PCR products from each fraction collected from the CsTFA gradient. RNA extracts from T. repens were amplified by RT-PCR specific for AM fungi, using samples before (A0), immediately after (A1), and 5 h after (A2) the 5-h 13CO2 labeling period.
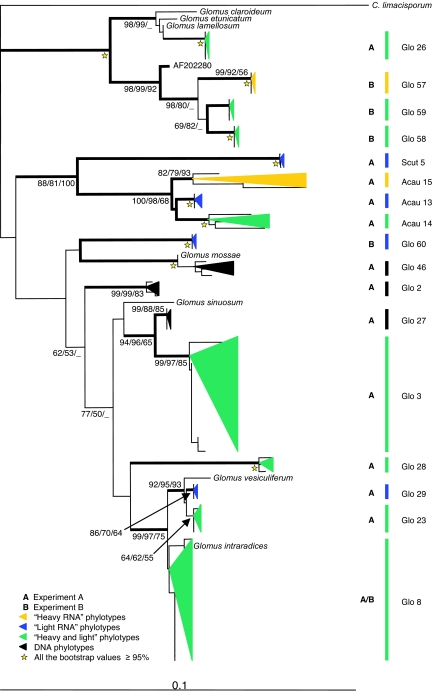
Cloning and sequencing of the various PCR products identified 17 phylotypes (phylogenetically related sequences) of AM fungi colonizing the roots (Fig. 3). By computing rarefaction curves using a random resampling procedure and bootstrap (17) from the 25 to 30 sequenced clones from each clone library, we demonstrate that we have not underestimated the diversity of AM fungal phylotypes in any of the RNA samples [supporting information (SI) Fig. 5]. In the case of the DNA samples, however, a greater sampling effort probably would have uncovered additional phylotypes because the corresponding curves have not reached their asymptotes (SI Fig. 5). These results were confirmed by using a Bayesian estimator of the diversity for noninvasive sampling (18). All of the sequences were related to recognized Glomus, Acaulospora, and Scutellospora species, although none was close enough to be assigned to them. Only 4 phylotypes have been reported from other field populations in previous studies, whereas 13 phylotypes (Glo26–Glo29, Glo46, Glo57–Glo60, Acau13–Acau15, and Scut5) were potentially new (Fig. 3). In experiment A, seven phylotypes were found exclusively within the AM fungal community colonizing A. capillaris and three for T. repens, whereas only three were shared by the two plants (SI Table 1). Despite the co-occurrence of the plant species in the same turf, the AM fungal community composition differed among host plants (SI Table 1), in agreement with previous studies (5–7).
Fig. 3.
Phylogenetic affinities of SSU rRNA representing Glomeromycota (AM fungi). Sequences amplified from T. repens, A. capillaris, and A. stolonifera roots are represented, along with representatives of the relevant known groups. The outgroup is a putative choanoflagellate Corallochytrium limacisporum (L42528). The tree was computed by NJ. (Scale bar: 0.1 substitutions per site.) The data were additionally analyzed by using MP and ML. Branches in bold were congruent among the three phylogenetic reconstructions (at least two bootstrap values of >70%). Bootstrap values of >50% are indicated at the nodes (NJ/MP/ML estimated from 1,000/500/100 iterations, respectively). Phylotypes are presented as colored triangles proportional to the number of clones found and their phylogenetic depth: black, DNA fraction; yellow, heavy RNA fraction; blue, light RNA fraction; green, sequences found in both RNA fractions. A, experiment A; B, experiment B.
For both plant species in experiment A, the heavy RNA corresponded to a subset of the diversity found in the light RNA or DNA (SI Table 1). In each T. repens sample, half of the AM fungal phylotypes were recovered from the heavy RNA and hence had received and metabolized 13C-labeled assimilates. The active AM fungi were different in the two root samples. Glomus Glo8, active in A1, is present in A2, but was not detected in the heavy RNA fraction, whereas Glomus Glo3 was apparently active in A2, but inactive in A1. It is likely that these Glomus species are using the same carbon assimilates. Hence, we can hypothesize that the observed variations in fungal communities are a consequence of competition among colonizers in the same ecological niche. We conclude that not only is the distribution of AM fungi among roots of T. repens highly heterogeneous, but so is their level of activity. Analyses of relative abundance from DNA studies, which have already revolutionized our view of the ecology of these key symbionts (4–7), may therefore give a misleading view of the active populations. A proposed explanation for the stability of the AM fungi–plant mutualistic association over 450 million years is a trade exchange of resources between partners enforced by embargo by one or both members of the symbiosis (19). Our results do not allow us to reject this hypothesis of sanction, but they also substantiate the hypothesis of host-plant preference and competition among AM fungal colonizers (5, 6, 20).
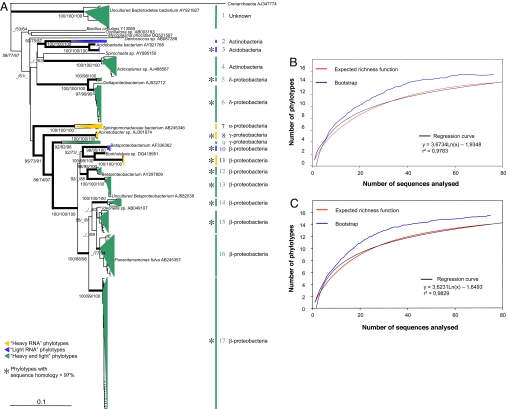
The A. stolonifera root-colonizing bacteria were analyzed from RNA in fraction 7 (unlabeled) and fraction 18 (heavily labeled) of experiment B. The statistical analyses of the bacterial phylotypes allow us to conclude that the sampling was large enough to describe the diversity for both light and heavy fractions (Fig. 4 B and C). The regression curves show that one or two additional phylotypes should be expected if the number of sequences were doubled. The G + C composition was similar for all RNA phylotypes from both light and heavy fractions and thus should not introduce any bias. The phylogenetic reconstructions segregate bacteria into 17 phylotypes (with ≤3% internal sequence divergence, except for 6 phylotypes that were slightly more heterogeneous). Five strongly supported phylotypes cannot be identified because they are too distant from any published sequences other than from environmental samples (Fig. 4). Phylotype 1 forms a particularly deep branch within the bacteria, whereas phylotypes 10–13 are within the class Betaproteobacteria. We can argue that these bacteria are not parasitic because (i) thorough examination of the plant roots indicated that they were healthy, and (ii) no long branches were observed in the phylogeny (typical of parasites because of higher mutation rate) (21). A number of sequences were close to the Burkholderiales, a group that includes both pathogens and symbionts. Several Burkholderia spp. have developed symbioses with plants and are found in roots, leaves, and stems (22). Furthermore, they colonize AM fungi as endosymbionts (23), and several are described as symbionts of Rhizopus, a fungus belonging to the Zygomycota, conferring on the fungus a certain level of pathogenicity toward plants (24). Another group of putative Burkholderiales sequences were close to those of nitrogen-fixing Ideonella sp. isolated from rice stems and roots (25).
Fig. 4.
Phylogenetic affinities of bacterial SSU rRNA sequences and statistical analyses of sampling effort. (A) Tree derived by ML analysis of all 151 bacterial sequences amplified from A. stolonifera roots, with representatives of major bacterial groups and the highest BLAST hit of each phylotype. The outgroup is an uncultured crenarchaeote (AJ347774). Branches in bold were congruent among MP, ML, and NJ phylogenetic reconstructions (at least two bootstrap values of >70%). Bootstrap values of >50% are indicated at the nodes (NJ/MP/ML estimated from 1,000/500/100 iterations, respectively). (Scale bar: 0.1 substitutions per site.) Phylotypes are presented as colored triangles proportional to the number of clones found and their phylogenetic depth: yellow, heavy RNA fraction; blue, light RNA fraction; green, sequences found in both RNA fractions. Asterisks indicate phylotypes in which all sequences have >97% identity. (B) Estimates of bacterial community diversity as a function of sampling effort for A. stolonifera root samples. Rarefaction curves are computed from 100 replicates of bootstrap (blue), and the expected richness function (red) is the number of phylotypes estimated (random sampling without replacement) from the sequences analyzed for heavy SSU RNA (i.e., 13C-labeled). (C) Estimates of bacterial community diversity for light SSU RNA.
The metagenomic approach using stable isotopes (26) has allowed us to describe the community diversity of AM fungi and bacteria and to relate this diversity to a functional trait, the consumption of newly formed photosynthates. An extension of our approach would probably reveal the presence of additional microorganisms, including archaea and a wider range of fungi (1). Because we have focused on highly labeled RNAs, we conclude that the microorganisms involved in the process are closely linked with their host plant and are likely to be plant-dependent. However, our results bring more questions than answers. We have much to learn about the ecology of microorganisms in roots, especially of the potentially new bacterial groups described herein.
Materials and Methods
Field Sites and Plant Growth.
In experiment A, six turfs (45 × 35 cm) were collected from the Natural Environment Research Council Soil Biodiversity field experiment at Sourhope near Kelso (Scotland), a seminatural grassland ecosystem (March 2002). Turfs were placed in plastic containers (45 × 35 cm) and acclimated for 3 months at 18°C lit 12 h per day before labeling.
In experiment B, three turfs of the same size were collected from a peatland experiment of the European research program RECIPE, located close to Baupte in Normandy (France) and acclimated similarly (March 2005).
13C Labeling and Total RNA Extraction.
In experiment A, a turf in its container was put into an acrylic air-flow chamber and labeled with 13CO2 (99 atom %) at atmospheric concentration. Air flow to the turf (≈5 liters/min) and CO2 delivery were controlled by measuring the concentration of CO2 in the vent gas, using an infrared gas analyzer. Core samples (4.1-cm diameter) were taken from the turf: A0, before the pulse labeling; A1, immediately after the 5-h labeling; and A2, A3, A4, and A5, 10, 16, 24, and 48 h after the start of the labeling, respectively. From each core, roots of T. repens and A. capillaris were immediately washed in tap water, three times in 0.1% Triton X-100, and five times in sterilized distilled water and then frozen. All roots appeared to be living and healthy (checked under binocular microscope). Total RNA was extracted from ≈30 mg (fresh weight) by using the RNeasy Plant Mini Kit (Qiagen, Valencia, CA). Experiment B was conducted similarly, using roots of A. stolonifera.
Isotopic Signature.
A sample of the remaining roots was dried. The 13C enrichment in root samples reported in δ 13C‰ (13C:12C ratio) was calibrated relative to internal gas standards and solid reference against Pee Dee Belemnite standard. All isotopic signatures (δ 13C) were determined by continuous-flow/combustion/isotope ratio mass spectrometry. Although the intensity of labeling might vary with root age, samples were large and representative enough to even out any such effect. Exponential regression curves were estimated by using Statistica 7.1.
Separation of Labeled RNA from Unlabeled RNA.
Before the experiment, the time, speed, and temperature of the ultracentrifugation and quantities of CsTFA and RNA were optimized to separate [13C]RNA from [12C]RNA. The isopycnic ultracentrifugation was performed using Quick Seal centrifugation tubes (Beckman, Fullerton, CA) filled with 13.5 ml of an aqueous solution of 1.9 g/ml CsTFA (Amersham Biosciences, Piscataway, NJ), to which 50 ng of RNA was added (optimal quantity). Tubes were spun at 48,000 rpm in a 90Ti rotor (Beckman) at 4°C for 48 h. We noticed that RNA degradation occurs (i.e., RNA is broken) when tubes are spun at speeds >55,000 rpm. Twenty-four fractions of the ultracentrifugation gradient were taken, starting from the top of the tube, by puncturing the wall every 0.1 in (2.54 mm), using a needle, syringe, and guide. RNA was precipitated with two volumes of isopropanol. After centrifugation, the pellet was washed in 75% ethanol, dried, and redissolved in 20 μl of ultrapure DNase- and RNase-free water (Sigma–Aldrich, St. Louis, MO). Densities of fractions from two gradients run under the same conditions as the experiments, but without nucleic acid, were estimated by weighing measured volumes.
RT-PCR.
RT-PCR was carried out on 5 μl of each of the 24 fractions from each sample (A0–5, B0–1) by using the Titan One Tube RT-PCR kit (Roche Molecular Systems, Alameda, CA), with primers NS31 and AM1 for the specific amplification of AM fungal SSU rDNA (4) and (for experiment B) the primers Eub_519f and Eub_1390r specific for bacteria (15) in the modified forms 5′-GTTTCAGCMGCCGCGGT-3′ and 5′-GTTTGACGGGGTGTGT-3′, respectively. For AM fungal sequences, reverse transcription at 51°C for 30 min was followed by PCR amplification with 2 min denaturation at 94°C, 35 cycles of 30 sec at 94°C, 1 min at 58°C or 52°C (AM fungi or bacteria, respectively), 50 sec at 68°C, and final extension for 10 min at 68°C.
Cloning and Sequencing.
The RT-PCR yield was low for the later samples (A3–5), so analysis was limited to A1, A2, and B1. The amplified fragments were cloned in pGEM-T vector (Promega, Madison, WI) and DH5α-competent cells (Gibco/BRL, Carlsbad, CA). From each clone library, 25 to 30 randomly selected positive clones were sequenced (ABI-PRISM Dye Terminator Cycle Sequencing Ready Reaction Kit; PerkinElmer, Beltsville, MD) for both strands by using primers T7 or SP6. The two sequences were aligned by using the programs Autoassembler (PerkinElmer) and Sequencher (GeneCodes, Ann Arbor, MI) for experiments A and B, respectively. No chimeric artifacts were found among the clones sequenced of AM fungi, whereas 53 artifacts were detected and deleted from the data set among 204 bacteria sequences by using CHIMERA_CHECK 2.7 (Ribosomal Database Project II; http://rdp.cme.msu.edu).
Diversity and Phylogenetic Analyses.
Rarefaction curves were computed for each data set. The number of species was quantified for 100 random combinations of 1 to N sequences and also by performing 100 bootstrap pseudoreplicates implemented in Estimates (17).
Multiple alignments, one for the AM fungal phylogenetic analyses containing all of the sequences plus a set of eight AM fungal sequences from GenBank/EMBL/DDBJ and one for the 151 bacterial sequences, were performed by using CLUSTALX 1.81 (27) and refined by eye. For the two data sets, the phylogenetic analyses were done as follows: CLUSTALX 1.81 was used for neighbor joining (NJ) phylogenies, with distance correction using K2P and complete omission of gaps. For AM fungi and bacteria, bootstrapping was repeated 200 and 1,000 times, respectively. PAUP 4.0β10 was used for maximum parsimony (MP) by using a heuristic tree search with 500 replicates (for AM fungi, 300 for bacteria) of random addition, tree bisection and reconnection as branching algorithm, and 15 random-addition swaps per replicate. For the bacteria, an identical likelihood score was found for the constructions by using tree bisection and reconnection or subtree pruning-regrafting as swapping algorithms. Then, the bacterial set was studied by using maximum-likelihood (ML) procedures under the HKY model with 200 bootstrap iterations. For the AM fungal data set, the ML phylogeny was constructed under the GTR + I + G model (100 bootstrap iterations). Modeltest 3.7 software (28) was used to select the model.
Supplementary Material
Acknowledgments
We thank Ouest Génopole and Allen Mould for the sequencing and S. Diquelou and S. Lemauviel for labeling of experiment B. This work was supported by the Soil Biodiversity Thematic Program of the Natural Environment Research Council and the Ecosphère Continentale–Fonctionnement et Dynamique de la Biosphère Continentale Program of the Centre National de la Recherche Scientifique.
Abbreviations
- AM
arbuscular mycorrhizal
- CsTFA
cesium trifluoroacetate
- ML
maximum likelihood
- MP
maximum parsimony
- NJ
neighbor joining
- SIP
stable isotope probing
- SSU
small subunit.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AF481533–AF481698, experiment A; AY273534–AY273619, experiment B; EF040887–EF041036, bacteria; EF041037–EF041100, AM fungi).
This article contains supporting information online at www.pnas.org/cgi/content/full/0705902104/DC1.
References
- 1.Vandenkoornhuyse P, Baldauf SL, Leyval C, Straczek J, Young JPW. Science. 2002;295:2051. doi: 10.1126/science.295.5562.2051. [DOI] [PubMed] [Google Scholar]
- 2.Chelius MK, Triplett EW. Microb Ecol. 2001;41:252–263. doi: 10.1007/s002480000087. [DOI] [PubMed] [Google Scholar]
- 3.Smith SE, Read DJ. Mycorrhizal Symbiosis. 2nd Ed. San Diego: Academic; 1997. [Google Scholar]
- 4.Helgason T, Daniell TJ, Husband R, Fitter AH, Young JPW. Nature. 1998;394:431. doi: 10.1038/28764. [DOI] [PubMed] [Google Scholar]
- 5.Vandenkoornhuyse P, Husband R, Daniell TJ, Watson IJ, Duck JM, Fitter AH, Young JPW. Mol Ecol. 2002;11:1555–1564. doi: 10.1046/j.1365-294x.2002.01538.x. [DOI] [PubMed] [Google Scholar]
- 6.Vandenkoornhuyse P, Ridgway KP, Watson IJ, Fitter AH, Young JPW. Mol Ecol. 2003;12:3085–3095. doi: 10.1046/j.1365-294x.2003.01967.x. [DOI] [PubMed] [Google Scholar]
- 7.Husband R, Herre EA, Turner SL, Gallery R, Young JPW. Mol Ecol. 2002;11:2669–2678. doi: 10.1046/j.1365-294x.2002.01647.x. [DOI] [PubMed] [Google Scholar]
- 8.Gollote A, van Tuinen D, Atkinson D. Mycorrhiza. 2004;14:111–117. doi: 10.1007/s00572-003-0244-7. [DOI] [PubMed] [Google Scholar]
- 9.Bago B, Pfeffer PE, Shachar-Hill Y. Plant Physiol. 2000;124:949–957. doi: 10.1104/pp.124.3.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van der Heijden MGA, Klironomos JN, Ursic M, Moutoglis M, Streitwolf-Engel R, Boller T, Wiemken A, Sanders I. Nature. 1998;396:69–72. [Google Scholar]
- 11.Manefield M, Whiteley AS, Griffiths RI, Bailey MJ. Appl Environ Microbiol. 2002;68:5367–5373. doi: 10.1128/AEM.68.11.5367-5373.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rangel-Castro I, Killham K, Ostle N, Nicol GW, Anderson IC, Scrimgeour CM, Ineson P, Meharg A, Prosser J. Environ Microbiol. 2005;7:828–838. doi: 10.1111/j.1462-2920.2005.00756.x. [DOI] [PubMed] [Google Scholar]
- 13.Padmanabhan P, Padmanabhan S, Derito C, Gray A, Gannon D, Snape JR, Tsai CS, Park W, Jeon C, Madsen EL. Appl Environ Microbiol. 2003;69:1614–1622. doi: 10.1128/AEM.69.3.1614-1622.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnson D, Leake JR, Ostle N, Ineson P, Read DJ. New Phytol. 2002;153:327–334. [Google Scholar]
- 15.Orphan VJ, Taylor LT, Hafenbradl D, Delong EF. Appl Environ Microbiol. 2000;66:700–711. doi: 10.1128/aem.66.2.700-711.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Whiteley AS, Thomson B, Lueders T, Manefield M. Nat Protoc. 2007;2:838–844. doi: 10.1038/nprot.2007.115. [DOI] [PubMed] [Google Scholar]
- 17.Colwell RK, Mao CX, Chang J. Ecology. 2004;85:2717–2727. [Google Scholar]
- 18.Petit E, Valière N. Conserv Biol. 2006;20:1062–1073. doi: 10.1111/j.1523-1739.2006.00417.x. [DOI] [PubMed] [Google Scholar]
- 19.Kiers ET, van der Heijden MGA. Ecology. 2006;87:1627–1636. doi: 10.1890/0012-9658(2006)87[1627:msitam]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 20.Fitter AH. New Phytol. 2006;172:3–6. doi: 10.1111/j.1469-8137.2006.01861.x. [DOI] [PubMed] [Google Scholar]
- 21.Gribaldo S, Philippe H. Theor Popul Biol. 2002;61:391–408. doi: 10.1006/tpbi.2002.1593. [DOI] [PubMed] [Google Scholar]
- 22.Coenye T, Vandamme P. Environ Microbiol. 2003;5:719–729. doi: 10.1046/j.1462-2920.2003.00471.x. [DOI] [PubMed] [Google Scholar]
- 23.Bianciotto V, Bandi C, Minerdi D, Sironi M, Tichy HV, Bonfante P. Appl Environ Microbiol. 1996;62:3005–3010. doi: 10.1128/aem.62.8.3005-3010.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Partida-Martinez LP, Hertweck C. Nature. 2005;437:823–824. doi: 10.1038/nature03997. [DOI] [PubMed] [Google Scholar]
- 25.Elbeltagy A, Nishioka K, Sato T, Suzuki H, Ye B, Hamada T, Isawa T, Mitsui H, Minamisawa K. Appl Environ Microbiol. 2001;67:5285–5293. doi: 10.1128/AEM.67.11.5285-5293.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dumont MG, Murrell JC. Nat Rev Microbiol. 2005;3:499–504. doi: 10.1038/nrmicro1162. [DOI] [PubMed] [Google Scholar]
- 27.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Posada D, Crandall KA. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.