Abstract
Insights into the structural basis of protein–protein recognition have come principally from the analysis of proteins such as antibodies, hormone receptors, and proteases that bind their ligands with relatively high affinity (Ka ≈ 109 M−1). In contrast, few studies have been done on the very low affinity interactions mediating cell adhesion and cell–cell recognition. As a site of protein–protein recognition, the ligand binding face of the T lymphocyte cell–cell recognition molecule, CD2, which binds its ligands 104- to 105-fold more weakly than do antibodies and proteases, is unusual in being both very flat and highly charged. An analysis of the effect of mutations and ionic strength on CD2 binding to its ligand, CD48, indicates that these charged residues contribute little, if any, binding energy to this interaction. However, the loss of these charged residues is shown to markedly reduce ligand-binding specificity. Thus, the charged residues increase the specificity of CD2 binding without increasing the affinity. This phenomenon is likely to result from a requirement for electrostatic complementarity between charged binding surfaces to compensate for the removal, upon binding, of water interacting with the charged residues. It is proposed that this mode of recognition is highly suited to biological interactions requiring a low affinity because it uncouples increases in specificity from increases in affinity.
The T-lymphocyte cell surface antigen, CD2, binds to the structurally related molecules CD48 in mice and rats and to CD58 in humans (1). Studies of the interactions of these molecules are providing important insights into the structural basis of protein recognition at the cell surface (1, 2).
In the absence of crystal structures of complexes of these pairs of molecules, it was proposed (3) that homophilic, twofold rotationally symmetric crystal lattice contacts seen initially in crystals of rat soluble CD2 (sCD2) and, subsequently, in crystals of human sCD2 (4), represent a suitable model for the interactions of CD2 with its ligands. A complementary mutagenesis study of the interactions of rat CD2 and CD48 strongly supported this contention (5). The crystal lattice contact is mediated by the GFCC′C" face of domain 1 of CD2 which, as a site of protein–protein interactions, has two unusual properties (4). First, the solvent-exposed surface of the GFCC′C" β-sheet is much flatter than the ligand-binding sites of other molecules involved in protein recognition, such as antibodies. It is generally thought that the surface–shape complementarity of the largely nonplanar interfaces of most protein complexes plays a central role in generating both binding specificity and high affinity (6). Second, the GFCC′C" face is notable for its unusually high density of charged residues: 45 and 70% of the residues buried in the rat and human sCD2 crystal contacts are charged, respectively. In comparison, on average, only 29% of the residues constituting the solvent-exposed surfaces of proteins are charged (7). In general, the composition of the ligand-binding sites of the best characterized protein–protein complexes, i.e., those binding with relatively high affinity, is not significantly different from that of the average protein surface and may in fact exhibit a slight tendency toward increased hydrophobicity (8, 9). The exclusion of water from hydrophobic residues in the interface is an important source of the energy driving complex formation (6).
An important property of the interaction of CD2 with its ligands is that binding is very weak. Surface plasmon resonance-based analyses have indicated that the very low affinities of these interactions (Ka = 104–105 M−1) are characterized by extremely fast off-rates (koff > 4 s−1) (10, 11). More recent analyses of the interactions of the leukocyte cell surface antigens CD80 (12) and CD62L (13) with their respective ligands suggest that low affinities and very fast binding kinetics may be general features of the molecular interactions mediating leukocyte cell–cell recognition. This presumably reflects the fact that specific cell–cell recognition generally involves the simultaneous engagement of hundreds, if not thousands, of molecules (2).
In the present study, an analysis of the effects of mutations and ionic strength on the binding of CD2 to CD48 has been undertaken to establish a link between the unusual structural properties of the ligand-binding face of CD2 and its ability to mediate weak but specific protein recognition at the cell surface. The results suggest that the clustering of energetically neutral charged residues in the ligand-binding face may ensure that protein recognition by CD2 has a high degree of specificity while remaining very weak.
MATERIALS AND METHODS
Mutant Construction.
Most of the CD2 mutants were generated after subcloning the domain 1-encoding region of the rat CD2 gene into M13 and mutating this sequence with a commercially available kit (Sculptor, Amersham). The mutants were expressed and purified in the form of fusion proteins consisting of glutathione S-transferase fused with the amino terminus of CD2 domain 1 (CD2d1-GST) as described (14). In a few cases (R31Y, S52E, T86A, and N90K), the mutations were introduced into a soluble chimeric protein (sCD2-CD4) comprising the entire extracellular portion of rat CD2 fused to domains 3 and 4 of rat CD4 (15). Sequence encoding sCD2-CD4 (16) was excised from the pEE14 vector using HindIII and EcoRI, blunt-ended, and subcloned by blunt-ended ligation into the XbaI site of the phagemid-eukaryotic expression vector pEF-BOS (17). CD2 mutants were generated directly in the sCD2-CD4 construct by using the Muta-Gene Phagemid Mutagenesis Kit v. 2 (Bio-Rad). For CD48 mutants, DNA encoding a soluble rat CD48-CD4 chimeric protein (sCD48-CD4) in pEF-BOS was mutated as described (5). Mutant forms of sCD2-CD4 and sCD48-CD4 were expressed in COS-7 cell tissue culture supernatants also as described (5). All of the mutants were checked by dideoxy sequencing of the entire CD2 or CD48 sequence using the Applied Biosystems model 373A DNA sequencing system. Binding to mAbs was determined as described (18). All of the mutants bound both of the domain 1 specific antibodies OX34 and OX55 except D28K, D28A, E41R, and K43E, which did not bind OX34, and R70E, which did not bind OX55.
Binding Studies.
All Biacore experiments were performed on BIACORE 1000 or BIACORE 2000 instruments (Biacore AB, Stevenage, Herts, UK) at 25°C in the running buffer HBS (150 mM NaCl/1 mM CaCl2/1 mM MgCl2/10 mM Hepes, pH 7.4), and 0.005% Surfactant P-20 (Biacore AB). The mAbs OX55 and OX68 were coupled covalently to CM5 research grade sensor chips (Biacore AB) via primary amines using the standard Amine Coupling Kit (Biacore AB). For coupling, OX55 was injected at 50 μg/ml in 10 mM sodium acetate (pH 5) and OX68 was injected at 80 μg/ml in the same buffer. Following coupling, OX55 was regenerated with 50 mM sodium hydroxide whereas OX68 was regenerated with 0.1 M Gly/hydrochloric acid (pH 2.5). Immobilization levels were typically 11,000 response units (range 8,500–14,000).
RESULTS
Defining the Ligand-Binding Site of CD2.
In the absence of a structure of the complete rat CD2/ligand (CD48) complex, the ligand-binding GFCC′C" face of domain 1 of rat CD2 was mutated exhaustively to define the full extent of the ligand-binding site. Amino acid substitutions intended to maximally alter the chemical properties of solvent-exposed, putative interface residues were chosen at 23 sites on the GFCC′C" face (Table 1, Fig. 1A). Mutations at five additional sites, H12, N17, E56, R70, and D94, were used to confirm that the GFCC′C" face is the only CD48-binding site on domain 1 of CD2. All of the mutants used for this analysis, which were generated by in vitro mutagenesis of DNA constructs encoding CD2 fusion proteins or chimeras, were expressed at levels similar to that of the equivalent wild-type proteins, were highly soluble, and bound at least one anti-CD2 mAb. According to these criteria, each of the mutants was judged to be folded correctly.
Table 1.
Effect of nonconservative or alanine mutations of CD2 residues on CD48 binding
| Residue | Nonconservative mutation
|
Alanine mutation
|
||
|---|---|---|---|---|
| Change | CD48 binding* | Kd† | ΔKd | |
| H12 | D | ++ | nd‡ | nd |
| N17 | D | ++ | nd | nd |
| D26 | K | ++ | 0.8 | nd |
| D28 | K | — | >20 | >20 |
| E29 | R | — | >20 | >20 |
| R31 | Y | — | >20 | >20 |
| E33 | R | — | >20 | 1.5 |
| R34 | D | ++ | 0.4 | 0.9 |
| S36 | E | ++ | 1.2 | nd |
| T37 | K | ++ | 0.5 | nd |
| L38 | Y | — | >20 | >20 |
| E41 | R | — | nd | 1.1 |
| K43 | E | — | nd | 2.0 |
| K45 | E | ++ | 0.5 | nd |
| M46 | Y | ++ | 0.8 | nd |
| K47 | D | ++ | 1.1 | nd |
| F49 | R | — | >20 | >20 |
| K51 | E | ++ | 2.9 | 1.3 |
| S52 | E | ++ | 2.2 | nd |
| E56 | R | ++ | 1.2 | 1.2 |
| R70 | E | ++ | nd | nd |
| T79 | E | ++ | 2.3 | nd |
| Y81 | S | — | >20 | >20 |
| T83 | D | ++ | 2.1 | nd |
| T86 | D | + | >20 | 0.7 |
| R87 | E | — | >20 | 0.2 |
| N90 | K | ++ | 2.4 | nd |
| D94 | K | ++ | 1.9 | nd |
Binding was measured as in Fig. 2 and quantified as follows: ++, binding >60% of wild-type level; +, binding detectable but <5% of wild-type level; — no detectable binding.
Dissociation constant measured by equilibrium binding as in Fig. 3; the figures represent the fold increase in the Kd of sCD48 binding to the mutant compared with wild-type CD2 measured in the same experiment. Where no or little binding was detected at 1.5 mM sCD48 the Kd was assumed to be >1.5 mM. The Kd for sCD48 binding wild-type CD2 was 80 ± 6 (mean ± SD, n = 4).
nd, not done.
Figure 1.
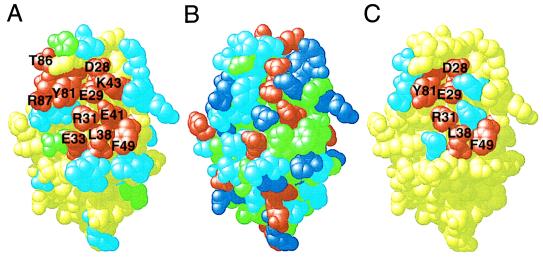
Mutagenesis of domain 1 of CD2. The crystal structure of domain 1 of CD2 (residues 1–99) is shown in Corey, Pauling, and Koltun format drawn using rasmol (31). In each panel, the view is approximately perpendicular to the ligand-binding GFCC′C" β-sheet surface. (A) Residues whose nonconservative substitution significantly interfered with, or had no effect on, ligand (CD48) binding by CD2 are colored red and light blue, respectively. All of the unmutated residues are colored yellow, except for the sites of N-glycosylation (N67, N77, and N84) which are colored green. (B) The chemical composition of domain 1 of CD2 is indicated by coloring acidic residues red, basic residues dark blue, uncharged polar residues light blue, and hydrophobic residues green. (C) Residues whose substitution with alanine reduced ligand-binding affinity >20-fold are colored red and those for which the reduction in affinity was twofold or less are colored light blue. The details of the substitutions, and their effects, are given in Table 1.
The ability of each mutant to bind sCD48 was determined by surface plasmon resonance; representative sensograms are shown in Fig. 2 for the ligand-binding and nonbinding mutants K47D and E41R, respectively. In total, eleven of the 23 mutants of the GFCC′C" face failed to bind ligand strongly (Table 1). The eight charged or polar residues (D28, E29, R31, E33, E41, K43, T86, and R87), two aromatic residues (Y81 and F49), and single aliphatic residue (L38) that were altered in these mutants, and which are concluded on this basis to constitute the ligand-binding site, form a contiguous surface which extends diagonally from the FG loop to the CC′ loop and includes residues in β-strands F, C, C′, and C" (Fig. 1 A and B). This surface, or “structural epitope” as defined by Wells and colleagues (19, 20), is very similar to the protein surfaces forming the conserved crystal contacts mediated by the GFCC′C" face observed in rat and human sCD2 crystals (3, 4) and virtually identical to the putative ligand-binding site identified on the basis of line-width perturbation and chemical shifts observed when two-dimensional NMR spectra of CD2 domain 1 are recorded in the presence of sCD48 (21). A similar mutational analysis has shown that the ligand (CD2)-binding face of CD48 is also highly charged (the structural epitope of CD48 consists of six charged residues and single polar, aliphatic, and aromatic residues) (5).
Figure 2.
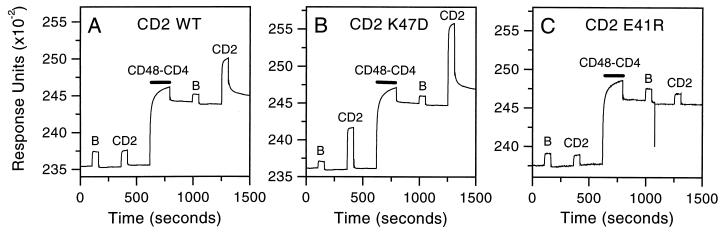
Analysis of the binding of CD2 mutants to CD48. The chimeric protein sCD48-CD4 was bound to the Biacore sensor surface via its CD4 portion by injecting it (0.5 mg/ml for 3 min, black bar) over a sensor surface to which the anti-CD4 antibody OX68 had been covalently coupled. The control protein BSA (B) and wild-type CD2d1-GST (CD2) protein or the indicated mutants of this protein were injected at 0.5 mg/ml for 1 min over the sensor surface both before (as a negative control) and after the sCD48-CD4 had been immobilized. A background response caused by the high protein concentration is seen with injection before sCD48-CD4 immobilization, but the response with wild-type CD2 clearly is increased when injected over immobilized sCD48-CD4 (A), indicating binding. Similarly, the CD2 mutant K47D also binds sCD48-CD4 (B), whereas the mutant E41R does not (C). These experiments were performed at a buffer flow rate of 5 μl/min.
Energetic Contribution of Charged Residues to Ligand Binding.
In other systems, most notably the human growth hormone–receptor interaction (19, 20), alanine-scanning mutagenesis has been used to identify small subsets of binding site residues which contribute the bulk of the ligand-binding energy (defined as the “functional epitope”). To characterize the energetic basis of ligand recognition by CD2, each of the 11 residues forming the structural epitope of the GFCC′C" face of CD2 was substituted with alanine and the affinities of these mutants for monomeric sCD48 were determined (as shown for mutant E41A in Fig. 3). Alanine substitution of five of the eight charged or polar residues forming the structural epitope lead only to modest increases (up to fourfold) or slight decreases (up to twofold) in affinity (Figs. 1C and 3; Table 1). Twentyfold or greater decreases in ligand-binding affinity were observed at only six of the eleven positions forming the structural epitope: D28, E29, R31, L38, F49, and Y81 (Figs. 1C and 3; Table 1).
Figure 3.
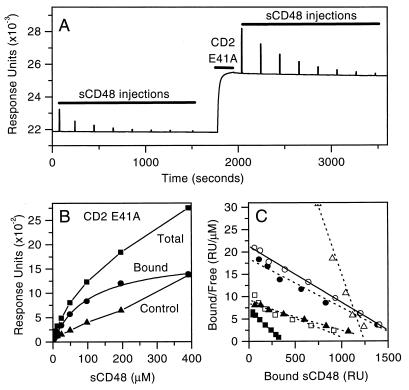
Affinity of CD48 binding to CD2 mutants. (A) The CD2d1-GST mutant E41A (CD2 E41A) was immobilized by injecting it (0.5 mg/ml, 3 min, short bar) at 5 μl/min over a sensor surface to which the anti-CD2 antibody OX55 had been covalently coupled. A range of concentrations (390 μM and seven, twofold dilutions thereof) of sCD48 (long bars) was injected briefly (6 s) at 20 μl/min before (for the background or control response) and after the immobilization of CD2 E41A. (B) The equilibrium response observed during injection of sCD48 at increasing concentrations, before (control, filled triangles) and after (total, filled squares) CD2 E41A immobilization, is plotted. Subtraction of the control from the total response gives the amount of sCD48 bound (filled circles). (C) Linear regression analysis of a Scatchard plot of this data (filled circles) yields a Kd of 91 μM. Also shown are Scatchard plots of the data for sCD48 binding wild-type CD2 (open circles, Kd 80 μM) and the mutants K43A (filled triangles, Kd 160 μM), R87A (open triangles, Kd 16 μM), E33A (open squares, Kd 120 μM), and T86A (filled squares, Kd 54 μM).
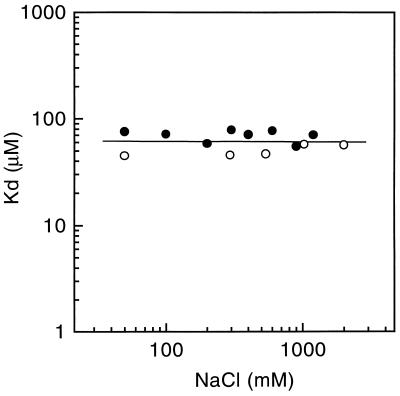
The results of the alanine-scanning mutagenesis raised the possibility that D28, E29, and R31 participate in electrostatically favorable interactions that contribute ligand-binding energy. If this were true, it would be expected that the binding of CD2 to CD48 would be sensitive to electrostatic screening at high salt concentrations. However, the equilibrium-binding affinity of sCD2 for sCD48 was found to be completely unaffected by varying the sodium chloride concentration from 0.05 to 2 M (Fig. 4). This indicates that the net electrostatic component of the ligand-binding energy is zero. Unless there are balancing favorable and unfavorable interactions involving D28, E29, and R31, it can be argued that only three of the 11 residues forming the structural epitope of CD2, i.e., L38, F49, and Y81, are the source of most of the ligand-binding energy. It seems likely that, rather than directly preventing the formation of electrostatically favorable contacts involving D28, E29, and R31, the substitution of alanine at these positions induces local changes to adjacent residues in the ligand-binding site, Y81 and L38 for example, that are unfavorable. It remains a formal possibility that the aliphatic regions of the side chains of these residues are involved in energetically favorable hydrophobic contacts. However, the crystal structure of sCD2 indicates that the aliphatic regions of residues D28, E29, and R31 are poorly exposed.
Figure 4.
Effect of varying the ionic strength on the affinity of CD48 for CD2. The CD48/CD2-binding affinity was measured in Hepes buffer (1 mM CaCl2/1 mM MgCl2/10 mM Hepes, pH 7.4) and 0.005% Surfactant P-20 (Biacore AB) containing the indicated final concentration of sodium chloride. The affinity was measured as in Fig. 3 except that sCD2 was coupled directly to the sensor surface, as described (10), and sCD48 was injected over a blank surface to measure the background (control) response. The open and closed circles represent data from two independent experiments.
Role of Charged Residues in Restricting the Specificity of Ligand Binding.
The mutational and electrostatic-screening data indicate that the charged residues clustered in the ligand-binding site of CD2 contribute little or no energy to ligand binding. It is significant, however, that many of these residues are highly conserved between species. For example, all of the charged residues constituting the structural epitope are conserved in the rat and mouse CD2 protein sequences whereas, overall, only 66% of residues are conserved throughout the entire extracellular domains of rat and mouse CD2 (22). This observation strongly implies that these residues have an important role in ligand binding.
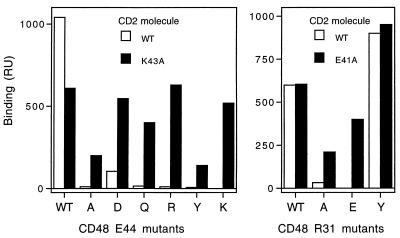
The possibility that the charged residues determine ligand-binding specificity was therefore tested by examining the effects on specificity of mutating E41 and K43 to alanine. Complementary mutagenesis has shown previously that E41 and K43 of CD2 interact with the oppositely charged residues R31 and E44 of CD48, respectively (5). As shown in Fig. 5, wild-type CD2 only binds strongly to wild-type CD48 and the mutant R31Y; binding to E44D is considerably weaker. In contrast, the E41A and K43A mutants of CD2 bind wild-type CD48 and tolerate a series of nine nonconservative substitutions at R31 or E44 of CD48, respectively. These results indicate that CD2 residues E41 and K43 have a significant impact on ligand-binding specificity without contributing to ligand-binding energy.
Figure 5.
Analysis of the specificity of CD2 mutants. Wild-type sCD48-CD4 or mutant forms of this protein with the indicated substitutions at positions 44 (Left) and 31 (Right) in domain 1 of CD48 were immobilized to the sensor surface via their CD4 portions by injecting tissue culture supernatant containing the proteins over a sensor surface to which the anti-CD4 antibody OX68 had been covalently coupled. Wild-type CD2d1-GST (WT) and the CD2 mutants K43A (Left) and E41A (Right) were injected (0.1 mg/ml for 1 min) over the sensor surface before (for control response) and after the binding of wild-type sCD48-CD4 or the sCD48-CD4 mutants. Binding responses were measured and are shown with the control responses subtracted. The CD2 mutants K43A and E41A bound to wild-type sCD48-CD4 and all nine of the immobilized sCD48-CD4 mutants tested whereas the wild-type CD2d1-GST protein only bound strongly to wild-type sCD48-CD4 and the sCD48-CD4 R31Y mutant and weakly to the E44D mutant. These experiments were performed at a buffer flow rate of 3 μl/min.
DISCUSSION
This first analysis of the structural basis of a low affinity interaction typical of cell–cell recognition molecules suggests the following mode of ligand binding by CD2: interactions involving two aromatics and one aliphatic residue generate sufficient energy to allow for weak binding, and the eight charged or polar residues completing the flat ligand-binding site generate a high degree of binding specificity without increasing the affinity of the interaction. Consistent with the proposed binding mechanism, an analysis of the heat capacity of the interaction of CD2 with CD48 has indicated that hydrophobic effects, presumably mediated principally by L38 and the highly conserved residues, F49 and Y81 (22), generate a significant proportion of the total ligand-binding energy (R. O’Brien, P.A.v., S.J.D., and J. E. Ladbury, unpublished data).
An alternative explanation for the clustering of charged residues in the ligand-binding site of CD2 is that these residues maximize association rates through long-range electrostatic interactions as proposed for the interactions of some proteins in solution (23). However, this seems unlikely given that the affinity of CD48 binding to CD2 is unaffected by substantial changes in ionic strength and that the net charge of the ligand-binding site of CD2 is close to zero at physiological pH. In any event, such effects are unlikely to be important for interactions between cell–cell recognition molecules because the association rates for interactions of membrane-bound molecules such as CD2 are limited by slow lateral diffusion in the plane of the membrane (24, 25).
It has recently been pointed out that electrostatic interactions (including salt bridges) can confer specificity on protein–protein interactions and protein folding without necessarily being energetically favorable (26). The underlying reason for this is that charged residues interact favorably with polar solvents such as water. The burial of charged residues upon ligand binding requires disruption of these favorable interactions (desolvation). This unfavorable effect can be overcome if the charged residues form energetically favorable-electrostatic interactions with the ligand-binding surface. The net result frequently will be energetically neutral. Thus, by introducing a requirement for electrostatic complementarity on the adjacent ligand-binding surface, these charged residues increase the specificity of binding without increasing the affinity. This mode of recognition is distinct from the paradigm that has emerged from the analysis of high affinity interactions in which binding specificity and affinity are both largely dependent on surface–shape complementarity between relatively hydrophobic surfaces (6).
The results presented here suggest that CD2 exploits energetically neutral-electrostatic interactions to combine a high degree of specificity with a low affinity. Electrostatic complementarity may be used to generate specificity in low affinity interactions because, unlike surface–shape complementarity, it uncouples increases in specificity from increases in affinity. The question therefore arises as to whether or not other low affinity protein–protein interactions at the cell surface exploit electrostatic interactions in this way. Crystal structures have recently been solved for complexes involving homotypic [N-cadherin (27) and peripheral nerve myelin P0 (28)] and heterotypic [T cell antigen receptor (TCR) with HLA-A2 plus peptide antigen (29) and CD8 with HLA-A2 (30)] cell–cell recognition molecules, and it is clear that charged residues are not clustered at the interfaces of these molecules to the extent observed for CD2. Additional work is required to elucidate the structural basis of the binding affinity and specificity of these interactions. However, a striking example of the contribution of a charged residue to binding specificity has recently emerged from a comparative analysis of the binding specificity of two TCRs bearing either a charged (Glu) or neutral (Gly) residue in a particular position in the α-chain CDR3 region (P. A. Reay, J. L. Jorgensen, R. M. Kantor, and M. M. Davis, unpublished data). The TCR with Glu was highly specific, only recognizing the antigenic peptide if it had a Lys in a particular position. In contrast, the TCR with Gly was highly promiscuous, able to tolerate 18 of 20 possible amino acids in the same position on the antigenic peptide. Although steric factors are likely to play a role in this instance, the analysis strongly suggests that the presence of the charged residue dramatically increases the specificity of the TCR while not contributing substantially to its affinity.
Acknowledgments
The authors thank A. N. Barclay, D. L. Bodian, and D. I. Stuart for helpful comments on the manuscript and P. A. Reay for sharing data before publication. This work was supported by the U.K. Medical Research Council, the Human Frontier Science Program, and The Wellcome Trust.
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
Abbreviation: TCR, T cell antigen receptor.
References
- 1.Davis S J, van der Merwe P A. Immunol Today. 1996;17:177–187. doi: 10.1016/0167-5699(96)80617-7. [DOI] [PubMed] [Google Scholar]
- 2.van der Merwe P A, Barclay A N. Trends Biochem Sci. 1994;19:354–358. doi: 10.1016/0968-0004(94)90109-0. [DOI] [PubMed] [Google Scholar]
- 3.Jones E Y, Davis S J, Williams A F, Harlos K, Stuart D I. Nature (London) 1992;360:232–239. doi: 10.1038/360232a0. [DOI] [PubMed] [Google Scholar]
- 4.Bodian D L, Jones E Y, Harlos K, Stuart D I, Davis S J. Structure. 1994;2:755–766. doi: 10.1016/s0969-2126(94)00076-x. [DOI] [PubMed] [Google Scholar]
- 5.van der Merwe P A, McNamee P N, Davies E A, Barclay A N, Davis S J. Curr Biol. 1995;5:74–84. doi: 10.1016/s0960-9822(95)00019-4. [DOI] [PubMed] [Google Scholar]
- 6.Creighton T E. Proteins: Structures and Molecular Properties. 2nd Ed. New York: Freeman; 1993. [Google Scholar]
- 7.Miller S, Janin J, Lesk A M, Chothia C. J Mol Biol. 1987;196:641–656. doi: 10.1016/0022-2836(87)90038-6. [DOI] [PubMed] [Google Scholar]
- 8.Janin J, Chothia C. J Biol Chem. 1990;265:16027–16030. [PubMed] [Google Scholar]
- 9.Jones S, Thornton J M. Proc Natl Acad Sci USA. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van der Merwe P A, Brown M H, Davis S J, Barclay A N. EMBO J. 1993;12:4945–4954. doi: 10.1002/j.1460-2075.1993.tb06188.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van der Merwe P A, Barclay A N, Mason D W, Davies E A, Morgan B P, Tone M, Krishnam A K, Ianelli C, Davis S J. Biochemistry. 1994;33:10149–10160. doi: 10.1021/bi00199a043. [DOI] [PubMed] [Google Scholar]
- 12.van der Merwe P A, Bodian D L, Daenke S, Linsley P, Davis S J. J Exp Med. 1997;185:393–403. doi: 10.1084/jem.185.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nicholson M W, Barclay A N, Singer M S, Rosen S D, van der Merwe P A. J Biol Chem. 1998;273:763–770. doi: 10.1074/jbc.273.2.763. [DOI] [PubMed] [Google Scholar]
- 14.Driscoll P C, Cyster J G, Campbell I D, Williams A F. Nature (London) 1991;353:762–765. doi: 10.1038/353762a0. [DOI] [PubMed] [Google Scholar]
- 15.Brown M H, Barclay A N. Protein Eng. 1994;7:515–521. doi: 10.1093/protein/7.4.515. [DOI] [PubMed] [Google Scholar]
- 16.Brown M H, Preston S, Barclay A N. Eur J Immunol. 1995;25:3222–3228. doi: 10.1002/eji.1830251204. [DOI] [PubMed] [Google Scholar]
- 17.Mizushima S, Nagata S. Nucleic Acids Res. 1990;18:5322. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Davis S J, Davies E A, van der Merwe P A. Biochem Soc Trans. 1995;23:188–194. doi: 10.1042/bst0230188. [DOI] [PubMed] [Google Scholar]
- 19.Cunningham B C, Wells J A. J Mol Biol. 1993;234:554–563. doi: 10.1006/jmbi.1993.1611. [DOI] [PubMed] [Google Scholar]
- 20.Clackson T, Wells J A. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 21.McAlister M S, Mott H R, van der Merwe P A, Campbell I D, Davis S J, Driscoll P C. Biochemistry. 1996;35:5982–5991. doi: 10.1021/bi952756u. [DOI] [PubMed] [Google Scholar]
- 22.Tavernor A S, Kydd J H, Bodian D L, Jones E Y, Stuart D I, Davis S J, Butcher G W. Eur J Biochem. 1994;219:969–976. doi: 10.1111/j.1432-1033.1994.tb18579.x. [DOI] [PubMed] [Google Scholar]
- 23.Schreiber G, Fersht A R. Nat Struct Biol. 1996;3:427–431. doi: 10.1038/nsb0596-427. [DOI] [PubMed] [Google Scholar]
- 24.Bell G I. Science. 1978;200:618–627. doi: 10.1126/science.347575. [DOI] [PubMed] [Google Scholar]
- 25.Dustin M L. J Biol Chem. 1997;272:15782–15788. doi: 10.1074/jbc.272.25.15782. [DOI] [PubMed] [Google Scholar]
- 26.Honig B, Nicholls A. Science. 1995;268:1144–1149. doi: 10.1126/science.7761829. [DOI] [PubMed] [Google Scholar]
- 27.Shapiro L, Fannon A M, Kwong P D, Thompson A, Lehmann M S, Grubel G, Legrand J F, Als-Nielsen J, Colman D R, Hendrickson W A. Nature (London) 1995;374:327–337. doi: 10.1038/374327a0. [DOI] [PubMed] [Google Scholar]
- 28.Shapiro L, Doyle J P, Hensley P, Colman D R, Hendrickson W A. Neuron. 1996;17:435–439. doi: 10.1016/s0896-6273(00)80176-2. [DOI] [PubMed] [Google Scholar]
- 29.Garboczi D N, Ghosh P, Utz U, Fan Q R, Biddison W E, Wiley D C. Nature (London) 1996;384:134–141. doi: 10.1038/384134a0. [DOI] [PubMed] [Google Scholar]
- 30.Gao G F, Tormo J, Gerth U C, Wyer J R, McMichael A J, Stuart D I, Bell J I, Jones E Y, Jakobsen B K. Nature (London) 1997;387:630–634. doi: 10.1038/42523. [DOI] [PubMed] [Google Scholar]
- 31.Sayle R A, Milner-White E J. Trends Biochem Sci. 1995;20:374–376. doi: 10.1016/s0968-0004(00)89080-5. [DOI] [PubMed] [Google Scholar]