Abstract
Small, volatile hydrocarbons, including trichloroethylene, vinyl chloride, carbon tetrachloride, benzene, and chloroform, are common environmental pollutants that pose serious health effects. We have developed transgenic poplar (Populus tremula × Populus alba) plants with greatly increased rates of metabolism and removal of these pollutants through the overexpression of cytochrome P450 2E1, a key enzyme in the metabolism of a variety of halogenated compounds. The transgenic poplar plants exhibited increased removal rates of these pollutants from hydroponic solution. When the plants were exposed to gaseous trichloroethylene, chloroform, and benzene, they also demonstrated superior removal of the pollutants from the air. In view of their large size and extensive root systems, these transgenic poplars may provide the means to effectively remediate sites contaminated with a variety of pollutants at much faster rates and at lower costs than can be achieved with current conventional techniques.
Keywords: CYP2E1, P450, poplar, trichloroethylene, carbon tetrachloride
Phytoremediation is the use of plants for the treatment of environmental pollutants (1, 2). Plants act as solar-powered pump-and-treat systems as they take up water-soluble contaminants through their roots and transport/translocate them through various plant tissues where they can be metabolized, sequestered, or volatilized. Other types of contaminant remediation, including microbial bioremediation, usually require inputs of chemicals and energy; however, phytoremediation is largely self-maintaining (i.e., autotrophic), is less expensive, and has greater public approval. Phytoremediation also yields other benefits including carbon sequestration, soil stabilization, and the possibility of biofuel or fiber production. Many plant species have an inherent ability to metabolize a variety of environmental pollutants. The genus Populus contains many fast-growing tree species that are well suited for phytoremediation because of their rapid growth, extensive root systems, high water uptake, and amenability to transformation. Poplar trees can remove trichloroethylene (TCE) and carbon tetrachloride from solution, degrading it using a pathway similar to that in mammals (3, 4).
Although plants are capable of reducing the concentrations of some organic environmental pollutants, the activity is often too slow to be of practical value. Because phytoremediation proceeds primarily only during the growing season, substantial remediation must be achieved during a limited time period. The effectiveness of phytoremediation can be greatly enhanced by introducing genes known to be involved in metabolism of pollutants in other organisms (5). For example, the nitroaromatic explosives TNT and RDX are phytotoxic and cannot be effectively treated by using conventional phytoremediation. By introducing bacterial genes involved in the metabolism of TNT and RDX, the tolerance and uptake of these pollutants by transgenic plants were considerably improved (6, 7). In addition, phytoremediation of herbicides has been enhanced by using transgenic plants expressing GST (8) or cytochrome P450 genes (9, 10). Development of transgenic plants for enhanced phytoremediation of metals has also been successful (11), including plants have been developed to detoxify and remove mercury (12), lead and cadmium (13), and selenium from polluted soils (14).
Low-molecular-weight volatile compounds such as TCE, vinyl chloride, chloroform, carbon tetrachloride, and benzene are serious environmental pollutants that are proven or probable human carcinogens, neurotoxins, and hepatotoxins. TCE, a heavily used industrial degreaser, is the most common pollutant at Superfund sites in the United States because of improper disposal practices. Vinyl chloride is a proven human carcinogen and is commonly found in TCE-contaminated sites as a result of microbial dehalorespiration of TCE. Chloroform, a byproduct of the disinfection process used to treat drinking water in the United States, is a nearly ubiquitous environmental pollutant. Carbon tetrachloride was used routinely as a solvent and is now also a common pollutant at Superfund sites. Benzene, another proven human carcinogen, is a common pollutant associated with petroleum. Current engineering methods to remove these pollutants are expensive and intensive, annually costing billions of dollars worldwide (1).
Cytochrome P450 2E1 is a mammalian enzyme with broad specificity for environmental pollutants including TCE, chloroform, carbon tetrachloride, benzene, vinyl chloride, and others (15). In a proof-of-concept project, introduction of a mammalian CYP2E1 gene into tobacco plants resulted in plants with increased metabolism of TCE (16). In this study we report on the development of the first transgenic trees with enhanced phytoremediation capabilities for removing and degrading several of the most widespread and dangerous pollutants from water and air.
Results
Increased Metabolism of TCE in Poplar.
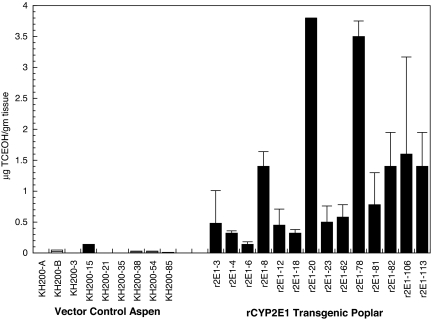
By using Agrobacterium tumefaciens-mediated plant transformation, we developed hybrid poplar plants that strongly express an enzyme capable of TCE metabolism. An early metabolite of TCE after oxidation is trichloroethanol, a compound that is found in both plants and mammals. We exposed small cuttings taken from the apical stem of plants containing either the transgene, CYP2E1, or a null vector to TCE in 40-ml sealed vials. We then monitored the appearance of the early metabolite trichloroethanol in plant tissues. The CYP2E1-containing transgenic cuttings had an average increased TCE metabolism that was nearly 45-fold greater than in the control cuttings (Fig. 1). Two (lines 78 and 20) of the CYP2E1 transgenic lines (from independent transgenic events) had TCE metabolism rates that were >100-fold higher than in controls. The transgenic cuttings grew normally and did not display any adverse reaction to the TCE or its metabolites.
Fig. 1.
Metabolism of TCE by hybrid poplar cuttings. Hybrid poplar was transformed with a vector containing CYP2E1 or the null vector control, pKH200, and independent transformed lines were regenerated and propagated. Cuttings were surface-sterilized and transferred to 40-ml vials, and the medium was dosed to a level of 50 μg/ml TCE. After 1 week of exposure, plants were frozen with liquid nitrogen, and the TCE metabolites were extracted with methyl tert-butyl ether. The extracts were analyzed with a GC-ECD. Trichloroethanol was quantified by using external standards.
Expression of CYP2E1 Transgene.
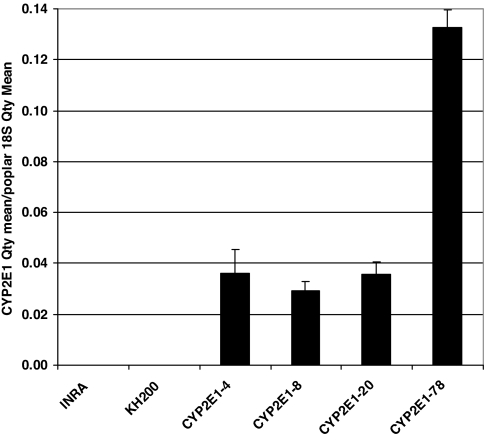
Using quantitative RT-PCR to evaluate the expression levels of the transgene CYP2E1, we found the gene to be highly expressed in all transgenic plants. There was no expression of CYP2E1 in the wild-type or null vector plants. The transgenic plant line CYP2E1-78 showed 3.7–4.6 times higher expression levels of CYP2E1 when compared with other transclones (Fig. 2). This clone consistently exhibited the highest level of TCE metabolism in our studies. The high expression of the transgene in the CYP2E1-78 plant is in accordance with the highest rate of TCE removal by this plant line (described below).
Fig. 2.
Expression of the CYP2E1 transgene in wild-type, null vector, and CYP2E1 transgenic hybrid poplar lines. RNA was extracted from leaf tissues of control and transgenic hybrid poplars, and then quantitative RT-PCR was performed, and expression was normalized by using 18S as the housekeeping gene.
Removal of Pollutants from Hydroponic Solution.
To study whether the increased metabolism of TCE in the poplar cuttings led to increased uptake from solution, we monitored the concentration of TCE in the liquid growth medium. Poplar cuttings were surface-sterilized and incubated in hydroponic solutions containing TCE. Cuttings from the CYP2E1 transgenic plants removed TCE from solution at much greater rates than the cuttings from the plants that had been transformed with the null vector (Table 1). After 1 week, the null vector and unplanted controls removed <3% of the TCE from solution whereas the CYP2E1 transgenics removed from 51% to 91%. Analysis of the percent removal of TCE from solution showed the controls to be significantly different from all CYP2E1 transgenic poplar lines, with the most significant difference being between line 78 and the unplanted and null vector controls (α = 0.05, P < 0.0001 and P < 0.001, respectively). The best performing transgenic line, line 78, removed TCE 53 times faster than did the vector control plants, showing the highest statistical difference in uptake rate (micrograms TCE day−1·g−1 plant wet weight) between line 78 and the unplanted and null vector controls (α = 0.05, P < 0.003 and P < 0.008, respectively). All statistical analysis was performed by using one-way ANOVA with Tukey's honestly significant difference post hoc analysis.
Table 1.
Removal of TCE from hydroponic solution in 1 week by poplar cuttings in sealed vials
| Plant line | % removal | Rate* |
|---|---|---|
| Unplanted | 0.8 ± 1.1 | 0.14 ± 0.11 |
| KH200-61 | 2.5 ± 0.4 | 0.38 ± 2.81 |
| CYP2E1 line 4 | 90.9 ± 15.7 | 18.6 ± 2.50 |
| CYP2E1 line 8 | 51.4 ± 6.2 | 12.3 ± 2.81 |
| CYP2E1 line 20 | 79.0 ± 29.6 | 12.5 ± 5.76 |
| CYP2E1 line 78 | 87.2 ± 11.1 | 20.3 ± 4.60 |
Cuttings in triplicate were dosed with 708 μg of TCE in sealed 40-ml VOA vials. TCE-dosed, unplanted vials were used as a control. After 1 week of TCE exposure, plants were weighed, and the hydroponic solution was sampled. TCE was extracted by using a salt-hexane solution and analyzed by GC-ECD. Percent removal was calculated by comparing total TCE concentration after 4 h and after 1 week.
*Micrograms of TCE per day per gram of fresh weight ± SEM.
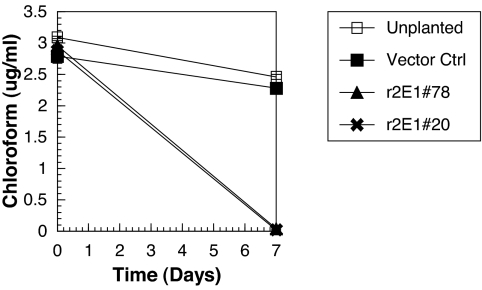
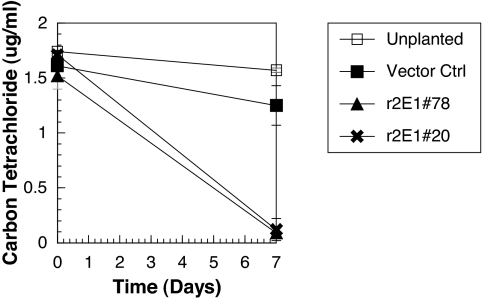
Because cytochrome P450 2E1 has a broad substrate specificity that includes other important environmental pollutants, we determined the removal rates of chloroform and carbon tetrachloride by transgenic lines 78 and 20 compared them with null vector control plants. In 1 week, the transgenic plants removed 99% of the chloroform compared with only 18–20% removed by the controls (Fig. 3). The removal rate was increased from 0.02 μg of chloroform h−1·g−1 plant wet weight by the vector control plants to an average of 0.17 by the CYP2E1 transgenic lines, a 9-fold improvement. The removal rate of carbon tetrachloride was also increased. After 1 week of exposure, the CYP2E1 plants removed 92–94% of the carbon tetrachloride compared with 10–22% by the controls (Fig. 4). Therefore, increased metabolism of these P450 2E1 substrates in poplar resulted in an increased removal of the pollutant from solution.
Fig. 3.
Removal of chloroform from hydroponic solution in 1 week. Cuttings of null vector control poplar and two independent plant lines transformed with CYP2E1 were incubated in 40-ml glass vials containing chloroform. After 7 days, chloroform was extracted with hexane. The extracts were analyzed with GC-ECD by using external standards. Standard errors are presented by the SEM.
Fig. 4.
Removal of carbon tetrachloride from hydroponic solution in 1 week. Cuttings of null vector control poplar and two independent plant lines transformed with CYP2E1 were incubated in 40-ml glass vials containing chloroform. After 7 days, carbon tetrachloride was extracted by using hexane, and the concentration was determined by GC-ECD. Standard errors are presented by the SEM.
Vinyl chloride is a proven human carcinogen that is produced by the dehalorespiration of TCE by soil microorganisms. We tested for removal of this important pollutant by transgenic line 78. Cuttings from control and CYP2E1 plants were exposed to gaseous vinyl chloride. In 5 days, an average of 49% of the vinyl chloride was removed by the CYP2E1 cuttings compared with an average of 29% by the vector control cuttings. The CYP2E1 transgenic poplar cuttings (line 78) had an initial uptake rate of 1.9 ± 0.4 μg of vinyl chloride h−1·g−1 plant wet weight, compared with a rate of 0.61 ± 0.16 μg for the control cuttings. This 3-fold increased rate of vinyl chloride by the CYP2E1 line 78 cuttings was statistically significant (P < 0.01).
After 7 days of exposure to vinyl chloride, all of the CYP2E1 poplar cuttings exhibited significant blackening of the mesophyllic (non-vein) leaf tissue; this was not apparent with the control cuttings. When control and modified cuttings were placed in the same vials and exposed to vinyl chloride, leaf blackening was limited only to the modified poplars. Human cytochrome P450 2E1 catalyzes the oxidation of vinyl chloride into transient reactive metabolites such as chloroethylene oxide and 2-chloroacetaldehyde, which can bind protein and DNA (15, 17). These results suggest that production of toxic vinyl chloride metabolites may result in toxicity within the tissues of CYP2E1 transgenics. However, environmental levels of vinyl chloride rarely approach the concentrations used in these experiments, so metabolic toxicity should rarely be a problem in field applications.
Removal of Pollutants from Air.
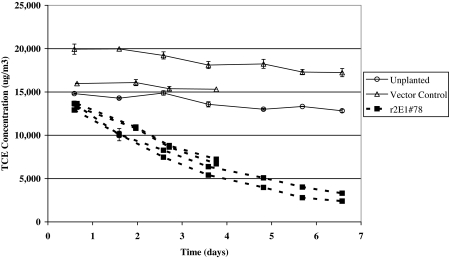
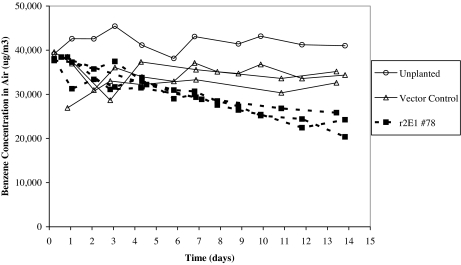
To examine whether the CYP2E1 transgenic poplar had increased air pollutant removal rates, transgenic line 78 plants and control plants were exposed to volatile forms of TCE and benzene in desiccators. The TCE experiments used whole plants; benzene experiments used plant cuttings in hydroponics because preliminary experiments indicated that soil bacteria metabolized benzene. The CYP2E1 plants removed 79% of the TCE from the air. During the same period, null vector control plants did not show any statistically significant uptake of TCE compared with the unplanted soil control (Fig. 5). Benzene was also removed by these transgenic cuttings at an increased rate (Fig. 6). The CYP2E1 cuttings removed 0.28 μg of benzene h−1·g−1 plant wet weight compared with 0.025 for the null vector control plants. In 2 weeks, the control plants removed an average of only 13% of the benzene from air compared with 36% and 46% by two independent CYP2E1 plant lines.
Fig. 5.
Removal of volatile TCE by transgenic poplar. Small plants in 4-inch pots of soil were incubated in 10-inch-diameter Pyrex glass desiccators containing an initial concentration of ≈15,000 μg·m−3 TCE. TCE concentration in air was measured daily by analyzing air samples on GC-ECD and quantifying with external standards. Each line represents an individual plant sealed in a desiccator system. Points are averages of triplicate daily samples with error bars indicating ± 1 SEM.
Fig. 6.
Removal of volatile benzene from air. Cuttings of null vector control plants and transformed poplar CYP2E1-78 were placed in hydroponic solution, incubated in desiccators, and exposed to volatile benzene with an initial concentration of ≈40,000 μg·m−3. Benzene concentration in the air was analyzed daily by GC–flame ionization detector and quantified compared with external standards. Each line represents an individual cutting in a desiccator system. Data points are averages of triplicate daily samples with error bars indicating ± 1 SEM.
Discussion
To our knowledge, this work represents the first development of transgenic trees for increased removal of a broad range of serious environmental pollutants from water and air. The use of transgenic plants can overcome many of the obstacles of traditional phytoremediation. One limitation with some forms of phytoremediation is that plants can move pollutants from the ground and transpire them unaltered into the air. Plants that can metabolize the pollutants to inert forms would therefore be most desirable. Furthermore, environmental pollutants are often phytotoxic, rendering phytoremediation impossible without the introduction of genes into the plant required for the breakdown of the pollutant. Some plant species with an inherent ability to hyperaccumulate metals are too small or have too limited a root system to remove significant amounts of the polluting metal; thus, transfer of the genes from a small hyperaccumulating plant to a species of greater biomass and longevity could make the phytoremediation of metals throughout the soil profile more likely to succeed. Furthermore, polluted environments are often contaminated with multiple chemicals; therefore, plants that contain genes enabling them to remediate a wider range of pollutants are desirable. The use of transgenic plants can therefore be used in a variety of ways to increase the effectiveness of phytoremediation.
Expression of the CYP2E1 gene was sufficient to confer greatly increased removal of TCE, vinyl chloride, carbon tetrachloride, chloroform, and benzene, all of which are substrates of cytochrome P450 2E1. Because cytochrome P450 enzymes use NADPH-cytochrome P450 oxidoreductase and cytochrome b5 for electron transfer, we can infer that the poplar homologs of these enzymes are apparently similar enough to the mammalian enzymes for full function of P450 2E1 in plants. Because the initial oxidation of pollutants catalyzed by cytochrome P450 enzymes is frequently rate-limiting, the strategy of overexpressing the responsible P450 gene could be widely applicable for increasing the removal of a variety of pollutants by plants.
Because it has been previously demonstrated that hybrid cottonwood, another kind of poplar, can remove TCE from hydroponic solution (3, 18, 19), the poor performance of the nontransgenic hybrid poplar in this study was unexpected. However, the hybrid cottonwood (Populus trichocarpa × Populus deltoides) studies were done by using rooted cuttings in flasks with the foliage in open air or in field trials with full-grown trees, whereas this study was conducted with unrooted poplar (Populus tremula × Populus alba) cuttings in sealed vials. This particular clone, 717-1B4, is difficult to root in hydroponics. Therefore, the lack of TCE removal by the control poplar could have been due to the differences between the different poplar genotypes or the experimental designs. Our data clearly indicate that overexpression of CYP2E1 leads to enhanced removal of pollutants compared with vector controls. To determine whether the improvements seen in these laboratory studies will ultimately lead to enhanced phytoremediation, field studies are needed with CYP2E1 clone 78 and nontransgenic control trees.
Another unexpected result was that our transgenic poplar did not demonstrate increased removal of ethylene dibromide (data not shown), a substrate of CYP2E1 in hepatic microsomes, which was substantially metabolized in our previous study with transgenic tobacco plants expressing human CYP2E1 (16). Because the transgenic poplar expressing rabbit CYP2E1 in this study did not behave in this way, it is possible that the rabbit and human homologs of CYP2E1 have slightly different substrate specificities.
The use of transgenic plants can reduce the known risk of having carcinogens and other hazardous pollutants in the environment. Apart from intellectual property and other business considerations, commercial use of these trees requires federal regulatory approval and monitoring, and regulations are becoming increasingly strict for transgenic plants intended for biopharmaceutical or industrial purposes, including phytoremediation (see www.aphis.usda.gov/brs/pdf/Pharma_Guidance.pdf). Additional studies are needed to ensure that plant tissues do not cause unacceptable impacts on non-target organisms. Bioremediation transgenes are not expected to improve plant fitness and, thus, should not become common in wild or feral populations, an assumption that will be verified via early field studies. Nonetheless, high levels of containment of the added transgenes may be needed for regulatory and marketplace acceptance. This can be readily accomplished in trees by harvest before the onset of flowering. Local spread by vegetative propagation can be monitored and manually controlled. Additionally, there are a large number of genetic flowering control systems for trees under development that can prevent, or drastically reduce, transgene dispersal via pollen or seeds (20). By a combination of management and mitigation technologies, the environmental benefits of using transgenic trees for bioremediation should be within reach in the near future.
Materials and Methods
Construction of Transgenic Poplar Lines.
The plasmid pSLD50-6, containing the rabbit CYP2E1 cDNA under the control of the cauliflower mosaic virus 35S promoter, is described by Banerjee et al. (21). The poplar hybrid clone INRA 717-1B4 (P. tremula × P. alba) was transformed with A. tumefaciens strain C58C1 (pGV3850, pSLD50-6) using standard protocols (22). Null vector control plants were derived from transformations using pKH200 (23) instead of pSLD50-6. Independent lines of both types were verified by PCR and propagated by cuttings in Murashige and Skoog tissue culture medium (24) (Caisson Labs) and in soil.
Expression of CYP2E1 Transgene in Hybrid Poplar.
Leaf samples of ≈100 mg were taken from hybrid poplar plants grown in soil in a plant growth room: INRA 717-1B4 wild type, KH200 null vector, CYP2E1-4, CYP2E1-8, CYP2E1-20, and CYP2E1-78. RNA extractions from these fresh leaf samples were performed by using the RNeasy Plant Mini Kit (Qiagen). The RNA was then used to synthesize cDNA by using the Bio-Rad iScript kit with SYBR Green labeling agent, and quantification of transgene via RT-PCR was performed by using SYBRGreen-labeled probes of 18S as the housekeeping gene and CYP2E1.
TCE Metabolism Experiments.
The transgenic lines were screened in duplicate for enhanced metabolism of TCE. Cuttings were surface-sterilized with 10% commercial bleach and 1% iodophor and placed in sterile 40-ml clear Volatile Organics Analysis (VOA) vials containing 14 ml of 1/2× Hoagland's solution (25). Cuttings of this hybrid clone (717-1B4) form roots at low efficiency; therefore, all of these experiments were conducted with unrooted cuttings. The VOA vials were sealed with septum valve caps (Mininert) to prevent escape of volatile TCE. TCE (99.5% purity; Sigma–Aldrich) was added to a level of ≈50 μg/ml. After 1 week of exposure, the plants were flash-frozen in liquid nitrogen and ground to a powder, and the metabolites were extracted as described (3). A standard curve of pure trichloroethanol (98% purity; Lancaster) was used to quantify the metabolite by using a PerkinElmer gas chromatograph equipped with an electron capture detector (GC-ECD).
TCE Uptake Experiments.
Cuttings of ≈8 cm in height, of null vector control lines (pKH200) and rCYP2E1 lines (pSLD50-6), in triplicate, were surface-sterilized with 0.1% mercuric chloride, placed into vials, and dosed with TCE as with the metabolism experiment. Initial samples were taken from the medium in each vial after 4 h of equilibration. The plants were incubated at ≈25°C for 7 days with a 16-h photoperiod. A second sampling of the medium was taken after 7 days. The TCE was extracted by using hexane and sodium chloride, analyzed by GC-ECD, and quantified by using standards in hexane. The average weights of the plants were 1.1 g for the control lines, 0.9 g for CYP2E1 line 4, 0.7 g for line 8, 1.17 g for line 20, and 0.74 g for line 78.
Chloroform Uptake Experiments.
Cuttings with four to six leaves on average taken from the apical stem of plants of ≈8 cm in height from control plant lines and CYP2E1 plant lines 78 and 20 were surface-sterilized and transferred to vials as described for the TCE uptake experiments. Chloroform (Baker) was added to the hydroponic solution to a level of ≈3 μg/ml. Chloroform was extracted from the hydroponic solution by using hexane and sodium chloride. The concentration of chloroform was analyzed by GC-ECD and quantified with external standards. A total of nine plants and three media samples were analyzed in this experiment, which was replicated twice. The average fresh weights of the plants at the end of the experiment were 1.3 g for the controls, 1.1 g for CYP2E1 line 78, and 1.0 g for line 20.
Carbon Tetrachloride Uptake Experiments.
Cuttings with four to six leaves on average from the apical stem of plants between 7 cm and 8 cm in height were taken and treated as described for the chloroform uptake experiments, but carbon tetrachloride (99.9% purity; Sigma–Aldrich) was added to the hydroponic solution to an approximate final concentration of 1.6 μg/ml. The average weights of the plants were 1.0 g for the KH200 controls, 1.4 g for line 78, and 1.6 g for line 20. Carbon tetrachloride was extracted from the hydroponic solution by using hexane and sodium chloride and analyzed by GC-ECD. A total of nine plants and three media samples were analyzed in this experiment, and the experiment was replicated twice.
Volatile TCE Experiments.
Small rooted poplar plants in 4-inch pots containing 1:1 ProMix (Premier Horticulture, Quakertown, PA) and perlite (Therm-O-Rock, Chandler, AZ) were incubated in 10-inch-diameter Pyrex glass desiccators. The desiccators were sealed with Fluorolube Grease GR-290 (Fisher Scientific). The sample ports were valved and additionally sealed with rubber septa. The sample ports were closed such that the septa were exposed only during sample collection. Eighty-two microliters of TCE-saturated water in 10-ml glass beakers was placed in the desiccators before sealing. Sealed desiccators were incubated at room temperature with a 16-h photoperiod. The TCE concentration was measured daily by withdrawing 500 μl of air from the sample port by using a gas-tight syringe (Hamilton, Reno, NV) and manually injecting into a PerkinElmer GC-ECD (Supelco PTE-5 Capillary Column); daily analysis was performed in triplicate. TCE was quantified by using external standards. The desiccators remained sealed for the entire incubation period. A total of four CYP2E1 line 78 and three KH200 cuttings were evaluated, each in its own sealed dessicator.
Volatile Benzene Experiments.
Plant cuttings from apical stems were placed in ≈225 ml of sterile hydroponic solution (1/2× Hoagland's solution) in 250-ml flasks. The mouth of the flask was plugged with cotton around the plant stems, and the entire flask and the cotton were covered with aluminum foil. The cuttings were transferred into desiccators. Approximately 200 μl of benzene-saturated water was placed in a 10-ml beaker inside the desiccator. Sealed desiccators were incubated at room temperature with a 16-h photoperiod. Dessicator air was analyzed as in volatile TCE experiments except that a 500-μl sample was injected into an SRI 8610C GC-flame ionization detector (Alumina Sulfate Plot column; Supelco). Average mass was 1.5 g for the KH200 control cuttings and 1.6 g for the CYP2E1 line 78 cuttings. A total of four CYP2E1 line 78 and three KH200 cuttings were evaluated.
Vinyl Chloride Experiments.
Cuttings of the apical ends were taken from nontransgenic poplar (717-1B4) and transgenic CYP2E1 poplar line 78 and sterilized as described above. The plants were transferred to VOA vials containing 10 ml of 1/2× Murashige and Skoog broth and capped with Mininert valves. Each vial was dosed with ≈350 μg of vinyl chloride (Aldrich Chemical). The gas was allowed to equilibrate for at least 12 h before sampling. Sealed vials were incubated at room temperature with a 16-h photoperiod. Vial headspace was analyzed by withdrawing 150 μl with a gas-tight syringe and injecting into an SRI 8610 GC–flame ionization detector. Injections were performed in triplicate. Each experimental set was performed with three control and three transgenic cuttings; each set was performed twice. Average mass was 2.1 g for the control cuttings and 1.4 g for the CYP2E1 line 78 cuttings.
Acknowledgments
This paper is dedicated to the late Prof. Milton P. Gordon (Univeristy of Washington) who initiated this project. For more than a dozen years, he championed the idea of enhancing phytoremediation using transgenics in the hope that it would be used more widely to reduce environmental pollution. We thank Prof. Tom Hinckley and Neil Bruce for critical reading of the manuscript. We also thank undergraduate student helpers Hannah Borja, Gregory Osborn, Amanda Thornton, and Matthew Weintraub for their assistance. This work was financially supported by Department of Energy Grant DE-FG02-03ER63663 and the University of Washington Superfund Basic Research Program sponsored by the National Institute on Environmental Health Sciences through Grant P42ES004696. C.A.J. was supported by the Valle Fellowship and the Exchange Program at the University of Washington.
Abbreviations
- TCE
trichloroethylene
- GC-ECD
gas chromatograph equipped with an electron capture detector.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Pilon-Smits EAH, Freeman JL. Front Ecol Environ. 2006;4:203–210. [Google Scholar]
- 2.Suresh B, Ravishankar GA. Crit Rev Biotechnol. 2004;24:97–124. doi: 10.1080/07388550490493627. [DOI] [PubMed] [Google Scholar]
- 3.Shang QT, Doty SL, Wilson AM, Howald WN, Gordon MP. Phytochemistry. 2001;58:1055–1065. doi: 10.1016/s0031-9422(01)00369-7. [DOI] [PubMed] [Google Scholar]
- 4.Wang X, Gordon MP, Strand SE. Biodegradation. 2002;13:297–305. doi: 10.1023/a:1022397121365. [DOI] [PubMed] [Google Scholar]
- 5.Cherian S, Oliveira MM. Environ Sci Technol. 2005;39:9377–9390. doi: 10.1021/es051134l. [DOI] [PubMed] [Google Scholar]
- 6.Hannink N, Rosser SJ, French CE, Basran A, Murray JAH, Nicklin S, Bruce NC. Nat Biotechnol. 2001;19:1168–1172. doi: 10.1038/nbt1201-1168. [DOI] [PubMed] [Google Scholar]
- 7.Rylott EL, Jackson RG, Edwards J, Womack GL, Seth-Smith HM, Rathbone DA, Strand SE, Bruce NC. Nat Biotechnol. 2006;24:216–219. doi: 10.1038/nbt1184. [DOI] [PubMed] [Google Scholar]
- 8.Karavangeli M, Labrou NE, Clonis YD, Tsaftaris A. Biomol Eng. 2005;22:121–128. doi: 10.1016/j.bioeng.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 9.Kawahigashi H, Hirose S, Ohkawa H, Ohkawa Y. J Agric Food Chem. 2006;54:2985–2991. doi: 10.1021/jf052610u. [DOI] [PubMed] [Google Scholar]
- 10.Kawahigashi H, Hirose S, Ohkawa H, Ohkawa Y. J Agric Food Chem. 2005;53:9155–9160. doi: 10.1021/jf0511143. [DOI] [PubMed] [Google Scholar]
- 11.Kramer U, Chardonnens AN. Appl Microbiol Biotechnol. 2001;55:661–672. doi: 10.1007/s002530100631. [DOI] [PubMed] [Google Scholar]
- 12.Rugh CL, Senecoff JF, Meagher RB, Merkle SA. Nat Biotechnol. 1998;16:925–928. doi: 10.1038/nbt1098-925. [DOI] [PubMed] [Google Scholar]
- 13.Song WY, Sohn EJ, Martinoia E, Lee YJ, Yang YY, Jasinski M, Forestier C, Hwang I, Lee Y. Nat Biotechnol. 2003;21:914–919. doi: 10.1038/nbt850. [DOI] [PubMed] [Google Scholar]
- 14.LeDuc DL, AbdelSamie M, Montes-Bayon M, Wu CP, Reisinger SJ, Terry N. Environ Pollut. 2006;144:70–76. doi: 10.1016/j.envpol.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 15.Guengerich FP, Kim DH, Iwasaki M. Chem Res Toxicol. 1991;4:168–179. doi: 10.1021/tx00020a008. [DOI] [PubMed] [Google Scholar]
- 16.Doty SL, Shang QT, Wilson AM, Tangen J, Westergreen A, Newman LA, Strand SE, Gordon MP. Proc Natl Acad Sci USA. 2000;97:6287–6291. doi: 10.1073/pnas.97.12.6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.elGhissassi F, Barbin A, Bartsch H. Biochem Pharmacol. 1998;55:1445–1452. doi: 10.1016/s0006-2952(97)00645-x. [DOI] [PubMed] [Google Scholar]
- 18.Newman L, Strand S, Choe N, Duffy J, Ekuan G, Ruszaj M, Shurtleff BB, Wilmoth J, Heilman P, Gordon MP. Environ Sci Technol. 1997;31:1062–1067. doi: 10.1289/ehp.98106s41001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newman L, Wang X, Muiznieks I, Ekuan G, Ruszaj M, Cortellucci R, Domroes D, Karscig G, Newman T, Crampton RS, et al. Environ Sci Technol. 1999;33:2257–2265. [Google Scholar]
- 20.Brunner AM, Li J, Shevchenko O, Mohamed R, Montgomery B, Elias A, Van Wormer, DiFazio SP, Strauss SH. Tree Genet Genomes. 2006;3(2):75–100. [Google Scholar]
- 21.Banerjee S, Shang QT, Wilson AM, Moore AL, Strand SE, Gordon MP, Doty SL. Biotechnol Bioeng. 2002;77:462–466. doi: 10.1002/bit.10151. [DOI] [PubMed] [Google Scholar]
- 22.Han K-H, Meilan R, Ma C, Strauss SH. Plant Cell Rep. 2000;19:315–320. doi: 10.1007/s002990050019. [DOI] [PubMed] [Google Scholar]
- 23.Han K-H, Ma C, Strauss SH. Transgenic Res. 1997;6:415–420. [Google Scholar]
- 24.Murashige T, Skoog F. Physiol Plant. 1962;15:473–497. [Google Scholar]
- 25.Hoagland DR, Arnon DI. The Water Culture Method for Growing Plants Without Soil. Berkeley, CA: Calif Agric Experiment Stn; 1950. [Google Scholar]