Abstract
Huntington's disease (HD) is a neurodegenerative disorder caused by expansion of a CAG repeat in the huntingtin (Htt) gene. HD is autosomal dominant and, in theory, amenable to therapeutic RNA silencing. We introduced cholesterol-conjugated small interfering RNA duplexes (cc-siRNA) targeting human Htt mRNA (siRNA-Htt) into mouse striata that also received adeno-associated virus containing either expanded (100 CAG) or wild-type (18 CAG) Htt cDNA encoding huntingtin (Htt) 1–400. Adeno-associated virus delivery to striatum and overlying cortex of the mutant Htt gene, but not the wild type, produced neuropathology and motor deficits. Treatment with cc-siRNA-Htt in mice with mutant Htt prolonged survival of striatal neurons, reduced neuropil aggregates, diminished inclusion size, and lowered the frequency of clasping and footslips on balance beam. cc-siRNA-Htt was designed to target human wild-type and mutant Htt and decreased levels of both in the striatum. Our findings indicate that a single administration into the adult striatum of an siRNA targeting Htt can silence mutant Htt, attenuate neuronal pathology, and delay the abnormal behavioral phenotype observed in a rapid-onset, viral transgenic mouse model of HD.
Keywords: gene delivery, gene silencing, Huntington's disease, neurodegenerative disease, RNAi
Huntington's disease (HD) is an autosomal dominant disease caused by a CAG repeat expansion in the Htt gene (1). Mutant Htt causes neuronal death, dementia, and movement dysfunction; there is no effective treatment. In an inducible transgenic mouse model of HD, turning off transgene expression reversed neuropathology and motor deficits (2). Lowering mutant Htt gene expression in brain may treat HD. In mice, viral vector delivery of short hairpin RNAs (shRNAs) against mutant Htt gene exon 1 or genes that cause other neurodegenerative disorders reduced neuropathology and motor deficits (3–10). Brain delivery of adeno-associated virus (AAV)-shRNA against mutant Htt improved signs of disease in HD transgenic models (7, 11). In the inaugural study on RNAi targeting Htt in vivo, shRNA against Htt in AAV2, delivered to the N171–82Q transgenic model of HD, improved ambulation at 4 months and rotarod performance at 10 and 18 weeks after injection (7). Five and one-half months after shRNA administration, quantitative RT-PCR revealed a 50% reduction in striatal Htt mRNA. Statistical changes in quantification of Htt protein reduction and inclusions were not reported. AAV5 delivery of shRNA against Htt in the R6/1 murine model of HD showed a 25% decrease in Htt protein and an 80% reduction in Htt mRNA 10 weeks after shRNA injection (10). The shRNA delayed onset of clasping by 2 weeks (20–22 weeks), and treated mice had fewer clasps. No difference in rotarod performance was detected. Inclusion size and number decreased in the striatum, but not in the cortex, compared with the corresponding contralateral brain regions. The authors provided an important caveat that one of the shRNAs had off-target effects; the cause of the off-target effects was not established. shRNA in AAV2 or AAV5 was used to target EGFP to knock down EGFP-Htt in another transgenic model of HD (11). shRNA reversed pathology after the onset of pathologic changes; however, behaviors were not studied. Administration of large amounts of siRNA against Htt in a Lipofectamine 2000 suspension into the lateral ventricle of newborn R6/2 transgenic mice (exon 1 of Htt) reduced whole-brain levels of mutant Htt in two mice and Htt mRNA up to 7 days posttreatment, delayed the onset of clasping, rotarod, and open-field phenotypes, and improved survival (12). Statistical quantification of neuropathology was not reported. Thus, prior studies examining RNAi against Htt provided the groundwork for therapeutic gene silencing in HD. Most of the studies used viral delivery of shRNA, and the study using siRNA required liposome delivery to newborns, with the potential liposome neuronal toxicity.
Caveats attend the use of shRNAs, which can be toxic when integrated into the host genome (13, 14), in part because shRNA production is unregulated. Long siRNAs (>29 nt) and shRNAs are prone to activate off-target gene expression (15). For patient safety, shRNA will need to be able to be switched off, currently a hurdle in viral delivery systems.
An alternative strategy for HD therapy is the use of small-interfering RNAs (siRNAs), ≈21-nt RNA duplexes. siRNA has been administered into cerebroventricles, vasculature, intrathecal space, and parenchyma (16–20). siRNAs were found effective and safe when introduced into mice and non-human primates (19, 21, 22). Several limitations impede progress in using siRNAs as a treatment for HD: entry and effectiveness in adult neurons without the use of potentially toxic transfection reagents; a clear demonstration that gene silencing reduces protein expression; and an improvement in behavioral deficits and neuropathology, especially neuron survival. Because bioactive molecules conjugated to cholesterol have improved cellular uptake in vitro (23), LDL receptors have been detected in brain (24), and cholesterol conjugation enhances siRNA uptake in cells outside of the central nervous system (16), we speculated that cholesterol-conjugated (cc) siRNA might enter neurons in vivo.
An in vivo, rapid-onset model of HD would be optimal to test gene silencing in brain. The current rapid mouse model of HD shows mutant Htt-induced pathology after 2 months. Transgenic mice expressing exon 1 of mutant Htt [R6/2 (25)] develop nuclear inclusions throughout the brain at 2 months of age and exhibit a rapidly progressing, severe phenotype. Other transgenic or knock-in mice expressing mutant Htt exhibit late-onset, mild phenotypes, often after 6 months of age (26, 27), and lack prominent neuronal loss. Neither model is ideal to test transient effects of a single injection of siRNA against Htt introduced directly to the striatum. We therefore developed an acute, in vivo, HD mouse model tailored to addressing the efficacy of siRNA. Here, we used AAV to deliver a 1,395-nt cDNA fragment of human mutant Htt into mouse striatum to evaluate the effectiveness of an siRNA targeting human Htt.
Results
Unconjugated and cc-siRNAs Enter Medium Spiny Striatal Neurons.
To confirm that the siRNA introduced into the striatum entered neurons, we examined striata of mice injected with Cy3-labeled siRNA-Htt. Cholesterol conjugation improves siRNA delivery in hepatocytes (16), so we compared delivery of cc-siRNA and unconjugated siRNA in neurons. Brain tissue was processed 1–5 days after injection and examined for immunoreactive localization of DARPP32, which is selectively expressed by medium size striatal spiny neurons. At the level of the striatal injection, Cy3 immunofluorescent bodies were present in the cytoplasm of the majority of DARPP32-positive striatal neurons at all concentrations tested, regardless of cholesterol conjugation (Fig. 1 a and b). Quantitative analysis showed that 86% and 88% of DARPP32-positive neurons exposed to Cy3 cc-siRNA (10 μM) and unconjugated Cy3 siRNA (10 μM) contained Cy3-labeled bodies (range 76–100% in both). These findings indicate that siRNA had entered the majority of striatal neurons. We typically observed a higher level of Cy3 fluorescence in the neuropil surrounding DARPP32-labeled cell bodies after injection of Cy3 cc-siRNA than unconjugated Cy3 siRNA (Fig. 1 c and d), although the difference did not achieve statistical significance (Fig. 1e; median; 59% versus 33%, P ≤ 0.18, respectively). These results suggested, but do not prove, that neuronal processes take up the cc-siRNA more readily than the unconjugated siRNA. We therefore used cc-siRNA in our subsequent studies.
Fig. 1.
Cholesterol-conjugated and unconjugated siRNA-Htt enters neurons. (a and b) Laser confocal images of DARPP32-labeled striatal neurons (green) treated with Cy3-siRNA-Htt. The striatum was injected with 10 μM unconjugated (a) or cc (b) Cy3-siRNA-Htt. Cy3 fluorescence appears in the neuronal cytoplasm as distinct bodies (red-orange color at arrows). Images were from striata 5 days (a) and 1 day (b) after injection. (c and d) Cy3 fluorescence in the striatal neuropil surrounding the DARPP32-labeled neurons. (e) Percent of mice with high Cy3 fluorescence in neuropil after injection of cc-siRNA-Htt (n = 17) or unconjugated siRNA-Htt (n = 18); not significant in Fisher's exact test. Bars indicate the confidence intervals.
Single Intrastriatal Injection of siRNA-Htt Protects Striatal Neurons from Mutant Htt Neuropathology.
Htt knock-in mice manifest neuropathology and motor deficits late and then sporadically at 10 months of age. Transgenic models vary in the extent of gene expression, brain regions involved, neuropathology, and behaviors (28). We therefore developed a rapid-onset murine model of HD, in which timed neuropathology and motor impairments were predictable, robust, and experimentally testable. AAV vectors are known to be effective for the delivery of exogenous genes into the rodent brain (29). Normal mice received a unilateral striatal injection of AAVHtt18Q or AAVHtt100Q (Htt cDNA coding for amino acids 1–400 of Htt, with either 18 or 100 CAG repeats, respectively). Two independent preparations of AAVHtt100Q virus were used (group 1 and group 2). Immunoperoxidase labeling with anti-Htt revealed intense Htt immunoreactivity in neurons within the dorsal half of the striatum, deep layers of the cortex, and septal area ipsilateral to the injection 2 weeks after injection (Fig. 2a). In AAVHtt100Q-infected mice, degenerating and shrunken Htt-labeled neurons appeared in cortex layers 5 and 6 and in the dorsal striatum. In the striatum of some mice, neurons were markedly depleted from a core region around the site of AAVHtt100Q injection (Fig. 2a), resulting in a region of diminished Htt immunoreactivity. No such loss of neurons was evident in mice injected with AAVHtt18Q. Measurements of somal cross-sectional area showed that neurons expressing AAVHtt100Q were significantly smaller compared with neurons expressing AAVHtt18Q (Fig. 2 b and c Left; mean ± SD; AAVHtt18Q versus AAVHtt100Q, 149 ± 10.4 versus 113 ± 10.1; P ≤ 0.003) or neurons with wild-type Htt in the noninjected striata (not shown). However, the size of striatal neurons expressing AAVHtt100Q was not different in mice cotreated with cc-siRNA-Htt (113 ± 10.1) compared with mice cotreated with control cc-siRNA against luciferase (siRNA-Luc; 119 ± 6; Fig. 2c Right).
Fig. 2.
Cholesterol-conjugated siRNA-Htt reversed neuropathology in the AAV HD mouse model. (a) Brain sections from normal mice that received a unilateral striatal injection of AAVHtt18Q (Left) or AAVHtt100Q (Right). Sections were immunoperoxidase-labeled by using anti-Htt antisera. Exogenous human wild-type and mutant Htt expression is visible in dorsal striatum (asterisks), septal nuclei (arrows), and deep layers of the cortex (arrowheads). (b) Htt-labeled striatal cells infected with AAVHtt100Q are smaller than cells expressing Htt18Q or neurons in the noninjected striatum. Insets show examples of Htt-labeled cells at higher magnification. (c) Mean ± SD for the cross-sectional area of immunoreactive striatal neurons in mice infected with AAVHtt18Q or AAVHtt100Q (Left) and mice infected with AAVHtt100Q and cotreated with siRNA-Luc (Luc) or siRNA-Htt (Htt) (Right). Neuronal size is reduced in striatal cells expressing mutant Htt100Q compared with striatal cells expressing Htt18Q (Left; *, P ≤ 0.003; n = 8 mice per group; 50 cells per mouse; Student's t test) but is not changed by cotreatment with cc-siRNA-Htt (Right; n = 4 per group). (d) Number of neurons determined by stereology in Nissl-stained sections in noninjected and AAVHtt100Q-injected striatum. Bar graphs show mean ± SD for number of neurons. Group 1 (Left): *, P ≤ 0.01, Student's t test. Group 2 (Right): not significant.
Stereology in Nissl-stained sections showed that the number of neurons in AAVHtt100Q-injected regions of striatum was reduced compared with noninjected striata; neuronal number in the injected striata of group 2 mice was lower compared with mice in group 1 (Fig. 2d). AAVHtt100Q-infected striata of group 1 mice cotreated with cc-siRNA-Htt had significantly more neurons compared with those given cc-siRNA-Luc (Fig. 2d, mean ± SD; cc-siRNA-Htt versus cc-siRNA-Luc, 5.3 ± 0.6 versus 3.5 ± 0.9, P ≤ 0.01). Neuronal survival was not improved by cotreatment with cc-siRNA-Htt compared with cc-siRNA-Luc in group 2 mice, most likely because of the more significant neuronal loss caused by AAVHtt100Q in group 2 [Fig. 2d, mean ± SD; cc-siRNA-Luc versus cc-siRNA-Htt, 2.87 ± 1.1 versus 2.13 ± 0.7, not significant (29)].
Striatal and cortical neurons infected with AAVHtt100Q had strong diffuse nuclear labeling or intranuclear aggregates when immunolabeled with anti-Htt antisera (Fig. 2b Inset and Fig. 3 a and b). Nuclear inclusions were detected in striatal (Fig. 3c) and cortical neurons with EM48 antibody, which detects only aggregated Htt. cc-siRNA-Htt treatment significantly reduced the median size of nuclear inclusions in striatal neurons in AAVHtt100Q-infected mice and in cortical neurons in group 1 AAVHtt100Q-infected mice (Fig. 3d, medians; group 1: cortex, cc-siRNA-Luc versus cc-siRNA-Htt, 6.89 versus 5.86, P ≤ 0.01; striatum, cc-siRNA-Luc versus cc-siRNA-Htt, 9.08 versus 7.78, P ≤ 0.01; group 2: cortex, cc-siRNA-Luc versus cc-siRNA-Htt, 6.22 versus 6.29, not significant; striatum, cc-siRNA-Luc versus cc-siRNA-Htt, 8.7 versus 6.18, P ≤ 0.004). The reduction in inclusion size by cc-siRNA-Htt suggests that buildup of inclusions is related to levels of accumulation of mutant Htt. Striata injected with AAVHtt18Q showed robust cytoplasmic staining in neurons with anti-Htt antisera but lacked inclusions with EM48 or anti-Htt antisera (Fig. 2 b and b Inset). The density of anti-Htt-labeled striatal neurons with inclusions was significantly higher in cc-siRNA-Htt-treated mice than in cc-siRNA-Luc-treated mice (Fig. 3e, mean ± SD; group 1: striatum, cc-siRNA-Luc versus cc-siRNA-Htt, 258 ± 55.1 versus 340 ± 32.2, P ≤ 0.002). These data support results from stereology, which showed that administration of cc-siRNA-Htt increased survival of striatal neurons expressing mutant Htt (Fig. 2d, group 1).
Fig. 3.
Inclusion pathology was reduced in AAVHtt100Q mice treated with cc-siRNA-Htt. Immunoperoxidase labeling was done with anti-Htt antisera. (a and b) Neurons with AAVHtt100Q inclusions in striatum and cortex. (c) Nuclear inclusions in striatal neurons labeled with EM48 antisera. (d) Scatter plot showing size distribution of inclusions in the two groups of mice treated with AAVHtt100Q. Densitometry was performed with EM48-stained sections. Horizontal bars indicate the medians. cc-siRNA-Htt (Htt) treatment reduces median inclusion size compared with cc-siRNA-Luc (Luc). Group 1: n = 8, 8, 8, 8; cortex, P ≤ 0.01; striatum, P ≤ 0.01. Group 2: n = 5, 6, 5, 6. Cortex, not significant; striatum, P ≤ 0.004. One hundred cells per mouse were evaluated. Mann–Whitney U test; *, P ≤ 0.01 for cortex and striatum. (e) Density of Htt-labeled neurons with inclusions per 2,500 μm2. cc-siRNA-Htt treatment increased the number of Htt-labeled neurons with inclusions. Findings are significant for striatum (*, P ≤ 0.002; Student's t test; cc-siRNA-Luc, n = 9; cc-siRNA-Htt, n = 8).
In HD brain, mutant N-Htt fragments accumulate in degenerating neuronal processes called dystrophic neurites (30). In HD transgenic mice, analogous neuronal processes are evident as neuropil aggregates (30, 31). Neuropil aggregates were detected in striata infected with AAVHtt100Q (Fig. 4a) but not in striata infected with AAVHtt18Q (data not shown). There was a higher density of neuropil aggregates in group 1 mice compared with group 2 mice, a finding possibly due to the presence of more neurons in group 1 compared with group 2 (Fig. 2 c and d). Neuropil aggregates in striatum were significantly lower in all AAVHtt100Q mice treated with cc-siRNA-Htt compared with those treated with cc-siRNA-Luc (Fig. 4, mean ± SD; group 1: cc-siRNA-Luc versus cc-siRNA-Htt, 5.73 ± 2.45 versus 1.85 ± 1.42, P ≤ 0.002; group 2: cc-siRNA-Luc versus cc-siRNA-Htt, 2.69 ± 1.60 versus 1.03 ± 0.51, P ≤ 0.036). Reducing expression of mutant Htt reduced formation of neuropil aggregates.
Fig. 4.
Mutant Htt neuropil aggregates were reduced by cc-siRNA-Htt. (a and b) Htt labeling in neuropil aggregates (arrows) in the striatum from mice treated with AAVHtt100Q and cc-siRNA-Luc (a) or cc-siRNA-Htt (b). The aggregates are small structures (a), which are faintly visible in b. The boxes in a and b denote the brain region in the Insets. (c and d) Bar graphs showing number of neuropil aggregates in group 1 and group 2 AAVHtt100Q mice treated with cc-siRNA-Luc (Luc) or cc-siRNA-Htt (Htt). Group 1: n = 8 per treatment; *, P ≤ 0.002. Group 2: cc-siRNA-Luc, n = 5; cc-siRNA-Htt, n = 6; *, P ≤ 0.036. Student's t test; one ×40 field per mouse.
In summary, introduction of N-terminal mutant Htt fragment into the mouse striatum by AAVHtt delivery recapitulated neuropathological features of HD, and cotreatment with siRNA targeting Htt ameliorated neuropathology.
Single Intrastriatal Injection of siRNA-Htt Ameliorated Mutant Htt-Induced Motor Dysfunction.
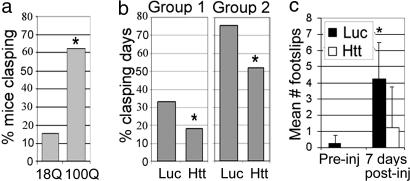
When suspended by the tail, neurologically impaired mice retract their limbs, a response called clasping. We found that more AAVHtt100Q mice clasped 14 days after injection compared with AAVHtt18Q mice (Fig. 5a; 62.5% versus 15%, respectively, P ≤ 0.02). The frequency of clasping in AAVHtt100Q mice was significantly reduced by cotreatment with cc-siRNA-Htt compared with cc-siRNA-Luc (Fig. 5b; cc-siRNA-Luc versus cc-siRNA-Htt; group 1: 33% versus 18%, P ≤ 0.01; group 2: 75.6% versus 51.9%, P ≤ 0.02). The percent of clasping days in the two groups of AAVHtt100Q mice positively correlated with the extent of neuronal death observed in the striatum. Mice injected with AAVHtt100Q and coinjected with cc-siRNA-Luc showed an increased number of footslips on the beam 7 days after injection. In contrast, AAVHtt100Q mice coinjected with cc-siRNA-Htt did not show a significant increase in footslips during beam walking, relative to the number observed before injection (Fig. 5c; 7 days after injection; cc-siRNA-Luc, 4.25 ± 2.22, versus cc-siRNA-Htt, 1.25 ± 2.5, P ≤ 0.01). cc-siRNA-Htt protected mice from motor deficits caused by AAVHtt100Q.
Fig. 5.
AAVHtt100Q mice show reduced motor deficits in the presence of cc-siRNA-Htt. (a) Percent of mice clasping 14 days after injection with AAVHtt18Q and AAVHtt100Q. Mice with AAVHtt100Q had significantly more clasping days than mice infected with AAVHtt18Q. AAVHtt18Q, 15% clasping, n = 13; AAVHtt100Q, 62.5% clasping, n = 16 (P = 0.02). (b) AAVHtt100Q mice cotreated with cc-siRNA-Htt had fewer clasping days than mice coinjected with cc-siRNA-Luc (group 1: cc-siRNA-Luc, 33%, n = 9, versus cc-siRNA-Htt, 18%, n = 8; *, P = 0.01; group 2: cc-siRNA-Luc, 75%, n = 5, versus cc-siRNA-Htt, 52%, n = 6; *, P ≤ 0.02; Fisher's exact test). (c) Shown are mean ± SD footslips that occurred during beam walking for mice injected with AAVHtt100Q. Mean footslips were reduced in the presence of cc-siRNA-Htt compared with cc-siRNA-Luc. *, P ≤ 0.01; Student's t test; n = 4 per group.
Single Intrastriatal Injection of siRNA-Htt Decreases Human Htt Gene Expression in Mouse Brain.
To examine whether RNAi accounted for improvement in neuropathology and motor deficits, we measured human Htt in the brain at the location of the AAVHtt delivery 2.7 days after injection. Brain lysates from mice injected with AAVHtt18Q or AAVHtt100Q, and cc-siRNA-Htt or cc-siRNA-Luc, were analyzed by Western blotting. Densitometry showed that coinjection of cc-siRNA-Htt reduced levels of Htt-18Q and Htt-100Q protein by 56% and 66%, respectively, compared with levels observed with the cc-siRNA-Luc (Fig. 6; mean ± SD; AAVHtt18Q, cc-siRNA-Luc versus cc-siRNA-Htt, 0.68 ± 0.25 versus 0.38 ± 0.16, P ≤ 0.03; AAVHtt100Q, cc-siRNA-Luc versus cc-siRNA-Htt, 1.02 ± 0.19 versus 0.34 ± 0.31, P ≤ 0.01). We conclude that that the observed improvements in neuropathology and behavior reflect silencing of the AAVHtt100Q gene by the cc-siRNA-Htt.
Fig. 6.
Silencing human Htt mRNA reduced amount of exogenous wild-type and mutant Htt in the striatum. (a) Lysates were prepared from dorsal striatum 2.7 days after coinjection of AAVHtt18Q or AAVHtt100Q with either cc-siRNA-Luc or cc-siRNA-Htt. The Western blot was probed first with anti-Htt antibody (Upper) and then reprobed with anti-tubulin antibody (Lower). Shown is expression of Htt100Q in mice treated with cc-siRNA-Luc or cc-siRNA-Htt. (b) Bar graphs show mean Htt/tubulin ratios from densitometry of Western blot films for mice infected with AAVHtt18Q (n = 4) or AAVHtt100Q (n = 3) and treated with cc-siRNA-Luc or cc-siRNA-Htt. *, P = 0.03 for AAVHtt18Q; *, P = 0.01 for AAVHtt100Q.
Single Intrastriatal Injection of siRNA Does Not Stimulate a Specific Immunogenic Response.
siRNAs exceeding 23 bp and shRNAs have been associated with an immunogenic response (32). We examined reactive microglia (CD11B immunoreactivity) and reactive astrocytes (30) (GFAP immunoreactivity) in mice injected with PBS or cc-siRNA-Luc. siRNA concentrations were the same as those used for coinjection with AAV. Astrocytic and microglial labeling did not differ between PBS and cc-siRNA treatments [supporting information (SI) Figs. 7]. cc-siRNAs did not change animal weight or temperature (SI Table 1).
Discussion
RNAi offers a promising therapy for autosomal dominantly inherited neurodegenerative disease. In theory, RNAi can target the underlying cause of HD, by silencing expression of the mutant protein. AAV-shRNA delivered to brain improved signs of disease in transgenic models of HD (7, 11). Here, we describe a new virus-mediated transgenic model of HD in the adult mouse that mimics features of HD within 2 weeks and use this model to test the effects of gene silencing with siRNA targeting Htt mRNA. Our findings demonstrated that cc-siRNA-Htt entered adult striatal neurons and effected a reduction in mutant Htt protein that ameliorated both the neuropathology and the motor deficits caused by the virally delivered mutant Htt protein.
A single administration of cc-siRNA-Htt was sufficient to attenuate multiple neuropathological features and aberrant motor behaviors in the rapid-onset model of HD: the transgene silencing subdued expression of mutant Htt in the striatum for at least 3 days and sustained a benefit in motor behavior for 1 week. The improvement in motor behaviors correlated with improvement in neuropathology, indicating that gene silencing of mutant Htt ameliorated striatal neuronal dysfunction. Our results demonstrate that siRNA treatment with a clinically relevant formulation provides therapeutically meaningful benefit in an adult mouse model of HD. Together, these findings indicate that the RNAi effect is durable and that siRNA therapeutics might be deliverable in pulses rather than in continuous flow. The effect of a single siRNA injection appears to be widespread. Although our siRNA treatment was directed to the striatum, the size of inclusions in some mice was reduced in cortical neurons. siRNA administration correlated with an increase in the number of neurons with inclusions. This in vivo finding supports cellular HD models in which nuclear inclusion formation is dissociated with cell survival (33, 34) and could offer a survival advantage (35).
siRNA modified by cholesterol conjugation was used because of prior demonstration that cholesterol conjugation might improve siRNA access to tissues (16). It is not clear whether cholesterol conjugation increased Cy3-siRNA uptake into somata of DARPP32-labeled medium size spiny neurons in striatum. However, at low concentrations, a trend toward more Cy3-cc-siRNA than unconjugated siRNA uptake was apparent in regions surrounding cell bodies and in white matter, raising the possibility that neuronal processes, rather than cell bodies, preferentially take up cc-siRNA. It is possible that coinjection of siRNA with AAV-Htt improved the siRNA's neuronal delivery. Pilot studies have indicated that cc-siRNA enters primary striatal neurons in the absence of AAV and that two injections of siRNA-Htt can silence endogenous Htt (SI Figs. 8 and 9). Further study will be needed to assess optimal delivery of unconjugated and modified siRNAs in reducing levels of mutant Htt in the brain.
Safety and flexibility of small RNAs, either in shRNA or siRNA, are paramount for therapeutic gene silencing. The siRNA duplex used here, which targeted a sequence of Htt 5′ to the CAG repeat of Htt mRNA, was well tolerated in the brain. Based on the extent of reactive microglia and astrocytes, siRNA administration into the striatum did not elicit an inflammatory response greater than vehicle injection alone. The cc-siRNA did not change animal weight or temperature (SI Table 1).
Viral delivery of a disease gene presents an alternative approach to traditional transgenic models for understanding pathophysiology and testing potential therapeutics. Lentivirus has been used to generate a rat model of HD (36). Delivery of truncated mutant Htt in lentivirus produced neuropathological features of HD but not behavioral phenotypes. In our model, AAV-Htt spread through dorsal striatum and deep cortical layers, areas affected early in HD. Cortical neurons are thought to contribute to striatal neuron vulnerability, because of loss of brain-derived neurotrophic factor from cortical neurons projecting to striatum (37). This AAV-Htt transgenic model has compelling features that provide a useful system for testing therapeutic candidates for HD; the model shows the neuropathological changes found in HD, including neuronal loss and motor deficits, both of which occur with a rapid time course and are amenable to quantification. The model distinguishes between CAG repeat number in wild-type and mutant Htt gene expression, despite overexpression of both genes.
Treatment of HD and other autosomal dominant neurodegenerative diseases will require intervention for years, if not decades. siRNA has a distinct advantage in providing a predictable and finite term of action; therefore, siRNA treatment can be adjusted or stopped should side effects arise. shRNA delivered by either genomic or epigenetic incorporation would not be conducive to regulation and could be prone to activation of IFN-dependent pathways (32). However, in concept, viral delivery of shRNA offers long-term intraneuronal siRNA expression. The next challenge in testing siRNA therapeutics in brain disease will be the safe delivery of small RNAs to maximize target specificity, with the capacity to distinguish between mutant and wild-type alleles.
Methods
Plasmid DNA and AAV Preparation.
We cloned N-terminal Htt cDNA with 18 CAG or 100 CAG repeats into plasmid pAAV-CBA-W, which encodes a chimeric promoter bearing the CMV enhancer element and the chicken β-actin promoter. The Htt cDNA codes 365 aa apart from the polyglutamine repeats. The vector was packaged into AAV1/8 mosaic vector. Viral titers up to 1.05 × 1013 (for 18Q) and 1.8 × 1013 (for 100Q) were obtained.
Animals, Stereotaxic Injections, and Behavioral Assessment.
The animal protocol abided was approved by the University of Massachusetts Medical School (A-978). The right striata of mice were coinjected with 3 μl of virus delivering AAVHtt18Q or AAVHtt100Q and a cc-siRNA-Htt or cc-siRNA-Luc (0.5 μl of 1 mM) by using a micropump syringe. Animals were monitored twice daily for weight, temperature, clasping, and beam walking. Mice were killed at 2 weeks for immunohistochemical studies. A group of mice was injected intrastriatally with 2 μl of Cy3-labeled cc-siRNA-Htt or unconjugated siRNA-Htt at 10 μM. Animals were killed 1, 3, and 5 days after injection. A group of mice was injected with AAVHtt18Q (n = 4) or AAVHtt100Q (n = 4) and coinjected with cc-siRNA-Luc or siRNA-Htt. Mice were killed 3 days after injection, and the brains were harvested for biochemical analysis. Data were analyzed by using Student's t test or Fisher's exact test.
Immunohistochemistry.
Antisera included anti-Htt (against Htt 1–17 aa) and EM48 polyclonal antibody [gift of Steven Hersch (Massachusetts General Hospital); 1:5,000] to detect Htt aggregates; anti-DARPP32 antibody (Chemicon; 1:1,000) and anti-GFAP polyclonal antisera (Chemicon; AB5804; 1:1,000) for astrocytes; and rat monoclonal antibody (Chemicon; AB1387z; 1:500) for CD11B for microglia. For further details, see ref. 23.
Quantitative Microscopic Analysis.
To measure the density of Htt-labeled neurons, four adjacent microscopic fields in layer 6 of cortex and five fields in striatum bordering the corpus callosum were selected by using a ×20 objective lens. Total cells with inclusions (per 2,500 μm2) were determined for mice in each group: cc-siRNA-Luc (n = 9) or cc-siRNA-Htt (n = 9). Stereological methods were performed for neuronal number (26, 38). No significant differences in striatal volumes were obtained between injected and noninjected striata. Mean volume was used to calculate total neuronal number.
For inclusion size, three microscopic fields in cortex and striatum were evaluated at ×40 objective in EM48-immunolabeled sections. Images were captured by using a SPOT camera (Diagnostic Instruments), and the cross-sectional area of inclusion size was determined by using SigmaScan Pro software (Jandel Scientific). About 100 cells were measured per mouse per region (range from 50–140 cells), and median inclusion size was determined and compared by Mann–Whitney U. Neuropil aggregates in striatum were counted in ×40 fields of 40-μm sections labeled with anti-Htt antisera. The results were grouped by treatment (cc-siRNA-Luc and cc-siRNA-Htt), and the mean number of aggregates per area was determined. To examine neuronal cross-sectional area, sections labeled with anti-Htt antibody were observed at ×40 on the injected and noninjected side of the striatum and analyzed as above. The cross-sectional area of 50 Htt-labeled neurons per striata was determined by using SigmaScan Pro. Mean neuronal size was determined for each animal and grouped according to CAG repeat length and treatment condition.
Biochemical Analysis.
The dorsal striatum from injected and noninjected sides of the brain was dissected on ice and incubated in lysis buffer [50 mM Tris (pH 7.4), 1% Nonidet P-40, 250 mM NaCl, 5 mM EDTA plus protease inhibitor mixture (Roche) and pepstatin A, 1 μg/ml]. Five-microgram protein samples were analyzed by Western blot with anti-Htt antisera and tubulin as described in ref. 39. Densitometry was performed by using SigmaScan Pro to obtain the signal intensity of Htt-18Q, Htt-100Q, and tubulin.
Preparation of siRNAs.
The guide strand contained three phosphorothioate backbone modification—two at the 3′ end and one at 5′ end of the sequence to enhance nuclease stability and a 5′ unpaired end to promote its assembly into the RNA-induced silencing complex (40). The passenger strand contained 3′ cholesterol and three phosphorothioate linkages (13, 41). The guide strand was complementary to a region of Htt mRNA 5′ to the CAG repeats. cc-siRNA-Htt comprised guide 5′-UpsUC AUC AGC UUU UCC AGG GpsUpsC-3′ and passenger 5′-CpsCC UGG AAA AGC UGA UGA CGpsGps-chol-3′. Control cc-siRNA-Luc comprised guide 5′-UCG AAG uAC UcA GCGuA AGTps T-3′ and passenger 5′-cuu AcG cuG AGu Acu ucG ATpsTps-Chol-3′. (The lowercase letters represent nucleotides with 2′-O-methyl modifications, ps denotes phosphorothioate, and Chol denotes cholesterol.) To generate Cy3-labeled siRNAs, Qusar-570 (Cy3) was conjugated to the 5′ end of the passenger strand.
Supplementary Material
Acknowledgments
We thank Michael Hayden (University of British Columbia, Vancouver, BC, Canada) for the gift of YAC 128 transgenic mice. This work was supported by National Institutes of Health Grant NS38194 (to N.A., P.D.Z., and M.D.), the High Q Foundation (N.A. and P.D.Z.), a National Institutes of Health Endocrine Training Grant (to the University of Massachusetts Medical School), and the Diabetes Endocrine Research Center at the University of Massachusetts Medical School.
Abbreviations
- AAV
adeno-associated virus
- AAVHtt18Q
AAV containing Htt cDNA producing Htt with 18 polyglutamines
- HD
Huntington's disease
- Luc
luciferase
- shRNA
short hairpin RNA
- siRNA
small interfering RNA.
Footnotes
Conflict of interest statement: P.D.Z. is a cofounder and scientific advisory board member of Alnylam Pharmaceuticals. D.W.Y.S., M.M., K.G.R., and R.K.P. are employees of Alnylam Pharmaceuticals.
This article contains supporting information online at www.pnas.org/cgi/content/full/0708285104/DC1.
References
- 1.Huntington's Disease Collaborative Research Group. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 2.Yamamoto A, Lucas JJ, Hen R. Cell. 2000;101:57–66. doi: 10.1016/S0092-8674(00)80623-6. [DOI] [PubMed] [Google Scholar]
- 3.Singer O, Marr RA, Rockenstein E, Crews L, Coufal NG, Gage FH, Verma IM, Masliah E. Nat Neurosci. 2005;8:1343–1349. doi: 10.1038/nn1531. [DOI] [PubMed] [Google Scholar]
- 4.Raoul C, Abbas-Terki T, Bensadoun JC, Guillot S, Haase G, Szulc J, Henderson CE, Aebischer P. Nat Med. 2005;11:423–428. doi: 10.1038/nm1207. [DOI] [PubMed] [Google Scholar]
- 5.Xia H, Mao Q, Eliason SL, Harper SQ, Martins IH, Orr HT, Paulson HL, Yang L, Kotin RM, Davidson BL. Nat Med. 2004;10:816–820. doi: 10.1038/nm1076. [DOI] [PubMed] [Google Scholar]
- 6.Ding H, Schwarz DS, Keene A, Affar el B, Fenton L, Xia X, Shi Y, Zamore PD, Xu Z. Aging Cell. 2003;2:209–217. doi: 10.1046/j.1474-9728.2003.00054.x. [DOI] [PubMed] [Google Scholar]
- 7.Harper SQ, Staber PD, He X, Eliason SL, Martins IH, Mao Q, Yang L, Kotin RM, Paulson HL, Davidson BL. Proc Natl Acad Sci USA. 2005;102:5820–5825. doi: 10.1073/pnas.0501507102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller VM, Xia H, Marrs GL, Gouvion CM, Lee G, Davidson BL, Paulson HL. Proc Natl Acad Sci USA. 2003;100:7195–7200. doi: 10.1073/pnas.1231012100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ralph GS, Radcliffe PA, Day DM, Carthy JM, Leroux MA, Lee DC, Wong LF, Bilsland LG, Greensmith L, Kingsman SM, et al. Nat Med. 2005;11:429–433. doi: 10.1038/nm1205. [DOI] [PubMed] [Google Scholar]
- 10.Rodriguez-Lebron E, Denovan-Wright EM, Nash K, Lewin AS, Mandel RJ. Mol Ther. 2005;12:618–633. doi: 10.1016/j.ymthe.2005.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Machida Y, Okada T, Kurosawa M, Oyama F, Ozawa K, Nukina N. Biochem Biophys Res Commun. 2006;343:190–197. doi: 10.1016/j.bbrc.2006.02.141. [DOI] [PubMed] [Google Scholar]
- 12.Wang YL, Liu W, Wada E, Murata M, Wada K, Kanazawa I. Neurosci Res. 2005;53:241–249. doi: 10.1016/j.neures.2005.06.021. [DOI] [PubMed] [Google Scholar]
- 13.Snove O, Jr, Rossi JJ. Nat Methods. 2006;3:689–695. doi: 10.1038/nmeth927. [DOI] [PubMed] [Google Scholar]
- 14.Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 15.Aleman LM, Doench J, Sharp PA. RNA. 2007;13:385–395. doi: 10.1261/rna.352507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soutschek J, Akinc A, Bramlage B, Charisse K, Constien R, Donoghue M, Elbashir S, Geick A, Hadwiger P, Harborth J, et al. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- 17.Song E, Zhu P, Lee SK, Chowdhury D, Kussman S, Dykxhoorn DM, Feng Y, Palliser D, Weiner DB, Shankar P, et al. Nat Biotechnol. 2005;23:709–717. doi: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]
- 18.Lewis DL, Wolff JA. Methods Enzymol. 2005;392:336–350. doi: 10.1016/S0076-6879(04)92020-4. [DOI] [PubMed] [Google Scholar]
- 19.Dorn G, Patel S, Wotherspoon G, Hemmings-Mieszczak M, Barclay J, Natt FJ, Martin P, Bevan S, Fox A, Ganju P, et al. Nucleic Acids Res. 2004;32:e49. doi: 10.1093/nar/gnh044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thakker DR, Natt F, Husken D, Maier R, Muller M, van der Putten H, Hoyer D, Cryan JF. Proc Natl Acad Sci USA. 2004;101:17270–17275. doi: 10.1073/pnas.0406214101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heidel JD, Hu S, Liu XF, Triche TJ, Davis ME. Nat Biotechnol. 2004;22:1579–1582. doi: 10.1038/nbt1038. [DOI] [PubMed] [Google Scholar]
- 22.Li BJ, Tang Q, Cheng D, Qin C, Xie FY, Wei Q, Xu J, Liu Y, Zheng BJ, Woodle MC, et al. Nat Med. 2005;11:944–951. doi: 10.1038/nm1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng K, Ye Z, Guntaka RV, Mahato RI. J Pharmacol Exp Ther. 2006;317:797–805. doi: 10.1124/jpet.105.100347. [DOI] [PubMed] [Google Scholar]
- 24.Hofmann SL, Russell DW, Goldstein JL, Brown MS. Proc Natl Acad Sci USA. 1987;84:6312–6316. doi: 10.1073/pnas.84.17.6312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davies SW, Turmaine M, Cozens BA, DiFiglia M, Sharp AH, Ross CA, Scherzinger E, Wanker EE, Mangiarini L, Bates GP. Cell. 1997;90:537–548. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- 26.Laforet GA, Sapp E, Chase K, McIntyre C, Boyce FM, Campbell M, Cadigan BA, Warzecki L, Tagle DA, Reddy PH, et al. J Neurosci. 2001;21:9112–9123. doi: 10.1523/JNEUROSCI.21-23-09112.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin CH, Tallaksen-Greene S, Chien WM, Cearley JA, Jackson WS, Crouse AB, Ren S, Li XJ, Albin RL, Detloff PJ. Hum Mol Genet. 2001;10:137–144. doi: 10.1093/hmg/10.2.137. [DOI] [PubMed] [Google Scholar]
- 28.Menalled LB. NeuroRx. 2005;2:465–470. doi: 10.1602/neurorx.2.3.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCown TJ. Curr Gene Ther. 2005;5:333–338. doi: 10.2174/1566523054064995. [DOI] [PubMed] [Google Scholar]
- 30.DiFiglia M, Sapp E, Chase KO, Davies SW, Bates GP, Vonsattel JP, Aronin N. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 31.Gutekunst CA, Li SH, Yi H, Mulroy JS, Kuemmerle S, Jones R, Rye D, Ferrante RJ, Hersch SM, Li XJ. J Neurosci. 1999;19:2522–2534. doi: 10.1523/JNEUROSCI.19-07-02522.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reynolds A, Anderson EM, Vermeulen A, Fedorov Y, Robinson K, Leake D, Karpilow J, Marshall WS, Khvorova A. RNA. 2006;12:988–993. doi: 10.1261/rna.2340906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saudou F, Finkbeiner S, Devys D, Greenberg ME. Cell. 1998;95:55–66. doi: 10.1016/s0092-8674(00)81782-1. [DOI] [PubMed] [Google Scholar]
- 34.Kim M, Lee HS, LaForet G, McIntyre C, Martin EJ, Chang P, Kim TW, Williams M, Reddy PH, Tagle D, et al. J Neurosci. 1999;19:964–973. doi: 10.1523/JNEUROSCI.19-03-00964.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- 36.de Almeida LP, Ross CA, Zala D, Aebischer P, Deglon N. J Neurosci. 2002;22:3473–3483. doi: 10.1523/JNEUROSCI.22-09-03473.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zuccato C, Liber D, Ramos C, Tarditi A, Rigamonti D, Tartari M, Valenza M, Cattaneo E. Pharmacol Res. 2005;52:133–139. doi: 10.1016/j.phrs.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 38.Hyman BT, Gomez-Isla T, Irizarry MC. J Neuropathol Exp Neurol. 1998;57:305–310. doi: 10.1097/00005072-199804000-00001. [DOI] [PubMed] [Google Scholar]
- 39.DiFiglia M, Sapp E, Chase K, Schwarz C, Meloni A, Young C, Martin E, Vonsattel JP, Carraway R, Reeves SA, et al. Neuron. 1995;14:1075–1081. doi: 10.1016/0896-6273(95)90346-1. [DOI] [PubMed] [Google Scholar]
- 40.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 41.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, Stoffel M. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.